Degradome Analysis of Tomato and Nicotiana benthamiana Plants Infected with Potato Spindle Tuber Viroid
Abstract
1. Introduction
2. Results
2.1. Analyses of Viroid-Derived Small RNAs in Tomato and N. benthamiana
2.2. Identification of Tomato mRNAs Cleaved by miRNA via RNA Silencing
2.3. Identification of Tomato mRNAs Cleaved by PSTVd-sRNAs via RNA Silencing
2.4. Evidence for Cleavage of the Bona Fide Targets by PSTVd-sRNAs
2.5. Correlation between Targeting by PSTVd-sRNAs and Steady State Level of Cleaved Host mRNAs
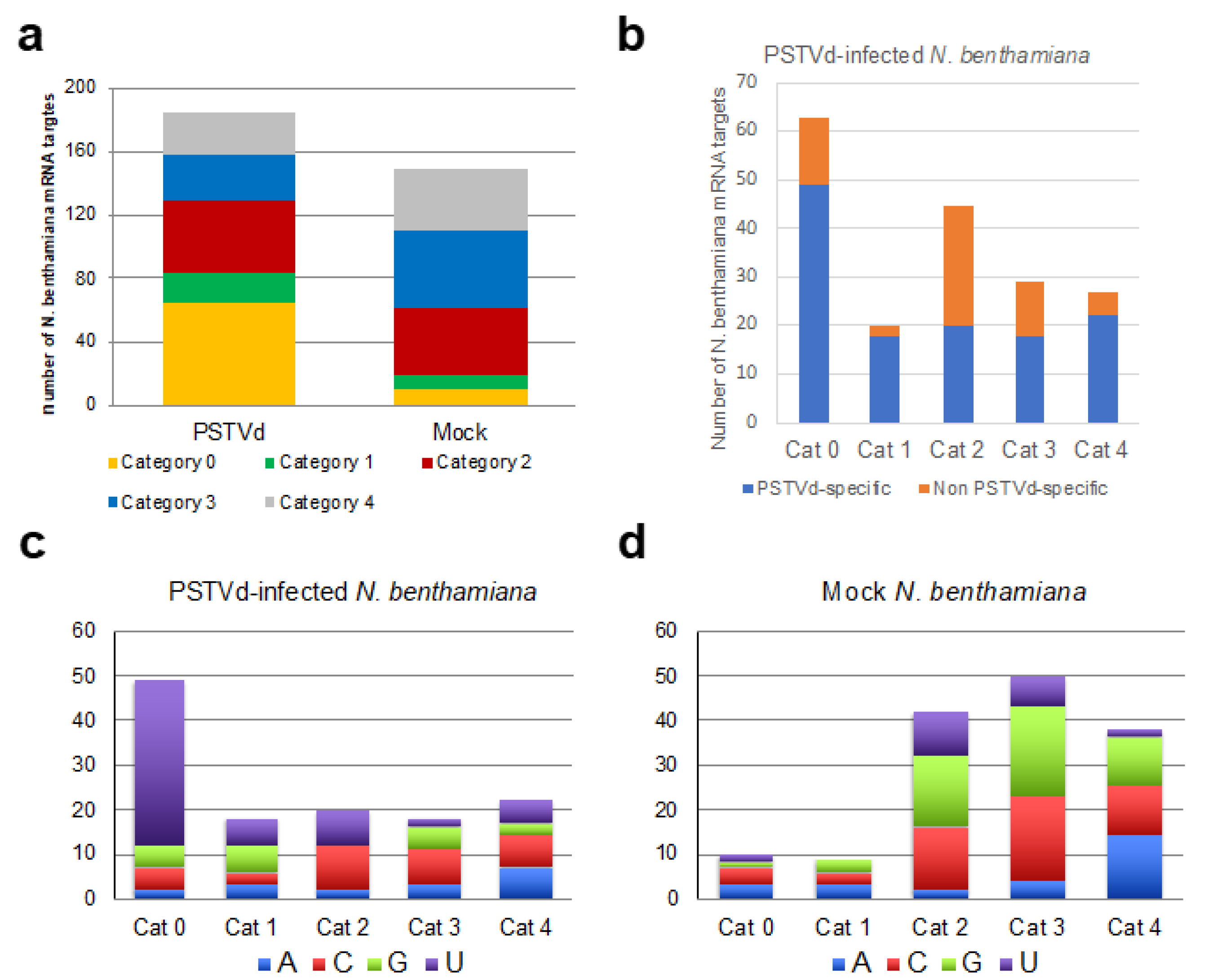
2.6. Identification of N. benthamiana mRNAs Potentially Cleaved by miRNAs and PSTVd-sRNAs via RNA Silencing
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Total RNA Isolation and Northern Blot Hybridization
4.3. Library Construction and High-Throughput Sequencing
4.4. Analysis of sRNA and Degradome Libraries
4.5. RNA Ligase Mediate-Rapid Amplification of CDNA Ends (5′ RLM-RACE)
4.6. Quantitative RT-PCR (RT-qPCR) for Gene Expression Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flores, R.; Minoia, S.; Carbonell, A.; Gisel, A.; Delgado, S.; López-Carrasco, A.; Navarro, B.; Di Serio, F. Viroids, the simplest RNA replicons: How they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015, 209, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Di Serio, F.; Owens, R.A.; Li, S.F.; Matoušek, J.; Pallás, V.; Randles, J.W.; Sano, T.; Verhoeven, J.T.J.; Vidalakis, G.; Flores, R. ICTV Report Consortium. ICTV Virus Taxonomy Profile. Pospiviroidae. J. Gen. Virol. 2020, in press. [Google Scholar] [CrossRef]
- Di Serio, F.; Li, S.F.; Matoušek, J.; Owens, R.A.; Pallás, V.; Randles, J.W.; Sano, T.; Verhoeven, J.T.J.; Vidalakis, G.; Flores, R. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Avsunviroidae. J. Gen. Virol. 2018, 99, 611–612. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Navarro, B.; Delgado, S.; Serra, P.; Di Serio, F. Viroid pathogenesis: A critical appraisal of the role of RNA silencing in triggering the initial molecular lesion. FEMS Microbiol. Rev. 2020, 44, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Adkar-Purushothama, C.R.; Perreault, J.P. Current overview on viroid-host interactions. Wiley Interdiscip. Rev. RNA 2020, 11, e1570. [Google Scholar] [CrossRef] [PubMed]
- Fukudome, A.; Fukuhara, T. Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant. Res. 2017, 130, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhang, X. Actions of plant Argonautes: Predictable or unpredictable? Curr. Opin. Plant Biol. 2018, 45, 59–67. [Google Scholar] [CrossRef]
- Itaya, A.; Folimonov, A.; Matsuda, Y.; Nelson, R.S.; Ding, B. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant -Microbe Interact. 2001, 14, 1332–1334. [Google Scholar] [CrossRef]
- Papaefthimiou, I.; Hamilton, A.J.; Denti, M.A.; Baulcombe, D.C.; Tsagris, M.; Tabler, M. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 2001, 29, 2395–2400. [Google Scholar] [CrossRef]
- Martínez de Alba, A.E.; Flores, R.; Hernández, C. Two chloroplastic viroids induce the accumulation of the small RNAs associated with post-transcriptional gene silencing. J. Virol. 2002, 76, 13094–13096. [Google Scholar] [CrossRef]
- Wang, M.B.; Bian, X.Y.; Wu, L.M.; Liu, L.X.; Smith, N.A.; Isenegger, D.; Wu, R.M.; Masuta, C.; Vance, V.B.; Watson, J.M.; et al. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 2004, 101, 3275–3280. [Google Scholar] [CrossRef]
- Minoia, S.; Carbonell, A.; Di Serio, F.; Gisel, A.; Carrington, J.C.; Navarro, B.; Flores, R. Specific ARGONAUTES bind selectively small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J. Virol. 2014, 88, 11933–11945. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Hernández, C.; Flores, R. Plus and minus RNAs of Peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 1992, 89, 3711–3715. [Google Scholar] [CrossRef]
- Malfitano, M.; Di Serio, F.; Covelli, L.; Ragozzino, A.; Hernández, C.; Flores, R. Peach latent mosaic viroid variants inducing peach calico (extreme chlorosis) contain a characteristic insertion that is responsible for this symptomatology. Virology 2003, 313, 492–501. [Google Scholar] [CrossRef]
- Rodio, M.E.; Delgado, S.; De Stradis, A.; Gómez, M.D.; Flores, R.; Di Serio, F. A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 2007, 19, 3610–3626. [Google Scholar] [CrossRef]
- Delgado, S.; Navarro, B.; Serra, P.; Gentit, P.; Cambra, M.A.; Chiumenti, M.; De Stradis, A.; Di Serio, F.; Flores, R. How sequence variants of a plastid-replicating viroid with one single nucleotide change initiate disease in its natural host. RNA Biol. 2019, 16, 906–917. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012, 70, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Adkar-Purushothama, C.R.; Brosseau, C.; Giguère, T.; Sano, T.; Moffett, P.; Perreault, J.P. Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 2015, 27, 2178–2194. [Google Scholar] [CrossRef]
- Adkar-Purushothama, C.R.; Iyer, P.S.; Perreault, J.P. Potato spindle tuber viroid infection triggers degradation of chloride channel protein CLC-b-like and Ribosomal protein S3a-like mRNAs in tomato plants. Sci. Rep. 2017, 7, 8341. [Google Scholar] [CrossRef]
- Adkar-Purushothama, C.R.; Perreault, J.P. Alterations of the viroid regions that interact with the host defense genes attenuate viroid infection in host plant. RNA Biol. 2018, 15, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, Y.; Ding, B.; Fei, Z. Comprehensive transcriptome analyses reveal that potato spindle tuber viroid triggers genome-wide changes in alternative splicing, inducible trans-acting activity of phased secondary small interfering RNAs, and immune responses. J. Virol. 2017, 91, e00247-17. [Google Scholar] [CrossRef]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef] [PubMed]
- German, M.A.; Pillay, M.; Jeong, D.H.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Di Serio, F.; Martínez de Alba, A.E.; Navarro, B.; Gisel, A.; Flores, R. RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus. J. Virol. 2010, 84, 2477–2489. [Google Scholar] [CrossRef]
- Adkar-Purushothama, C.R.; Perreault, J.P.; Sano, T. Analysis of small RNA production patterns among the two potato spindle tuber viroid variants in tomato plants. Genom. Data 2015, 6, 65–66. [Google Scholar] [CrossRef]
- Folkes, L.; Moxon, S.; Woolfenden, H.C.; Stocks, M.B.; Szittya, G.; Dalmay, T.; Moulton, V. PAREsnip: A tool for rapid genome-wide discovery of small RNA/target interactions evidenced through degradome sequencing. Nucleic Acids Res. 2012, 40, e103. [Google Scholar] [CrossRef]
- Moxon, S.; Jing, R.; Szittya, G.; Schwach, F.; Rusholme Pilcher, R.L.; Moulton, V.; Dalmay, T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008, 18, 1602–1609. [Google Scholar] [CrossRef]
- Itaya, A.; Bundschuh, R.; Archual, A.J.; Joung, J.G.; Fei, Z.; Dai, X.; Zhao, P.X.; Tang, Y.; Nelson, R.S.; Ding, B. Small RNAs in tomato fruit and leaf development. Biochim. Biophys. Acta 2008, 1779, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Wang, Q.; Jiang, F.; Cao, X.; Sun, M.; Liu, M.; Wu, Z. Identification of miRNAs and their targets in wild tomato at moderately and acutely elevated temperatures by high-throughput sequencing and degradome analysis. Sci. Rep. 2016, 6, 33777. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; van Haarst, J.C.; Maliepaard, C.; van de Geest, H.; Bovy, A.G.; Lammers, M.; Angenent, G.C.; de Maagd, R.A. Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J. Exp. Bot. 2013, 64, 1863–1878. [Google Scholar] [CrossRef]
- Luan, Y.; Wang, W.; Liu, P. Identification and functional analysis of novel and conserved microRNAs in tomato. Mol. Biol. Rep. 2014, 41, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Karim, M.R.; Harikrishna, J.A.; Shimizu, T.; Sasaya, T.; Omura, T.; Haque, M.A.; Hasan, S.M.; et al. NAC transcription factor family genes are differentially expressed in rice during infections with Rice dwarf virus, Rice black-streaked dwarf virus, Rice grassy stunt virus, Rice ragged stunt virus, and Rice transitory yellowing virus. Front. Plant Sci. 2015, 6, 676. [Google Scholar] [CrossRef]
- Mathew, I.E.; Agarwal, P. May the Fittest Protein Evolve: Favoring the Plant-Specific Origin and Expansion of NAC Transcription Factors. Bioessays 2018, 40, e1800018. [Google Scholar] [CrossRef]
- Jarad, M.; Mariappan, K.; Almeida-Trapp, M.; Mette, M.F.; Mithöfer, A.; Rayapuram, N.; Hirt, H. The Lamin-Like LITTLE NUCLEI 1 (LINC1) Regulates Pattern-Triggered Immunity and Jasmonic Acid Signaling. Front. Plant Sci. 2020, 10, 1639. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y.; Takagi, S. LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol. 2013, 54, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P. The origin recognition complex: From simple origins to complex functions. Genes Dev. 2002, 16, 659–672. [Google Scholar] [CrossRef]
- Collinge, M.A.; Spillane, C.; Köhler, C.; Gheyselinck, J.; Grossniklaus, U. Genetic interaction of an origin recognition complex subunit and the Polycomb group gene MEDEA during seed development. Plant Cell 2004, 16, 1035–1046. [Google Scholar] [CrossRef]
- Qin, Z.; Zhang, X.; Zhang, X.; Xin, W.; Li, J.; Hu, Y. The Arabidopsis transcription factor IIB-related protein BRP4 is involved in the regulation of mitotic cell-cycle progression during male gametogenesis. J. Exp. Bot. 2014, 65, 2521–2531. [Google Scholar] [CrossRef] [PubMed]
- Carol, R.J.; Takeda, S.; Linstead, P.; Durrant, M.C.; Kakesova, H.; Derbyshire, P.; Drea, S.; Zarsky, V.; Dolan, L. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 2005, 438, 1013–1016. [Google Scholar] [CrossRef]
- Shimotohno, A.; Umeda-Hara, C.; Bisova, K.; Uchimiya, H.; Umeda, M. The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell 2004, 16, 2954–2966. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, H.; Ohno, R.; Umeda, M. The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. Plant J. 2009, 59, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Swiderski, M.R.; Innes, R.W. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001, 26, 101–112. [Google Scholar] [CrossRef]
- Morandini, P.; Valera, M.; Albumi, C.; Bonza, M.C.; Giacometti, S.; Ravera, G.; Murgia, I.; Soave, C.; De Michelis, M.I. A novel interaction partner for the C-terminus of Arabidopsis thaliana plasma membrane H+-ATPase (AHA1 isoform): Site and mechanism of action on H+-ATPase activity differ from those of 14-3-3 proteins. Plant J. 2002, 31, 487–497. [Google Scholar] [CrossRef]
- Viotti, C.; Luoni, L.; Morandini, P.; De Michelis, M.I. Characterization of the interaction between the plasma membrane H-ATPase of Arabidopsis thaliana and a novel interactor (PPI1). FEBS J. 2005, 272, 5864–5871. [Google Scholar] [CrossRef] [PubMed]
- Baksa, I.; Nagy, T.; Barta, E.; Havelda, Z.; Várallyay, É.; Silhavy, D.; Burgyán, J.; Szittya, G. Identification of Nicotiana benthamiana microRNAs and their targets using high throughput sequencing and degradome analysis. BMC Genom. 2015, 16, 1025. [Google Scholar] [CrossRef]
- Diermann, N.; Matousek, J.; Junge, M.; Riesner, D.; Steger, G. Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol. Chem. 2010, 391, 1379–1390. [Google Scholar] [CrossRef]
- Tsushima, D.; Adkar-Purushothama, C.R.; Taneda, A.; Sano, T. Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild symptom-inducing isolates of potato spindle tuber viroid. J. Gen. Plant Pathol. 2015, 81, 49–62. [Google Scholar] [CrossRef]
- Owens, R.A.; Tech, K.B.; Shao, J.Y.; Sano, T.; Baker, C.J. Global analysis of tomato gene expression during potato spindle tuber viroid infection reveals a complex array of changes affecting hormone Signaling. Mol. Plant Microbe Int. 2012, 25, 582–598. [Google Scholar] [CrossRef]
- Martín, R.; Arenas, C.; Daròs, J.A.; Covarrubias, A.; Reyes, J.; Chua, N. Characterization of small RNAs derived from Citrus exocortis viroid (CEVd) in infected tomato plants. Virology 2007, 367, 135–146. [Google Scholar] [CrossRef]
- Morel, J.B.; Godon, C.; Mourrain, P.; Béclin, C.; Boutet, S.; Feuerbach, F.; Proux, F.; Vaucheret, H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 2002, 14, 629–639. [Google Scholar] [CrossRef]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef]
- Góra-Sochacka, A.; Więsyk, A.; Fogtman, A.; Lirski, M.; Zagórski-Ostoja, W. Root Transcriptomic Analysis Reveals Global Changes Induced by Systemic Infection of Solanum lycopersicum with Mild and Severe Variants of Potato Spindle Tuber Viroid. Viruses 2019, 11, 992. [Google Scholar] [CrossRef] [PubMed]
- Adkar-Purushothama, C.R.; Sano, T.; Perreault, J.P. Viroid-derived small RNA induces early flowering in tomato plants by RNA silencing. Mol. Plant Pathol. 2018, 19, 2446–2458. [Google Scholar] [CrossRef]
- Thibaut, O.; Bragard, C. Innate immunity activation and RNAi interplay in citrus exocortis viroid—Tomato pathosystem. Viruses 2018, 10, 587. [Google Scholar] [CrossRef]
- Štajner, N.; Radišek, S.; Mishra, A.K.; Nath, V.S.; Matoušek, J.; Jakše, J. Evaluation of disease severity and global transcriptome response induced by citrus bark cracking viroid, hop latent viroid, and their co-infection in hop (Humulus lupulus L.). Int. J. Mol. Sci. 2019, 20, 3154. [Google Scholar] [CrossRef]
- Takino, H.; Kitajima, S.; Hirano, S.; Oka, M.; Matsuura, T.; Ikeda, Y.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Mino, M. Global transcriptome analyses reveal that infection with chrysanthemum stunt viroid (CSVd) affects gene expression profile of chrysanthemum plants, but the genes involved in plant hormone metabolism and signaling may not be silencing target of CSVd-siRNAs. Plant Gene 2019, 18, 100181. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Qiu, Y.; Atta, S.; Zhou, C.; Cao, M. Global Transcriptomic Analysis Reveals Insights into the Response of ‘Etrog’ Citron (Citrus medica L.) to Citrus Exocortis Viroid Infection. Viruses 2019, 11, 453. [Google Scholar] [CrossRef] [PubMed]
- Lisón, P.; Tárraga, S.; López-Gresa, P.; Saurí, A.; Torres, C.; Campos, L.; Bellés, J.M.; Conejero, V.; Rodrigo, I. A noncoding plant pathogen provokes both transcriptional and posttranscriptional alterations in tomato. Proteomics 2013, 13, 833–844. [Google Scholar] [CrossRef]
- Cottilli, P.; Belda-Palazón, B.; Adkar-Purushothama, C.R.; Perreault, J.P.; Schleiff, E.; Rodrigo, I.; Ferrando, A.; Lisón, P. Citrus exocortis viroid causes ribosomal stress in tomato plants. Nucleic Acids Res. 2019, 47, 8649–8661. [Google Scholar] [CrossRef] [PubMed]
- Castellano, M.; Martinez, G.; Pallás, V.; Gómez, G. Alterations in host DNA methylation in response to constitutive expression of Hop stunt viroid RNA in Nicotiana benthamiana plants. Plant Pathol. 2015, 64, 1247–1257. [Google Scholar] [CrossRef]
- Castellano, M.; Pallás, V.; Gómez, G. A pathogenic long noncoding RNA redesigns the epigenetic landscape of the infected cells by subverting host Histone Deacetylase 6 activity. New Phytol. 2016, 211, 1311–1322. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.Q.; Liu, S.W.; Zhao, J.H.; Zhou, B.J.; Wang, S.P.; Guo, H.S.; Fang, Y.Y. Replication of a pathogenic non-coding RNA increases DNA methylation in plants associated with a bromodomain-containing viroid-binding protein. Sci. Rep. 2016, 6, 35751. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Castellano, M.; Tortosa, M.; Pallás, V.; Gómez, G. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res. 2014, 42, 1553–1562. [Google Scholar] [CrossRef]
- Torchetti, E.M.; Pegoraro, M.; Navarro, B.; Catoni, M.; Di Serio, F.; Noris, E. A nuclear-replicating viroid antagonizes infectivity and accumulation of a geminivirus by upregulating methylation-related genes and inducing hypermethylation of viral DNA. Sci. Rep. 2016, 6, 35101. [Google Scholar] [CrossRef]
- Carbonell, A.; Flores, R.; Gago, S. Trans-cleaving hammerhead ribozymes with tertiary stabilizing motifs: In vitro and in vivo activity against a structured viroid RNA. Nucleic Acids Res. 2011, 39, 2432–2444. [Google Scholar] [CrossRef] [PubMed]
- German, M.A.; Luo, S.; Schroth, G.; Meyers, C.; Green, P.J. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protocol. 2009, 4, 356–362. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]





| mRNA Target | Degradome Results | PSTVd-sRNAs | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. Thaliana Homolog | Alignment Score | mef (Kcal/mol) | PSTVd-sRNA:mRNA Hybrid | Cat | Cleavage Position | p-Value T3 | Number of Tags T3 | Normalized Weighted Number of Tags T3 | p-Value T4 | Number of Tags T4 | Normalized Weighted Number of Tags T4 | 5′Position | Polarity | PSTVd Variants * | Reads T3 | RPM T3 | Reads T4 | RPM T4 | |
| Solyc09g083420.2.1 | AT1G56290.1 (CWF19) | 2.5 | −37.4 |  | 0 | 1587 | 0.00 | 74 | 1.93 | 0.00 | 149 | 2.62 | 178 | (+) | Nb, Int, Mild, RG1, Lethal | 935 | 25.22 | 1330 | 35.89 |
| Solyc05g007450.2.1 | AT1G26840.1 (ORC6) | 3.0 | −29.6 |  | 0 | 645 | 0.00 | 113 | 2.95 | 0.00 | 259 | 4.55 | 179 | (+) | Nb, Int, mild, RG1, Lethal | 2037 | 54.97 | 2931 | 79.1 |
| Solyc11g012270.1.1 | AT3G07880.1 (SCN1) | 3.5 | −31.0 |  | 0 | 451 | 0.01 | 8 | 0.21 | 0.00 | 13 | 0.23 | 192 | (+) | Nb, Int, RG1, Lethal | 409 | 11.03 | 488 | 13.17 |
| Solyc03g045050.2.1 | AT1G67230.1 (LINC1) | 3.5 | −34.4 |  | 0 | 3520 | 0.00 | 196 | 5.12 | 0.00 | 316 | 5.55 | 238 | (+) | Nb, Int, Mild, RG1, Lethal | 2096 | 56.56 | 5535 | 149.4 |
| Solyc05g024290.2.1 | AT5G13160.1 (PBS1) | 3.0 | −29.6 |  | 0 | 1542 | 0.00 | 56 | 1.46 | 0.01 | 163 | 2.86 | 290 | (+) | Nb, Int, RG1, Lethal | 138 | 3.72 | 542 | 14.63 |
| Solyc12g007200.1.1 | AT4G28980.1 (CDKF;1) | 3.5 | −33.7 |  | 0 | 761 | 0.01 | 28 | 0.73 | 0.00 | 142 | 2.49 | 8 | (+) | Nb, Int, mild, RG1, Lethal | 277 | 7.47 | 354 | 9.553 |
| Solyc04g079940.2.1 | AT2G24430.1 (ANAC038) | 3.0 | −25.5 |  | 0 | 1339 | 0.00 | 33 | 0.86 | 0.02 | 38 | 0.67 | 70 | (−) | Nb, Int, mild, RG1, Lethal | 18 | 0.49 | 85 | 2.29 |
| Solyc07g063100.2.1 | AT4G27500.1 (PPI1) | 3.0 | −28.4 |  | 0 | 1675 | 0,00 | 637 | 16.65 | 0.00 | 2327 | 40.87 | 288 | (−) | Nb, Int, mild, RG1, Lethal | 319 | 8.61 | 458 | 12.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro, B.; Gisel, A.; Serra, P.; Chiumenti, M.; Di Serio, F.; Flores, R. Degradome Analysis of Tomato and Nicotiana benthamiana Plants Infected with Potato Spindle Tuber Viroid. Int. J. Mol. Sci. 2021, 22, 3725. https://doi.org/10.3390/ijms22073725
Navarro B, Gisel A, Serra P, Chiumenti M, Di Serio F, Flores R. Degradome Analysis of Tomato and Nicotiana benthamiana Plants Infected with Potato Spindle Tuber Viroid. International Journal of Molecular Sciences. 2021; 22(7):3725. https://doi.org/10.3390/ijms22073725
Chicago/Turabian StyleNavarro, Beatriz, Andreas Gisel, Pedro Serra, Michela Chiumenti, Francesco Di Serio, and Ricardo Flores. 2021. "Degradome Analysis of Tomato and Nicotiana benthamiana Plants Infected with Potato Spindle Tuber Viroid" International Journal of Molecular Sciences 22, no. 7: 3725. https://doi.org/10.3390/ijms22073725
APA StyleNavarro, B., Gisel, A., Serra, P., Chiumenti, M., Di Serio, F., & Flores, R. (2021). Degradome Analysis of Tomato and Nicotiana benthamiana Plants Infected with Potato Spindle Tuber Viroid. International Journal of Molecular Sciences, 22(7), 3725. https://doi.org/10.3390/ijms22073725








