Abstract
Wheat pre-harvest sprouting (PHS) causes serious losses in wheat yield. In this study, precise mapping was carried out in the chromosome segment substitution lines (CSSL) F2 population generated by a direct cross of Zhoumai 18 (PHS-sensitive) and Aegilops tauschii accession T093 (highly PHS-resistant). Three Ae. tauschii-derived quantitative trait loci (QTLs), QDor.3D.1, QDor.3D.2, and QDor.3D.3, were detected on chromosome 3DL using four simple sequence repeats (SSR) markers and 10 developed Kompetitive allele-specific PCR (KASP) markers. Alongside these QTL results, the RNA-Seq and qRT-PCR analysis revealed expression levels of TraesCS3D01G466100 in the QDor.3D.2 region that were significantly higher in CSSLs 495 than in Zhoumai 18 during the seed imbibition treatment. The cDNA sequencing results of TraesCS3D01G466100 showed two single nucleotide polymorphisms (SNPs), resulting in two changed amino acid substitutions between Zhoumai 18 and line 495, and the 148 nt amino acid substitution of TraesCS3D01G466100, derived from Ae. tauschii T093, which may play an important role in the functioning of ubiquitin ligase enzymes 3 (E3) according to the homology protein analysis, which could lead to differential PHS-resistance phenotypes. Taken together, our results may foster a better understanding of the mechanism of PHS resistance and are potentially valuable for marker-assisted selection in practical wheat breeding efforts.
1. Introduction
Wheat pre-harvest sprouting (PHS) is the precocious germination of grain in a moist environment long before a crop’s harvest. PHS may cause a decrease in the end-use quality of grain due to starch and protein degradation, leading to severe wheat yield losses [1], and it often occurs before the grain-ripening season in humid and warm areas such as southwestern China [2]. Annual losses caused by PHS to wheat farmers are likely to exceed 1 billion USD worldwide [3]. To accommodate the growing demands of the international and domestic markets, it is imperative that wheat PHS resistance be strengthened to ensure and augment its yield and quality.
The phenomenon of PHS is influenced by a number of plant physiological factors. Dormancy is currently presumed to be the chief factor involved [4]. Seed dormancy, which can enable seeds to survive in adverse environments, such as in a wrong season or regeneration niche, is a complex quantitative trait. The lack of dormancy directly results in PHS under particular humidity and temperature conditions. Besides seed dormancy, grain color, and chemical coat composition, structure and ear morphology, as well as hormonal factors, could also affect PHS [5,6]. Although PHS is influenced by multiple genes and environmental conditions [7,8,9], annual variation in the latter could modulate the effects of alleles contributing to wheat’s tolerance of PHS. Hence, genotype-based selection could still promote this tolerance trait independently of the environment [10,11].
Most often, PHS is evaluated in both field and controlled environments using three methods: visual observation, alpha-amylase testing, and the Hagberg-Perten Falling Numbers (FN) assay [12,13,14]. For such research, misting chambers with intact wheat heads or a weighted germination index are commonly used to evaluate (susceptibility in the laboratory. However, because evaluating the phenotype is time- and labor-consuming, marker-assisted selection (MAS) is a better way to evaluate PHS because not only is it faster and requires less time, but it also is more accurate than phenotype evaluations.
A quantitative trait locus (QTL) is a locus (section of DNA) that correlates with variation in a quantitative trait in the phenotype of a population of organisms [15]. Therefore, finding reliable markers closely linked to quantitative trait loci (QTLs) that confer PHS resistance is crucial for marker-assisted selection [9,16]. To date, numerous QTLs for PHS resistance have been successfully mapped onto hexaploidy wheat chromosomes [17,18,19,20,21,22,23,24,25,26,27]. The first QTL found for grain dormancy in wheat was reported in 1993 [17]. Since then, many QTLs associated with PHS resistance were discovered on 21 chromosomes of the wheat genome [20,21,22,28,29,30], and major QTLs for PHS resistance were also detected on chromosomes 2B [22,25], 3A [31], and 4A [21,32,33,34]. Yet, only a few QTLs have been consistently detected across all wheat varieties because the wheat materials are from or grown in different environments [6,8,16]. Because the markers are often useful in the region where the marker was designed, the choice of widely applicable markers, able to detect individual polymorphisms in genes that directly impact PHS, is essential [11].
Until recently, a few genes associated with PHS in wheat had been identified across multiple studies, including TaMFT/TaPHS1, TaMKK3, TaVp-1, Tamyb10, TaSdr, TaQSd, and TaDOG1 [1,35,36,37,38,39,40,41,42,43,44,45,46]. TaMFT/TaPHS1 was identified as a potential gene for a PHS QTL-QPhs.pseru-3A by Nakamura et al., 2018 and later confirmed by TaMFT that was found in a 3AS QTL, for which three single nucleotide polymorphisms (SNPs) were found in the promoter and third intron region, located at the −222 and 646/666 positions, respectively. Notably, those lines carrying alleles at −222 and 646/666 were distinguished as having high resistance to PHS [45,47]. TaMKK3 is another candidate gene in the Phs1-4AL QTL. It possibly encodes a mitogen-activated protein kinase kinase 3. The homologous gene MKK in Arabidopsis positively modifies abscisic acid (ABA) responsiveness [48]. So far, a resistance (R) gene has been identified in QTLs for both grain color and dormancy [28,34]. Recent studies suggest the R gene and TaVp-1 encode Tamyb10 transcription factors and embryo dormancy-related transcription factors, respectively [49]. In addition to those genes, others are considered candidate genes for PHS resistance because they are crucial participants in dormancy and/or germination in related or model plant species, including TaSdr, TaQSd, and TaDOG1 [38,42,43,50].
However, the narrow genetic basis of wheat varieties has limited further improvements to wheat yield and quality, while the wild relatives of wheat offer valuable genetic resources for wheat germplasm improvement. The introduction of this germplasm is a key way to broaden the genetic basis for variation in wheat grain yield and resolve its narrow wheat heredity. Ae. tauschii Cosson (DD, 2n = 2x = 14) is considered the progenitor of common wheat; because it contains abundant beneficial genes to resist PHS, disease, and stress, it is recognized as a potential gene donor for breeding wheat [51]. In the early stage of our study, chromosomal segment substitution lines (CSSLs) were obtained via direct hybridization between the Zhoumai 18 and Ae. tauschii T093 [52]. Fortunately, QDor-3D, an Ae. tauschii-derived stable QTL for PHS, was detected in the CSSLs and found located within the marker interval of Xgpw5094–Xcfd223, for which the value of phenotypic variation explained (PVE) was 11.77% [52]. From the resulting CSSL BC3F4, one of the individuals, 144148-1, had high resistance to PHS and was introgressed with the stable QTL, and so it could be used as key plant material for the precise mapping of Ae. tauschii genes controlling PHS in wheat. In the present study, 144148-1 was further hybridized with Zhoumai 18 to obtain the F2 segregation population, whose PHS resistance trait displayed significant phenotypic variance. However, the genetic and molecular mechanisms of differential PHS resistance in this F2 population were unclear. So, our objectives were threefold: to carry out the precise mapping of PHS resistance genes in the mapped F2 population, to develop user-friendly DNA markers, and to find candidate genes related to PHS for MAS.
2. Results
2.1. Phenotypic Characterization of PHS
For the advanced backcross 1884 F2 population, as well as the T093 and Zhoumai 18 wheat, their respective germination index values were calculated in 2016 to evaluate their PHS resistance in two experimental fields. The statistical analysis showed that T093 had significantly greater resistance to PHS than did Zhoumai 18. The results of ANOVA showed that the germination index (GI) did not differ significantly between environments (F = 0.398, p > 0.05). The trend in resistance to seed germination of the lines in the two experimental fields was basically the same. The germination index (GI) in the F2 population showed continuous variation (Figure 1), which suggested PHS was a typical quantitative trait. Further, the PHS resistance of the F2 population displayed significant phenotypic variance. The GI values of the population ranged from 0 to 1, its average being 0.54 (Figure 1).
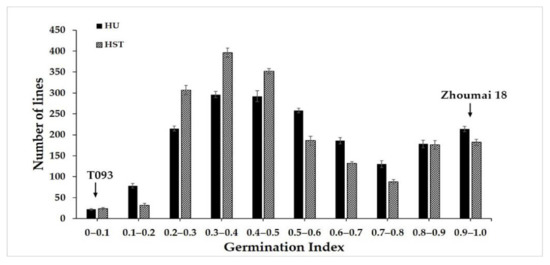
Figure 1.
Frequency distributions of the germination index values of the F2 population of wheat tested at two field sites. HU denotes Henan University; HST denotes the Henan Agriculture High-Technology Science and Technology Park.
2.2. Precise Mapping of Qphs-3D Using the SSR and KASP Markers
Our previous mapping revealed a major PHS QTL (Qdor.3D) within the marker interval of Xgpw5094–Xcfd223. In the present study, four polymorphic simple sequence repeats (SSR) primers were screened within this interval. Based on the bulk segregation analysis (BSA) and the wheat 660K array results, 10 SNPs were selected between Zhoumai 18 and T093 in the targeted QTL region to convert Kompetitive allele-specific PCR (KASP) markers, and their polymorphism between parents was confirmed using the BSA combined with the 660k SNP array. These KASP markers can be useful for identifying the alleles of T093 and Zhoumai 18. Subsequently, the 10 polymorphic KASP markers and four SSR markers were genotyped onto the F2 population and mapped in 3DL. The main peak of the logarithm of the odds (LOD) occurred in the region of 86 cM, situated between Xcfd223 and Xgwm383b (Figure 2). Three QTLs were identified in this interval, explaining 4.74–24.16% of the phenotypic variation in the F2 population (Table 1). The LODs of the three QTLs were 119.44, 70.28, and 22.17, respectively, and their corresponding additive effects were −16.61, −12.59, and −6.02. This indicated that all additive QTLs were derived from Ae. tauschii T093, which suggested these Ae. tauschii-derived loci had positive effects on PHS resistance. At the same time, a 157 cM linkage map on a 3D chromosome was constructed. The main QTL, named QDor-3D.1, having the highest LOD value (119.14), was mapped to an interval flanked by the SSR marker Xcfd223 and the KASP marker A009711. The physical distance between Xcfd223 and A009711 was 2.1 Mb, while the genetic distance between them was approximately 5.25 cM. QDor-3D.1 exerted a significant effect in that it explained 24.16% of the variance in the resistance phenotype (Table 1). QDor-3D.2, which was closely linked with QDor-3D.1, was mapped to an interval flanked by A009711 and A009180. The physical distance of QDor-3D.2 between KASP marker A009711 and KASP marker A009180 was 6.9 Mb, and this explained 15.49% of resistance phenotypic variance. Lastly, QDor-3D.3 was mapped to an interval flanked by the KASP marker A009716 and the SSR marker Xgwm383b. Of the three QTLs, QDor-3D.3 explained the least variance (4.74%) in the resistance phenotype.
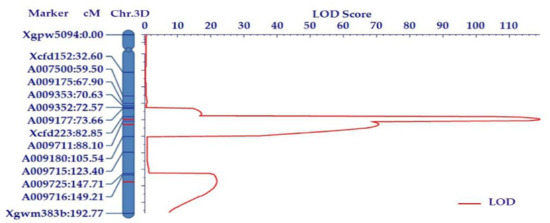
Figure 2.
Linkage groups assigned to the chromosome arms 3DL in the chromosomal segment substitution lines (CSSL) of Zhoumai 18 and T093. DNA markers and their positions are shown along the chromosome arms. The distance between adjacent markers is given in the cM units of Kosambi. The threshold logarithm of the odds (LOD) is 2.61.

Table 1.
Quantitative trait loci (QTL) peak location, left and right markers, LOD values, phenotypic variation explained (PVE), and additive effect of QTL on chromosome 3DL estimated in the F2 population in 2016.
2.3. RNA-Seq and qRT-PCR Assay
RNA-Seq was used to identify differentially expressed genes (DEGs) after wheat seeds’ imbibition. A total of 25.22 Gb of clean data was obtained for the 18 samples after filtering the low-quality reads. The percentage of bases with Q30 was at least 93.43%. The DEGs were detected using the R procedure in Bioconductor package EBseq_DESeq. The gene with |log2(FC)| ≥ 2 (FDR = 0.01) was considered to be differentially expressed. In comparing the genes at 0, 24, and 48 h of Zhoumai 18 and the CSSLs line 495 after germination, a total of 12 DEGs were identified and located in the PHS QTL interval on chromosome 3D: TraesCS3D01G461400, TraesCS3D01G462600, TraesCS3D01G464200, TraesCS3D01G466100, TraesCS3D01G467100, TraesCS3D01G467200, TraesCS3D01G467300, TraesCS3D01G467500, TraesCS3D01G467600, TraesCS3D01G468500, TraesCS3D01G470900, and TraesCS3D01G473200. The results showed that Zhoumai 18 is sensitive to PHS, whereas line 495 is resistant to PHS, whose respective germination index was 0.91 and 0.11 (Figure 3a,b). At the same time, according to the Swiss-protein and gene ontology (GO) annotations, TraesCS3D01G466100 functioned as an E3 ubiquitin-protein ligase in rice and participated in the seed dormancy process, making it a potential candidate gene. Dry seeds and imbibed seeds of Zhoumai 18 and line 495 were evaluated after 24 and 48 h, and their transcriptome and fluorescence quantitative PCR results are shown in Figure 3c,d. Both the fragments per kilobase of exon model per million mapped fragments (FPKM) and relative expression level of TraesCS3D01G466100 were always lower in Zhoumai 18 than in line 495, in all three stages. Therefore, its greater expression may promote dormancy and/or inhibit germination. In the target region, the gene TraesCS3D01G466100 was located at 569,781,286–569790580 bp, encoding a protein functionally similar to RING finger E3 ubiquitin ligase, whose function is the monoubiquitinating of histone H2B. Histone H2B monoubiquitylation plays an important role in seed dormancy regulation levels. Members of this RING finger E3 ubiquitin ligase family were identified as involved in the chromatin remodeling of the seed dormancy mechanism in Arabidopsis [53].
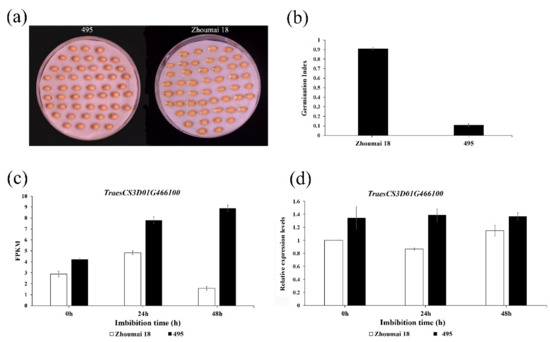
Figure 3.
Expression of TraesCS3D01G466100 promotes seed dormancy in wheat. (a) Germinated seeds of Zhoumai 18 and line 495. Photographs were taken at 48 h after imbibition. (b) Gemination index of Zhoumai 18 and line 495. (c) Fragments per kilobase of exon model per million mapped fragments (FPKM) of TraesCS3D01G466100 in the embryos of Zhoumai 18 and line 495. (d) Relative expression levels of TraesCS3D01G466100 in the embryos of Zhoumai 18 and line 495. Error bars show the standard deviation of three replications.
2.4. TraesCS3D01G466100 cDNA Sequencing and Protein BLAST
The full-length cDNA of TraesCS3D01G466100 amplified from seedlings of Zhoumai 18, T093, and line 495 were sequenced, from which a 2535-bp full-length cDNA was obtained by Geneious 4.8.5. This cDNA sequence encoded a functional peptide of 844 amino acids. In comparing the cDNA sequences of TraesCS3D01G466100 between Zhoumai 18 and line 495, nine SNPs were identified (Figure 4). Just two different amino acids, located at 148 nt (442 bp) and 412 nt (1234 bp), were identified in the predicted transcript (Figure 4 and Figure 5a). The 148 nt of the full-length cDNA sequence corresponded to His(H) in Zhoumai 18 but Tyr (Y) in line 495 and T093, while 412 nt was Gly (G) in Zhoumai 18 and T093 and Arg(R) in line 495. These discrepancies are nonetheless consistent with our results for the DNA sequence alignment. The corresponding cDNA sequence had C in Zhoumai 18 but T in line 495 at 442 bp, and likewise, G in Zhoumai 18 but C in line 495 at 1234 bp.
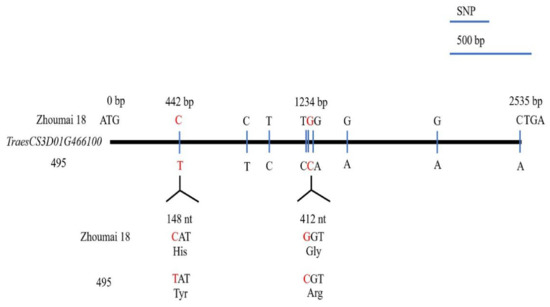
Figure 4.
Sequence variation in the TraesCS3D01G466100 cDNA structure of Zhoumai 18 and line 495 wheat for resistance to pre-harvest sprouting (PHS). The single nucleotide polymorphisms (SNPs) in the coding DNA sequence (CDS) are indicated by vertical lines; each SNP is indicated by the enlarged codons and their encoded amino acids.
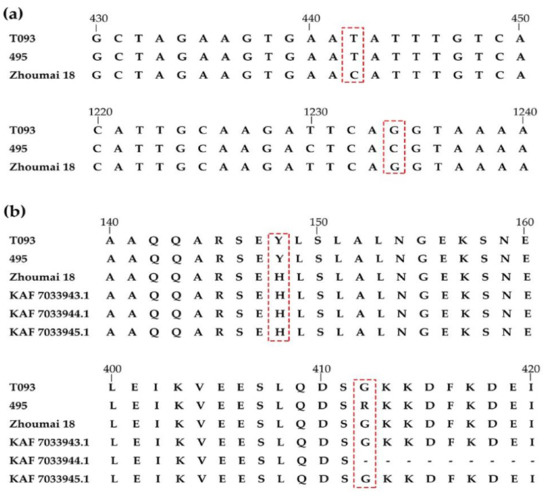
Figure 5.
Analysis of TraesCS3D01G466100 cDNA and alignment of protein sequences. (a) The TraesCS3D01G466100 cDNA structure and variations among the alleles from T093, line 495, and Zhoumai 18. The bases in virtual frames result in the change of amino acids in predicting protein sequence in T093, line 495, and Zhoumai 18. (b) Alignment of protein sequences among homologous TraesCS3D01G466100 from T093, line 495, and Zhoumai 18, and three other Triticum aestivum (KAF7033943.1, KAF7033944.1, and KAF7033945.1). The amino acids in virtual frames indicate the variation among line 495 and T093, Zhoumai 18, and three other Triticum aestivum.
3. Discussion
Using PHS-resistance genes is crucial for wheat breeding practices. The development of PHS resistance lines offers one basis for the enrichment of genetic resources in wheat breeding programs; accordingly, introducing genes from wheat-related species could provide an effective way to significantly improve wheat grain quality and its resistance to PHS. Liu et al. and Yu et al. both found that genes from T. turgidum and Ae. tauschii can be applied to enhance the PHS resistance of wheat [54,55]. A direct cross of Ae. tauschii with common wheat was reported in two studies [56,57]. Moreover, the CSSL population has been widely used for the fine mapping of complex traits in wheat, rice, maize, and other crops since 1987 [52,58,59,60]. In our previous study, we developed a population of CSSLs by relying on synthetic octaploid wheat as a bridge to identify a QDor-3D for PHS [52]. The direct hybridization of wheat and Ae. tauschii can effectively avoid interference from the A and B genomes, and the excellent genetic background of wheat makes the CSSLs more convenient for wheat breeding. Furthermore, in this study, an advanced F2 population was obtained from the individual 144148-1 of CSSLs backcrossed with Zhoumai 18 and then self-crossed. Therefore, only D genome introgression existed in the lines, and the chromosome would be further recombined via linkage and interchange processes. Moreover, any interference from genetic background variation was eliminated, which meant the reserved lines still retained better agronomic traits, and the QTL was guaranteed to be more accurate.
As it stands, most studies have indicated that when multiple genes are controlling the same trait, they are often closely linked, and so the QTL close linkage is a common phenomenon. For example, Fofana et al., 2009 identified three QTLs: QCL.crc-3A, QCL.crc-3B, and QCL.crc-3D associated with the red coloring of the seed coat. Work by Monna et al., 2002 produced high-resolution linkage mapping of Hd3, finding that Hd3a and Hd3b were tightly linked in that region. Fortunately, by expanding the population to generate a secondary segregation population, QTL mapping can more accurately decompose those QTL clusters under genetic background control. For example, Kumar et al. also identified three QTLs using GR and PG measures, namely QPhs.spa-3A, QPhs.spa-3B, and QPhs.spa-3D, which were localized to the same region as in the study of Fofana et al., mentioned above [61,62]. In our study, QDor-3D.1, QDor-3D.2, and QDor-3D.3 were all identified using a large segregating population derived from advanced backcross progeny. QDor-3D.1 and QDor-3D.2 were tightly linked in the QDor-3D region, which was positioned between Xcfd223 and A009180. More importantly, Xcfd223 was detected in both studies by Fofana et al. and Kumar et al. above, which suggests the QTL interval on 3DL figures prominently in the reduction of wheat germination. The co-located QTLs conferring PHS resistance could be found at analogous positions on 3D by respectively using CSSLs populations and the advanced F2 segregating population, thus providing strong evidence of the existence of nearby candidate genes for the PHS-resistance trait. Compared with previous studies, the PHS QTL region was saturated with more markers in our study, and so, it would be more convenient to further study the mechanism underpinning PHS resistance with these QTLs.
Several genes have been verified as associated with PHS in wheat in recent reports, including TaMFT/TaPHS1, TaMKKK3, Tamyb10, and TaVp1 in wheat and TaSdr, TaQSd, and TaDOG1 in related crops (i.e., barley, maize, rice) or model (Arabidopsis) organisms [1,35,36,37,38,39,40,41,42,43,44,45,46]. In recent years, RNA-Seq has emerged as a powerful technique for DEG identification at the transcriptome level, which could provide insights into the molecular mechanism(s) underpinning PHS resistance [63]. In addition, the quantitative analysis of DEGs can be further verified by real-time quantitative PCR. According to the DEGs in the QDor-3D.2 region detected by RNA-Seq, the expression level of TraesCS3D01G466100 differed significantly between Zhoumai 18 and line 495, and TraesCS3D01G466100 might be a RING finger-dependent E3 ubiquitin-protein ligase in rice that also participates in seed dormancy process according to its Swiss Prot and GO annotations. E3 ubiquitin-protein ligase homologous alignment with three other T. aestivum encoded on 3D chromosome indicated the locus 148 nt in line 495 was the same with his parent Ae. tauschii T093 yet different from Zhoumai 18 and three other T. aestivum (KAF 7033943.1, KAF 7033944.1, and KAF 7033945.1). The change in amino acid results from the sense mutation of the 442 positions. Here, the base was T at 442 bp in line 495, which was the same as in T093, which indicated that base T was most likely from T093 during introgression. The changed base causes the change in the amino acid, which then affects the function of E3, resulting in the significant QTLs related to PHS and the difference in the GI between Zhoumai 18 and line 495. Interestingly, in line 495, another base C at 1234 bp differs from both Zhoumai 18 and T093, which may be a mutation during introgression.
Ubiquitin-mediated protein degradation is an essential protein post-translational modification in eukaryotic cells, one related to the fine-scaled regulation of their biological growth and environmental adaptation by organisms. E3 (ubiquitin-ligating enzyme) along with E1 (ubiquitin-activating enzyme) and E2 (ubiquitin-conjugation enzyme) are required for target protein ubiquitination, which eventually leads to the degradation or relocation of that targeted protein. In recent years, mounting studies have shown that E3 plays a positive or negative regulatory role in a variety of disease resistance signaling pathways, mainly in the ABA signal transduction pathway. Surprisingly, however, the effect of E3 on PHS resistance is rarely reported on, especially in wheat. Liu et al.,2007 found that HISTONE MONOUBIQUITINATION1 (HUB1) encodes a C3HC4 RING finger protein. Because HUB1 probably regulates the monoubiquitinating of histone H2B that influenced PHS-related gene expression in Arabidopsis, it has been proposed that chromatin remodeling could be critical for the regulation of seed dormancy [53]. Zheng et al., 2018 also found that a RING-type ubiquitin E3-ligase RHA2b targets MYB30 for degradation to modulate ABA signaling in Arabidopsis [64]. Another study showed that OsHUB1/2 in rice was homologous to Arabidopsis HUB1/2 and that treatment with hormones (JA, SA, ET, or ABA) induced the transcription of OsHUB1/2. This way, OsHUB1/2 may interact with SPIN6 to regulate the SPL11-mediated rice immunity and cell death [65]. A database search revealed the sequence homology of HUB1 and HUB2 is based on a conserved gene that is required for H2B monoubiquitination to occur in different kinds of organisms, including Medicago truncatula (Mt_Bre1), rice (O. sativa; Os_Bre1A and Os_Bre1B), yeast (Sc_Bre1), and even humans (Hu_Bre1A and Hu_Bre1B) [66,67,68,69]. Nevertheless, this type of gene has never been found associated with PHS resistance in wheat until now (Figure 5b). We therefore postulate TraesCS3D01G466100 is a novel PHS resistance-related gene critical for the establishment of seed dormancy in wheat. This provides a new way forward for elucidating the mechanism of resistance to PHS in wheat. We anticipate it will be a valuable potential gene for breeding PHS-resistant wheat crops.
4. Materials and Methods
4.1. Plant Materials and Mapping Populations
In our previous study, the CSSLs were developed by direct hybridization between Ae. tauschii accession T093 that featured strong resistance to PHS and the recurrent parent Zhoumai 18, whose susceptibility to PHS is high [52,70,71]. Here, individual 144148-1 with stable PHS resistance in the CSSLs was backcrossed again with Zhoumai 18 and then self-crossed. In this way, a total of 1884 F2 plants were finally generated for further investigation in this study (Figure 6).

Figure 6.
Crossing scheme used to obtain the F2 population of wheat in this study. Dt denotes the genome of Aegilops tauschii.
4.2. Assessment of PHS in the Wheat F2 Population
To assess their PHS resistance, the F2 population, Zhoumai 18, and T093, were grown in 2015 in two places, Henan University (HU) and Henan Agriculture Hi-Tech Science and Technology Park (HST), respectively, both in China. The PHS resistance of the F2 population was evaluated in 2016. In the field experiment, all the lines were planted in 1.2-m-long single-row plots with 10 cm between plants and a 30-cm row gap. Twelve seeds were sown in each row. Five spikes were removed from each wheat plant after it attained physiological maturity. After all experimental plants were harvested, their spikes were dried for seven days at room temperature and stored in a refrigerator at −25 °C to maintain their dormancy until the germination test was conducted, following the methodology of Liu et al. [31]. Fifty seeds from each spikelet were sterilized with 5% sodium hypochlorite and then rinsed five times with sterile water until clean. These seeds were then placed in Petri plates, on moistened filter paper with sterilized water, at 22 °C in the dark. Over the next seven days, the germinating seeds were counted, and any germinated seeds found with radicle protrusions were immediately discarded. The phenotyping was conducted as described by James et al. [17]. The germination tests consisted of three replicates, each consisting of seeds from a different spike. The germination index (GI) was calculated as follows:
where n1 is the number of germinated seeds during the first day, n2 is the number of germinated seeds during the second day, and total seeds are the number of total observed seeds in each Petri plate. The one-way analysis of variance (ANOVA) of GI from two environments was conducted on the mean of the three replications using SPSS Statistics 19.0. The mean GI values of the two sites were used for the next QTL analysis.
GI = (7 × n1 + 6 × n2 +……. + 1 × n7)/(total seeds × 7)
4.3. DNA Extraction and Marker Analysis
The DNA of fresh, healthy leaves was isolated by the modified cetyltrimethylammonium bromide (CTAB) method [72] and dissolved in sterile distilled water. Both DNA purity and quality were measured with a nanophotometer (Implen Inc., Los Angeles, CA, USA).
Because our previous study found that a major PHS QTL (Qdor.3D) was located within the marker interval of Xgpw5094–Xcfd223 [52], this study further screened polymorphic SSR markers within this interval. Four polymorphic SSR primers, Xgpw5094, Xcfd223, Xcfd152, and Xgwm383b, were synthesized and designed based upon the genetic map of the D genome, in GrainGene 2.0 software (http://wheat.pw.usda.gov (accessed on 20 March 2021)), and the Ae. tauschii genome sequences [73,74] (Table 2). The PCR amplification for these SSR markers was conducted according to Röder and Bendich [75]. Specifically, the PCR cycles began with an initial denaturation at 95 °C for 3 min, then 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 45 s, and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. The ensuing PCR products were separated from 8% non-denaturing polyacrylamide gel sand and visualized with silver staining [76].

Table 2.
Primer sequences of the simple sequence repeats (SSR) assays developed in this study.
4.4. Bulk Segregation Analysis (BSA) with the 660K Chip Array and KASP Marker Development
The pooled genomic DNA from 30 F2 lines with high PHS resistance and likewise from 30 F2 lines with high PHS sensitivity, along with the DNA from T093 and Zhoumai 18, were used for the wheat 660K SNP chip analysis carried out by Beijing Golden Marker Co., Ltd. (Beijing, China). The data quality control was performed by Axiom Analysis Suite to provide a clean sample of SNP typing data. The calling and clustering of SNP genotypes were performed in Genome Studio Polyploid Clustering v 1.0 software. The correspondence of polymorphic SNPs in the BSA to chromosomes was based on the 660K physical map. The polymorphic SNPs between the high and low PHS-resistant bulks associated with the target QTLs on chromosome 3D in the marker interval Xgpw5094–Xcfd223 were successfully converted into 10 KASP markers for the fine mapping [77] (Table 3).

Table 3.
Primer sequences of the Kompetitive allele-specific PCR (KASP) assays developed in this study.
4.5. QTL Analysis
For the QTL analysis, QTL IciMapping v4.0 was used with its composite interval mapping function [78]. Four SSR markers and 10 KASP markers were assessed using the map function, with the logarithm of the odds (LOD) threshold set to 2.61. QTLs were detected if their LOD score was above 2.61. The nnTwoOpt algorithm was used for the markers in a given group, and then the rippling step was applied to obtain a linkage map of the F2 population. Finally, the PHS QTL regions were identified by running the BIP function using the inclusive composite interval mapping with an additive effect (ICIM-ADD). This QTL analysis was conducted for the PHS phenotypes of all 1884 members of the F2 population.
4.6. RNA Isolation and Sequencing
For RNA-Seq, dried seeds and treated seeds were sterilized. Seeds of Zhoumai 18 and line 495 were surface-sterilized as described above for the germination index, and these were imbibed between filter papers in Petri plates at room temperature under darkness. Dry seeds and imbibed seeds after 24 and 48 h were frozen in liquid nitrogen, then stored at −80 °C until their RNA extraction.
Total RNA was isolated with the Spin Column Plant Total RNA Purification Kit by following the manufacturer’s protocol (Sangon Biotech, Shanghai, China). RNA degradation was checked on 1% agarose, and contamination was checked by a nanophotometer spectrophotometer (Implen, Los Angeles, CA, USA). The quantification of RNA was performed using the Qubit RNA Assay Kit with a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA). Next, RNA integrity was checked using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA).
Sequencing libraries were generated for sequencing on the Illumina Novaseq 6000 platform (Illumina Inc., San Diego, CA, USA), by Biomarker Technology, in Beijing. The mRNA was purified from total RNA by poly-T oligo-attached magnetic beads. The cDNA was then synthesized by short mRNA fragments with PCR amplification used to enrich the cDNA templates. Finally, the libraries were sequenced. Three biological replicates were used for each of the sampling points.
4.7. Gene Expression Using Quantitative Real-Time PCR
According to the QTL mapping analysis, the gene related to PHS resistance was located in 564.6–573.6 Mb on chromosome 3D based on the IWGSC_RefSeq_v1.0 wheat genome. Combined with the transcriptome results, 12 DEGs were screened in the same interval. The high expression of TraesCS3D01G466100 was positively correlated with PHS resistance, and it was selected for the next experiment.
Real-time PCR was carried out to determine the expression levels of genes. The qRT-PCR was conducted on an ABI LightCyler QuantStudio 6 instrument, using the HiScript Reverse Transcriptase (RNase H) and 5 × HiScript Buffer (Vazyme, Nanjing, China). Each reaction was performed in a 20 µL mixture containing 10 µL of SYBR Green Master Mix; three biological replications were performed. The actin gene served as the internal control, and the specific primers were designed in Primer Premier 5.0 (Table 4). Data are presented here as relative transcript levels and calculated using the 2−ΔΔCt method [79].

Table 4.
Primers used for the qRT-PCR and cDNA cloning.
4.8. TraesCS3D01G466100 cDNA Sequencing and Protein BLAST
Total RNA was reversed to cDNA by a RevertAid First Stand cDNA Synthesis Kit (Fermentas). The primers for TraesCS3D01G466100 cDNA on the 3D chromosome were designed based on Ae. tauschii by a National Center for Biotechonology Information (NCBI) homologous BLAST, using Primer Premier 5.0 (Table 3). The cDNAs of Zhoumai 18, line 495, and T093 were used as templates to perform the PCR amplification. Next, the genes were cloned into a pEASY-Blunt simple vector and transformed into Trans-T1 Escherichia coli cells (TransGen, Beijing, China). Three positive clones were sequenced to ensure the accuracy of nucleotide sequences (Sunya, Zhengzhou, China). For their homology alignment, the multiple sequences were compared and analyzed using Geneious v4.8.5. These E3 ubiquitin-protein ligases were all derived from a 3D chromosome of T. aestivum.
5. Conclusions
An advanced F2 population was spawned from the individual 144148-1 of Ae. tauschii (T093)—T. aestivum (Zhoumai 18) CSSLs backcrossed with Zhoumai 18 and then self-crossed. QDor.3D.1, QDor.3D.2, and QDor.3D.3 were identified on the long arm of chromosome 3DL, using an advanced backcross F2 mapping population, for which QDor.3D.1 and QDor.3D.2 were tightly linked. Their additive effects revealed that these additive QTLs were derived from Ae. tauschii T093, which means these Ae. tauschii-derived loci positively influence PHS resistance. RNA-Seq and RT-PCR results indicated that TraesCS3D01G466100 was probably the candidate PHS-related gene. The functional annotation of TraesCS3D01G466100 pointed to an E3 ubiquitin-protein ligase, encoding a C3HC4 RING finger protein. The cDNA and predicted protein sequence analysis showed two amino acids altered between Zhoumai 18, line 495, and T093. Moreover, homology protein BLAST suggested the synonymous mutation site of 148 nt amino acids in line 495 was derived from the hybridization of Ae. tauschii. Because the RING finger-dependent E3 ubiquitin-protein ligase has not yet been found related to PHS resistance in wheat, we propose that the TraesCS3D01G466100 studied here is a novel PHS resistance-related gene in wheat. Both the QTLs and this newly identified TraesCS3D01G466100 should enrich the genetic resources available for wheat breeding aimed at mitigating PHS and could provide an important, fresh basis for agricultural practices promoting PHS resistance in cereal crops.
Author Contributions
Conceptualization, D.Z. and S.L.; data curation, J.H.; formal analysis, J.H., D.Z., X.C., M.H., and S.S.; funding acquisition, S.L. and Y.S.; investigation, J.H., D.Z., X.C. and Y.L.; methodology, J.H. and D.Z.; project administration, D.Z.; resources, S.L.; software, Q.S.; supervision Y.S. and S.L.; validation, S.L.; writing—original draft preparation, J.H. and Y.S.; writing—review and editing, J.H., Y.S., and S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 31871615, 31571649, and 31601297), China Postdoctoral Science Foundation (grant no. 2020M672218), and Kaifeng Key R & D and Promotion Project (grant no. 1902002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Oluwaseyi, S.; Nicholas, B.; James, S.; Simon, B.; Tina, H.; Peter, J.; Peter, W.; Tanja, G.; Duncan, S.; Barbara, B.; et al. The wheat Phs-A1 pre-harvest sprouting resistance locus delays the rate of seed dormancy loss and maps 0.3 cM distal to the PM19 genes in UK germplasm. J. Exp. Bot. 2016, 67, 4169–4178. [Google Scholar]
- Yang, J.; Tan, C.; Lang, J.; Tang, H.; Hao, M.; Tan, Z.; Yu, H.; Zhou, Y.; Liu, Z.; Li, M.; et al. Identification of qPHS.sicau-1B and qPHS.sicau-3D from synthetic wheat for pre-harvest sprouting resistance wheat improvement. Mol. Breed. 2019, 39, 132. [Google Scholar] [CrossRef]
- Depauw, R.M.; Hucl, P.; Knox, R.E.; Singh, A.K.; Fox, S.L.; Humphreys, D.G. Developing standardized methods for breeding preharvest sprouting resistant wheat, challenges and successes in Canadian wheat. Euphytica 2012, 188, 7–14. [Google Scholar] [CrossRef]
- Li, C.; Ni, P.; Francki, M.; Hunter, A.; Zhang, Y.; Schibeci, D.; Li, H.; Tarr, A.; Wang, J.; Cakir, M.; et al. Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct. Integr. Genom. 2004, 4, 84–93. [Google Scholar] [CrossRef]
- King, R.; Richards, R. Water uptake in relation to pre-harvest sprouting damage in wheat: Ear characteristics. Aust. J. Agric. Res. 1984, 35, 327–336. [Google Scholar] [CrossRef]
- Mares, D.J.; Mrva, K. Wheat grain preharvest sprouting and late maturity alpha-amylase. Planta 2014, 240, 1167–1178. [Google Scholar] [CrossRef]
- Paterson, A.H.; Sorrells, M.E.; Obendorf, R.L. Methods of evaluation for preharvest sprouting resistance in wheat breeding programs. Can. J. Plant Sci. 1989, 69, 681–689. [Google Scholar] [CrossRef]
- Flintham, J.; Adlam, R.; Bassoi, M.; Holdsworth, M.; Gale, M. Mapping genes for resistance to sprouting damage in wheat. Euphytica 2002, 126, 39–45. [Google Scholar] [CrossRef]
- Kulwal, P.; Ishikawa, G.; Benscher, D.; Feng, Z.; Zongyun, F.; Jadhav, A.; Mehetre, S.; Sorrells, M.E. Association mapping for pre-harvest sprouting resistance in white winter wheat. Theor. Appl. Genet. 2012, 125, 793–805. [Google Scholar] [CrossRef]
- Graybosch, R.A.; Amand, P.S.; Bai, G.H. Evaluation of genetic markers for prediction of preharvest sprouting tolerance in hard white winter wheats. Plant Breed. 2013, 132, 359–366. [Google Scholar] [CrossRef]
- Vetch, J.M.; Stougaard, R.N.; Martin, J.M.; Giroux, M.J. Review: Revealing the genetic mechanisms of pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.). Plant Sci. 2019, 281, 180–185. [Google Scholar] [CrossRef]
- Hareland, G.A. Effects of pearling on falling number and α-amylase activity of preharvest sprouted spring Wheat. Cereal Chem. J. 2003, 80, 232–237. [Google Scholar] [CrossRef]
- Bamard, A.; Bona, L. Sprout damage and falling number in South Mrican and Hungarian wheats. Cereal Res. Commun. 2004, 32, 259–264. [Google Scholar]
- Clarke, F.R.; Knox, R.E.; Depauw, R.M. Expression of dormancy in a spring wheat cross grown in field and controlled environment conditions. Euphytica 2005, 143, 297–300. [Google Scholar] [CrossRef]
- Miles, C.M.; Wayne, M. Quantitative Trait Locus (QTL) Analysis. Nat. Educ. 2008, 1, 208. [Google Scholar]
- Gao, X.; Hu, C.H.; Li, H.Z.; Yao, Y.J.; Meng, M.; Dong, J.; Zhao, W.C.; Chen, Q.J.; Li, X.Y. Factors affecting pre-harvest sprouting resistance in wheat (Triticum aestivum L.): A review. J. Anim. Plant Sci. 2013, 23, 556–565. [Google Scholar]
- Anderson, J.A.; Sorrells, M.E.S.; Tanksley, S.D. RFLP analysis of genomic regions associated with resistance to pre-harvest sprouting in wheat. Crop Sci. 1993, 33, 453–459. [Google Scholar] [CrossRef]
- Kato, K.; Nakamura, W.; Tabiki, T.; Miura, H.; Sawada, S. Detection of loci controlling seed dormancy on group 4 chromosomes of wheat and comparative mapping with rice and barley genomes. Theor. Appl. Genet. 2001, 102, 980–985. [Google Scholar] [CrossRef]
- Mares, D.; Mrva, K.; Cheong, J.; Williams, K.; Watson, B.; Storlie, E.; Sutherland, M.; Zou, Y. A QTL located on chromosome 4A associated with dormancy in white- and red-grained wheats of diverse origin. Theor. Appl. Genet. 2005, 111, 1357–1364. [Google Scholar] [CrossRef]
- Mori, M.; Uchino, N.; Chono, M.; Kato, K.; Miura, H. Mapping QTLs for grain dormancy on wheat chromosome 3A and the group 4 chromosomes, and their combined effect. Theor. Appl. Genet. 2005, 110, 1315–1323. [Google Scholar] [CrossRef]
- Ogbonnaya, F.C.; Imtiaz, M.; Ye, G.; Hearnden, P.R.; Hernandez, E.; Eastwood, R.F.; Van Ginkel, M.; Shorter, S.C.; Winchester, J.M. Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor. Appl. Genet. 2008, 116, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, J.D.; Tanaka, J.; Benscher, D.; Sorrells, M.E. Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor. Appl. Genet. 2009, 119, 1223–1235. [Google Scholar] [CrossRef]
- Rasul, G.; Humphreys, D.G.; Brûlé-Babel, A.; McCartney, C.A.; Knox, R.E.; Depauw, R.M.; Somers, D.J. Mapping QTLs for pre-harvest sprouting traits in the spring wheat cross ‘RL4452/AC Domain’. Euphytica 2009, 168, 363–378. [Google Scholar] [CrossRef]
- Liu, S.; Sehgal, S.K.; Li, J.; Lin, M.; Trick, H.N.; Yu, J.; Gill, B.S.; Bai, G. Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics 2013, 195, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Somyong, S.; Ishikawa, G.; Munkvold, J.D.; Tanaka, J.; Benscher, D.; Cho, Y.G.; Sorrells, M.E. Fine mapping of a preharvest sprouting QTL interval on chromosome 2B in white wheat. Theor. Appl. Genet. 2014, 127, 1843–1855. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Hayashi, K.; Tokui, M.; Mori, M.; Miura, H.; Onishi, K. Detection of QTLs for traits associated with pre-harvest sprouting resistance in bread wheat (Triticum aestivum L.). Breed. Sci. 2016, 66, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.L.; Wang, S.X.; Zhang, H.P.; Zhao, L.X.; Wu, Z.Y.; Jiang, H.; Cao, J.J.; Liu, K.; Qin, M.; Lu, J.; et al. Identification of major loci for seed dormancy at different post-ripening stages after harvest and validation of a novel locus on chromosome 2AL in common wheat. Mol. Breed. 2016, 36, 174. [Google Scholar] [CrossRef]
- Groos, C.; Gay, G.; Perretant, M.R.; Gervais, L.; Bernard, M.; Dedryver, F.; Charmet, G. Study of the relationship between preharvest sprouting and grain color by quantitative trait loci analysis in a white× red grain bread wheat cross. Theor. Appl. Genet. 2002, 104, 39–47. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, H.; Cheng, M.P.; Dankwa, K.O.; Chen, Z.X.; Li, Z.Y.; Gao, S.; Liu, Y.X.; Jiang, Q.T.; Lan, X.J.; et al. Genome-Wide association study for pre-harvest sprouting resistance in a large germplasm collection of Chinese wheat Landraces. Front. Plant Sci. 2017, 8, 401. [Google Scholar] [CrossRef]
- Martinez, S.A.; Godoy, J.; Huang, M.; Zhang, Z.W.; Carter, A.H.; Garland Campbell, K.A.; Steber, C.M. Genome-wide association mapping for tolerance to preharvest sprouting and low falling numbers in wheat. Front. Plant Sci. 2018, 9, 141. [Google Scholar] [CrossRef]
- Liu, S.; Cai, S.; Graybosch, R.; Chen, C.; Bai, G. Quantitative trait loci for resistance to pre-harvest sprouting in US hard white winter wheat Rio Blanco. Theor. Appl. Genet. 2008, 117, 691–699. [Google Scholar] [CrossRef]
- Chen, C.X.; Cai, S.B.; Bai, G.H. A major QTL controlling seed dormancy and pre-harvest sprouting resistance on chromosome 4A in a Chinese wheat landrace. Mol. Breed. 2007, 21, 351–358. [Google Scholar] [CrossRef]
- Tyagi, S.; Gupta, P.K. Meta-analysis of QTLs involved in pre-harvest sprouting tolerance and dormancy in bread wheat. Triticeae Genom. Genet. 2012, 3. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, D.; Liu, S.; Zhang, G.; Yu, J.; Fritz, A.K.; Bai, G. Genome-wide association analysis on pre-harvest sprouting resistance and grain color in U.S. winter wheat. BMC Genom. 2016, 17, 794. [Google Scholar] [CrossRef]
- McCarty, D.R.; Hattori, T.; Carson, C.B.; Vasil, V.; Lazar, M.; Vasil, I.K. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 1991, 66, 895–905. [Google Scholar] [CrossRef]
- Himi, E.; Mares, D.J.; Yanagisawa, A.; Noda, K. Effect of grain colour gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat. J. Exp. Bot. 2002, 53, 1569–1574. [Google Scholar] [CrossRef]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef]
- Sugimoto, K.; Takeuchi, Y.; Ebana, K.; Miyao, A.; Hirochika, H.; Hara, N.; Ishiyama, K.; Kobayashi, M.; Ban, Y.; Hattori, T.; et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 2010, 107, 5792–5797. [Google Scholar] [CrossRef]
- Liu, S.B.; Sehgal, S.K.; Lin, M.; Li, J.R.; Trick, H.N.; Gill, B.S.; Bai, G.H. Independent mis-splicing mutations in TaPHS1 causing loss of preharvest sprouting (PHS) resistance during wheat domestication. New Phytol. 2015, 208, 928–935. [Google Scholar] [CrossRef]
- Torada, A.; Koike, M.; Ogawa, T.; Takenouchi, Y.; Tadamura, K.; Wu, J.; Matsumoto, T.; Kawaura, K.; Ogihara, Y. A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr. Biol. 2016, 26, 782–787. [Google Scholar] [CrossRef]
- Feng, Y.; Qu, R.D.; Liu, S.M.; Yang, Y. Rich haplotypes of Viviparous-1 in Triticum aestivum subsp. spelta with different abscisic acid sensitivities. J. Agric. Food Chem. 2017, 97, 497–504. [Google Scholar]
- Onishi, K.; Yamane, M.; Yamaji, N.; Tokui, M.; Kanamori, H.; Wu, J.; Komatsuda, T.; Sato, K. Sequence differences in the seed dormancy gene Qsd1 among various wheat genomes. BMC Genom. 2017, 18, 497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Xia, X.C.; He, Z.H. The seed dormancy allele TaSdr-A1a associated with pre-harvest sprouting tolerance is mainly present in Chinese wheat landraces. Theor. Appl. Genet. 2017, 130, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.H.; Fu, L.; Wu, Q.H.; Chen, J.J.; Chen, Y.X.; Xie, J.Z.; Wang, Z.Z.; Wang, G.X.; Zhang, D.Y.; Liang, Y.; et al. QTL mapping revealed TaVp-1A conferred pre-harvest sprouting resistance in wheat population Yanda 1817× Beinong 6. J. Integr. Agric. 2017, 16, 435–444. [Google Scholar] [CrossRef]
- Jiang, H.; Zhao, L.X.; Chen, X.J.; Cao, J.J.; Wu, Z.Y.; Liu, K.; Zhang, C.; Wei, W.X.; Xie, H.Y.; Li, L.; et al. A novel 33-bp insertion in the promoter of TaMFT-3A is associated with pre-harvest sprouting resistance in common wheat. Mol. Breed. 2018, 38, 69. [Google Scholar] [CrossRef]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R.; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S. Grain dormancy genes responsible for preventing pre-harvest sprouting in barley and wheat. Breed. Sci. 2018, 68, 295–304. [Google Scholar] [CrossRef]
- Danquah, A.; De Zélicourt, A.; Boudsocq, M.; Neubauer, J.; Frey, N.F.D.; Leonhardt, N.; Pateyron, S.; Gwinner, F.; Tamby, J.P.; Ortiz-Masia, D.; et al. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 2015, 82, 232–244. [Google Scholar] [CrossRef]
- Bailey, P.C.; McKibbin, R.S.; Lenton, J.R.; Holdsworth, M.J.; Flintham, J.E.; Gale, M.D. Genetic map locations for orthologous Vp1 genes in wheat and rice. Theor. Appl. Genet. 1999, 98, 281–284. [Google Scholar] [CrossRef]
- Kole, C. Wild Crop Relatives Genomic and Breeding Resources Cereals; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Wang, J.R.; Luo, M.C.; Chen, Z.X.; You, F.M.; Wei, Y.M.; Zheng, Y.L.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937. [Google Scholar] [CrossRef]
- Zhang, D.L.; He, J.; Huang, L.Y.; Zhang, C.C.; Zhou, Y.; Su, Y.R.; Li, S.P. An advanced backcross population through synthetic octaploid wheat as a “bridge”: Development and QTL detection for seed dormancy. Front. Plant Sci. 2017, 8, 2123. [Google Scholar]
- Liu, Y.; Koornneef, M.; Soppe, W.J. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 2007, 19, 433–444. [Google Scholar] [CrossRef]
- Liu, D.C.; Lan, X.J.; Rong Wang, Z.R.; Zheng, Y.L.; Zhou, Y.H.; Yang, J.L.; Chi, Y. Evaluation of Aegilops tauschii Cosson for preharvest sprouting tolerance. Genet. Resour. Crop Evol. 1998, 45, 495–498. [Google Scholar]
- Yu, M.; Chen, G.Y.; Zhang, L.Q.; Liu, Y.X.; Liu, D.C.; Wang, J.R.; Pu, Z.E.; Zhang, L.; Lan, X.J.; Wei, Y.M.; et al. QTL Mapping for important agronomic traits in synthetic hexaploid wheat derived from Aegiliops tauschii ssp. tauschii. J. Integr. Agric. 2014, 13, 1835–1844. [Google Scholar] [CrossRef]
- Sehgal, S.K.; Kaur, S.; Gupta, S.; Sharma, A.; Kaur, R.; Bains, N.S. A direct hybridization approach to gene transfer from Aegilops tauschii Coss. to Triticum aestivum L. Plant Breed. 2010, 130, 98–100. [Google Scholar] [CrossRef]
- Olson, E.L.; Rouse, M.N.; Pumphrey, M.O.; Bowden, R.L.; Gill, B.S.; Poland, J.A. Simultaneous transfer, introgression, and genomic localization of genes for resistance to stem rust race TTKSK (Ug99) from Aegilops tauschii to wheat. Theor. Appl. Genet. 2013, 126, 1179–1188. [Google Scholar] [CrossRef]
- Gill, B.S.; Raupp, W.J. Direct genetic transfers from Aegilops squarrosa L. to hexaploid wheat. Crop. Sci. 1987, 27, 445–450. [Google Scholar] [CrossRef]
- Ma, X.; Han, B.; Tang, J.; Zhang, J.; Cui, D.; Geng, L.; Zhou, H.; Li, M.; Han, L. Construction of chromosome segment substitution lines of Dongxiang common wild rice (Oryza rufipogon Griff.) in the background of the japonica rice cultivar Nipponbare (Oryza sativa L.). Plant Physiol. Biochem. 2019, 144, 274–282. [Google Scholar] [CrossRef]
- Yang, H.; Wang, W.; He, Q.; Xiang, S.; Tian, D.; Zhao, T.; Gai, J. Identifying a wild allele conferring small seed size, high protein content and low oil content using chromosome segment substitution lines in soybean. Theor. Appl. Genet. 2019, 132, 2793–2807. [Google Scholar] [CrossRef]
- Fofana, B.; Humphreys, D.G.; Rasul, G.; Cloutier, S.; Brûlé-Babel, A.; Woods, S.; Lukow, O.M.; Somers, D.J. Mapping quantitative trait loci controlling pre-harvest sprouting resistance in a red × white seeded spring wheat cross. Euphytica 2009, 165, 509–521. [Google Scholar] [CrossRef]
- Kumar, S.; Knox, R.E.; Clarke, F.R.; Pozniak, C.J.; DePauw, R.M.; Cuthbert, R.D.; Fox, S. Maximizing the identification of QTL for pre-harvest sprouting resistance using seed dormancy measures in a white-grained hexaploid wheat population. Euphytica 2015, 205, 287–309. [Google Scholar] [CrossRef]
- Huh, S.M.; Hwang, Y.S.; Shin, Y.S.; Nam, M.H.; Kim, O.Y.; Yoon, I.S. Comparative transcriptome profiling of developing caryopses from two rice cultivars with differential dormancy. J. Plant Physiol. 2013, 170, 1090–1100. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, Z.; Ma, L.; Liao, C. The ubiquitin E3 ligase RHA2b promotes degradation of MYB30 in abscisic acid signaling. Plant Physiol. 2018, 178, 428–440. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, J.; He, F.; Wang, Z.; Ning, Y.; Wang, G.L. OsHUB1 and OsHUB2 interact with SPIN6 and form homo- and hetero-dimers in rice. Plant Signal. Behav. 2015, 10, e1039212. [Google Scholar] [CrossRef]
- Hwang, W.W.; Venkatasubrahmanyam, S.; Ianculescu, A.G.; Tong, A.; Boone, C.; Madhani, H.D. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 2003, 11, 261–266. [Google Scholar] [CrossRef]
- Wood, A.; Krogan, N.J.; Dover, J.; Schneider, J.; Heidt, J.; Boateng, M.A.; Dean, K.; Golshani, A.; Zhang, Y.; Greenblatt, J.F.; et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 2003, 11, 267–274. [Google Scholar] [CrossRef]
- Kim, J.; Hake, S.B.; Roeder, R.G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell 2005, 20, 759–770. [Google Scholar] [CrossRef]
- Zhu, B.; Zheng, Y.; Pham, A.D.; Mandal, S.S.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell 2005, 20, 601–611. [Google Scholar] [CrossRef]
- Su, Y.R.; Liao, P.A.; Song, D.Y.; Huang, S.Q.; He, J.; Gao, X.F.; Li, S.P. Application of Aegilops tauschii—Triticum aestivum recombinant inbred lines for grain protein content QTL detection and wheat improvement. Can. J. Plant Sci. 2020, 100, 1–31. [Google Scholar] [CrossRef]
- Zhang, D.L.; Zhou, Y.; Zhao, X.P.; Lv, L.L.; Zhang, C.C.; Li, J.H.; Sun, G.L.; Li, S.P.; Song, C.P. Development and utilization of introgression lines using synthetic octaploid wheat (Aegilops tauschii x hexaploid wheat) as donor. Front. Plant Sci. 2018, 9, 1113. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from Plant Tissues. In Plant Molecular Biology Manual; Gelvin, S.B., Ed.; Springer: Dordrecht, The Netherlands, 1988. [Google Scholar]
- Jia, J.Z.; Zhao, S.C.; Kong, X.Y.; Li, Y.R.; Zhao, G.Y.; He, W.M.; Appels, R.; Pfeifer, M.; Tao, Y.; Zhang, X.Y.; et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 2013, 496, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.H.; Leroy, P.; Ganal, M.W. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar] [PubMed]
- Sanguinetti, C.; Dias-Neto, E. RAPD silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques 1994, 17, 5. [Google Scholar]
- Semagn, F.K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Silva, L.D.C.E.; Wang, S.; Zeng, Z.B. Composite Interval Mapping and Multiple Interval Mapping: Procedures and Guidelines for using Windows QTL Cartographer. In Quantitative Trait Loci; Rifkin, S.A., Ed.; Humana Press: Totowa, NJ, USA, 2012. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).