Generation of a Novel Nkx6-1 Venus Fusion Reporter Mouse Line
Abstract
1. Introduction
2. Results and Discussion
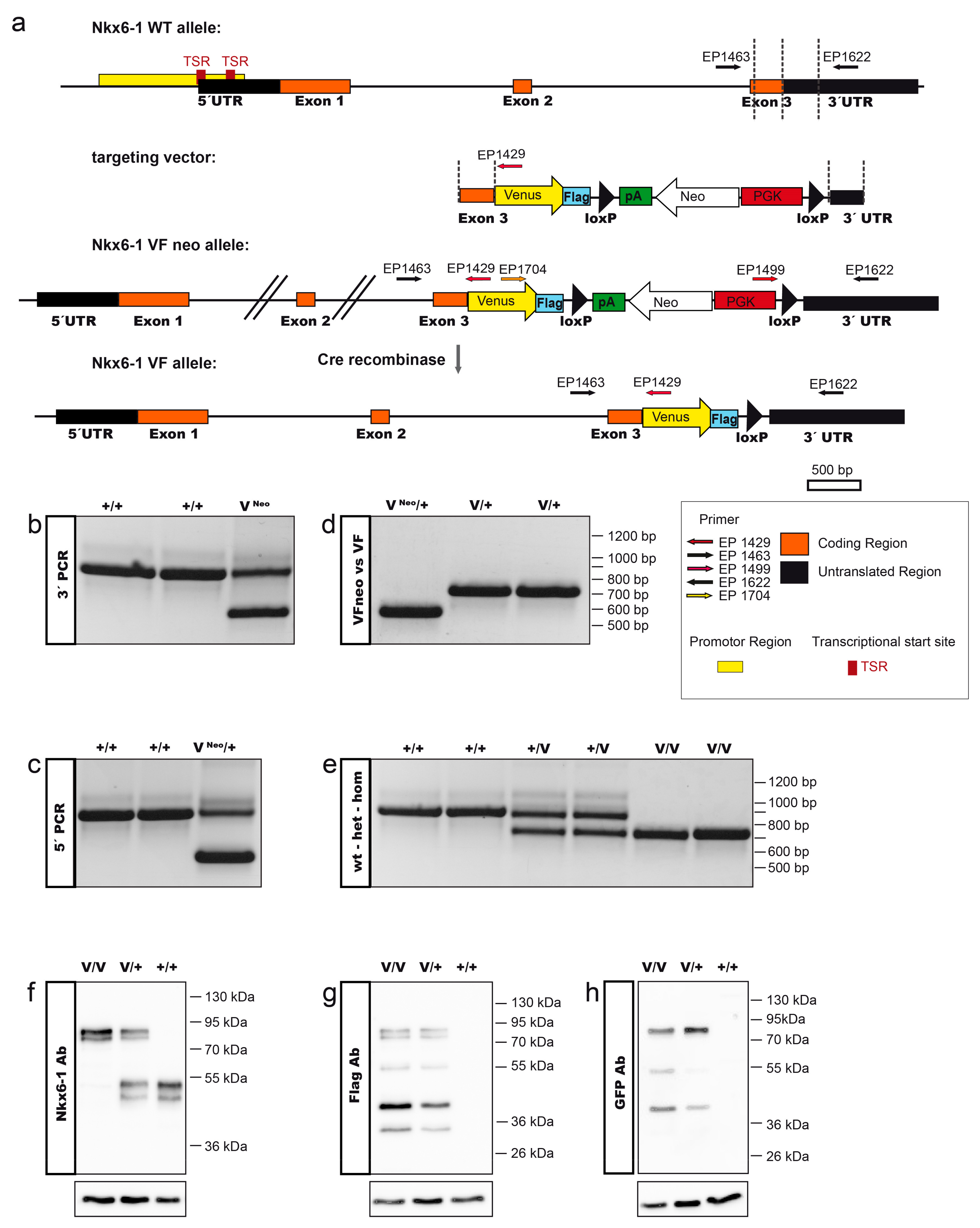
2.1. Generation of the Nkx6-1-VF Mouse Line
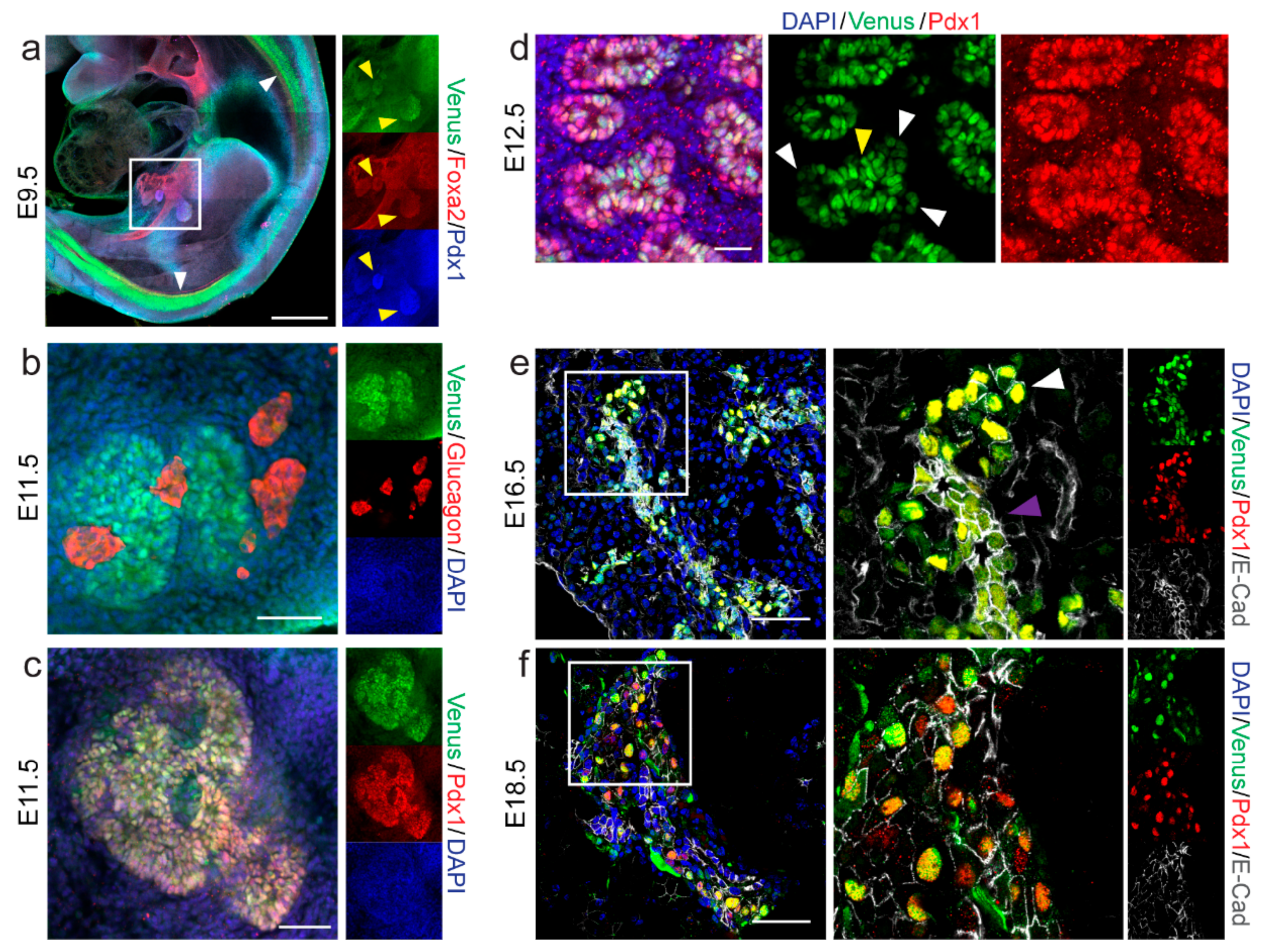
2.2. Spatio-Temporal Expression Pattern of Nkx6-1-VF Protein during Embryonic Pancreas Development
2.3. Nkx6-1-VF Marks the Endocrine Lineage during Secondary Transition
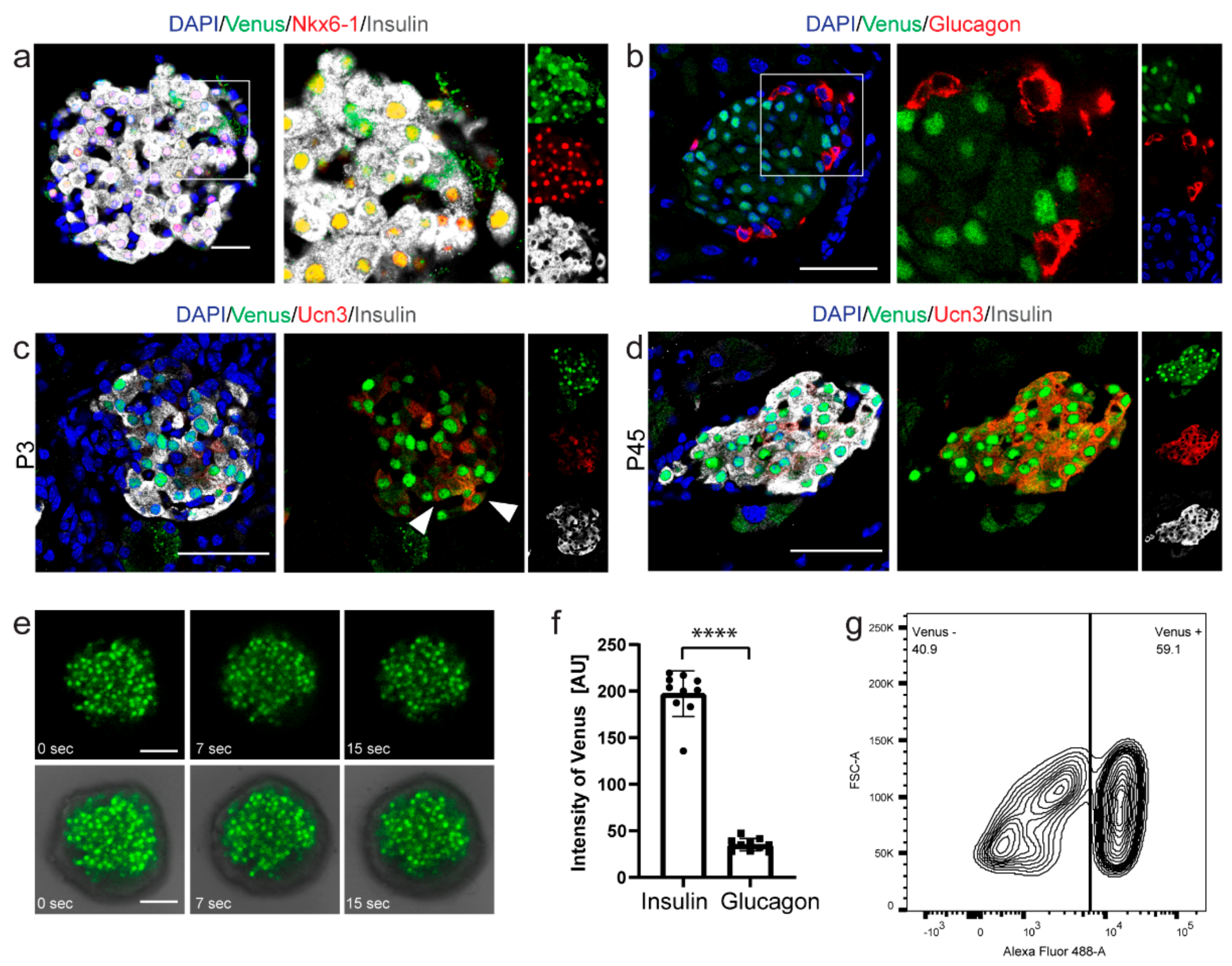
2.4. Nkx6-1-VF Expression Pattern in the Adult Pancreas
3. Materials and Methods
3.1. Generation of the Targeting Construct
3.2. Cell Culture and Homologous Recombination in ES Cells and Mouse Generation
3.3. Genotyping
3.4. Western Blot Analysis
3.5. Pancreas Dissection
3.6. Immunostaining of Sections
3.7. Islet Isolation
3.8. FACS Analysis
3.9. Time-Lapse Live Imaging
3.10. iDisco for Clearing of Mouse Embryos
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pan, F.C.; Wright, C. Pancreas organogenesis: From bud to plexus to gland. Dev. Dyn. 2011, 240, 530–565. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.P.; Wang, A.; Sander, M. Pancreas organogenesis: From lineage determination to morphogenesis. Annu. Rev. Cell Dev. Biol. 2013, 29, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Brissova, M.; Fowler, M.J.; Nicholson, W.E.; Chu, A.; Hirshberg, B.; Harlan, D.M.; Powers, A.C. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J. Histochem. Cytochem. 2005, 53, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Roscioni, S.S.; Migliorini, A.; Gegg, M.; Lickert, H. Impact of islet architecture on β-cell heterogeneity, plasticity and function. Nat. Rev. Endocrinol. 2016, 12, 695–709. [Google Scholar] [CrossRef]
- Bastidas-Ponce, A.; Scheibner, K.; Lickert, H.; Bakhti, M. Cellular and molecular mechanisms coordinating pancreas development. Development 2017, 144, 2873–2888. [Google Scholar] [CrossRef]
- Villasenor, A.; Chong, D.C.; Henkemeyer, M.; Cleaver, O. Epithelial dynamics of pancreatic branching morphogenesis. Development 2010, 137, 4295–4305. [Google Scholar] [CrossRef]
- Kesavan, G.; Lieven, O.; Mamidi, A.; Öhlin, Z.L.; Johansson, J.K.; Li, W.-C.; Lommel, S.; Greiner, T.U.; Semb, H. Cdc42/N-WASP signaling links actin dynamics to pancreatic cell delamination and differentiation. Development 2014, 141, 685–696. [Google Scholar] [CrossRef]
- Li, Y.; Tzatzalos, E.; Kwan, K.Y.; Grumet, M.; Cai, L. Transcriptional regulation of notch1 expression by Nkx6.1 in neural stem/progenitor cells during ventral spinal cord development. Sci. Rep. 2016, 6, 38665. [Google Scholar] [CrossRef]
- Prakash, N.; Puelles, E.; Freude, K.; Trümbach, D.; Omodei, D.; Di Salvio, M.; Sussel, L.; Ericson, J.; Sander, M.; Simeone, A.; et al. Nkx6-1 controls the identity and fate of red nucleus and oculomotor neurons in the mouse midbrain. Development 2009, 136, 2545–2555. [Google Scholar] [CrossRef]
- Cai, J.; Xu, X.; Yin, H.; Wu, R.; Modderman, G.; Chen, Y.; Qiu, M. Evidence for the differential regulation of Nkx-6.1 expression in the ventral spinal cord and foregut by Shh-dependent and-independent mechanisms. Genesis 2000, 27, 6–11. [Google Scholar] [CrossRef]
- Schaffer, A.E.; Freude, K.K.; Nelson, S.B.; Sander, M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev. Cell 2010, 18, 1022–1029. [Google Scholar] [CrossRef]
- Zhou, Q.; Law, A.C.; Rajagopal, J.; Anderson, W.J.; Gray, P.A.; Melton, D.A. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 2007, 13, 103–114. [Google Scholar] [CrossRef]
- Nelson, S.B.; Schaffer, A.E.; Sander, M. The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development 2007, 134, 2491–2500. [Google Scholar] [CrossRef]
- Sander, M.; Sussel, L.; Conners, J.; Scheel, D.; Kalamaras, J.; Cruz, F.D.; German, M. Homeobox gene Nkx6. 1 lies downstream of Nkx2. 2 in the major pathway of beta-cell formation in the pancreas. Development 2000, 127, 5533–5540. [Google Scholar] [PubMed]
- Schisler, J.C.; Fueger, P.T.; Babu, D.A.; Hohmeier, H.E.; Tessem, J.; Lu, D.; Becker, T.C.; Naziruddin, B.; Levy, M.; Mirmira, R.G.; et al. Stimulation of human and rat islet β-cell proliferation with retention of function by the homeodomain transcription factor Nkx6.1. Mol. Cell. Biol. 2008, 28, 3465–3476. [Google Scholar] [CrossRef] [PubMed]
- Tessem, J.; Moss, L.G.; Chao, L.C.; Arlotto, M.; Lu, D.; Jensen, M.V.; Stephens, S.B.; Tontonoz, P.; Hohmeier, H.E.; Newgard, C.B. Nkx6.1 regulates islet -cell proliferation via Nr4a1 and Nr4a3 nuclear receptors. Proc. Natl. Acad. Sci. USA 2014, 111, 5242–5247. [Google Scholar] [CrossRef] [PubMed]
- Øster, A.; Jensen, J.; Serup, P.; Galante, P.; Madsen, O.D.; Larsson, L.-I. Rat endocrine pancreatic development in relation to two homeobox gene products (Pdx-1 and Nkx 6.1). J. Histochem. Cytochem. 1998, 46, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hunter, C.S.; Du, A.; Ediger, B.; Walp, E.; Murray, J.; Stein, R.; May, C.L. Islet-1 regulates arx transcription during pancreatic islet α-cell development. J. Biol. Chem. 2011, 286, 15352–15360. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, A.E.; Taylor, B.L.; Benthuysen, J.R.; Liu, J.; Thorel, F.; Yuan, W.; Jiao, Y.; Kaestner, K.H.; Herrera, P.L.; Magnuson, M.A.; et al. Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic beta cell identity. PLoS Genet. 2013, 9, e1003274. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.; Serup, P.; Karlsen, C.; Nielsen, T.F.; Madsen, O.D. mRNA profiling of rat islet tumors reveals Nkx 6.1 as a β-cell-specific homeodomain transcription factor. J. Biol. Chem. 1996, 271, 18749–18758. [Google Scholar] [CrossRef]
- Taylor, B.L.; Liu, F.-F.; Sander, M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 2013, 4, 1262–1275. [Google Scholar] [CrossRef]
- Guo, L.; Inada, A.; Aguayo-Mazzucato, C.; Hollister-Lock, J.; Fujitani, Y.; Weir, G.C.; Wright, C.V.; Sharma, A.; Bonner-Weir, S. PDX1 in ducts is not required for postnatal formation of β-cells but is necessary for their subsequent maturation. Diabetes 2013, 62, 3459–3468. [Google Scholar] [CrossRef] [PubMed]
- Talchai, C.; Xuan, S.; Lin, H.V.; Sussel, L.; Accili, D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 2012, 150, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Spracklen, C.N.; Horikoshi, M.; Kim, Y.J.; Lin, K.; Bragg, F.; Moon, S.; Suzuki, K.; Tam, C.H.T.; Tabara, Y.; Kwak, S.-H.; et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nat. Cell Biol. 2020, 582, 240–245. [Google Scholar] [CrossRef]
- Suzuki, K.; Akiyama, M.; Ishigaki, K.; Kanai, M.; Hosoe, J.; Shojima, N.; Hozawa, A.; Kadota, A.; Kuriki, K.; Naito, M.; et al. Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population. Nat. Genet. 2019, 51, 379–386. [Google Scholar] [CrossRef]
- Yokoi, N.; Kanamori, M.; Horikawa, Y.; Takeda, J.; Sanke, T.; Furuta, H.; Nanjo, K.; Mori, H.; Kasuga, M.; Hara, K.; et al. Association studies of variants in the genes involved in pancreatic -cell function in type 2 diabetes in Japanese subjects. Diabetes 2006, 55, 2379–2386. [Google Scholar] [CrossRef]
- Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet 1999, 21, 70–71. [Google Scholar] [CrossRef]
- Genové, G.; Glick, B.S.; Barth, A.L. Brighter reporter genes from multimerized fluorescent proteins. Biotechniques 2005, 39, 814–822. [Google Scholar] [CrossRef]
- Copeland, N.G.; Jenkins, N.A.; Court, D.L. Recombineering: A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001, 2, 769–779. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, A.J.; Ran, F.A.; Konermann, S.M.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Hitz, C.; Wurst, W.; Kühn, R. Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference. Nucleic Acids Res. 2007, 35, e90. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Ponce, A.; Tritschler, S.; Dony, L.; Scheibner, K.; Tarquis-Medina, M.; Salinno, C.; Schirge, S.; Burtscher, I.; Böttcher, A.; Theis, F.J.; et al. Comprehensive single cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development 2019, 146, dev173849. [Google Scholar] [CrossRef] [PubMed]
- Burtscher, I.; Lickert, H. Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development 2009, 136, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Renier, N.; Wu, Z.; Simon, D.J.; Yang, J.; Ariel, P.; Tessier-Lavigne, M. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014, 159, 896–910. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burtscher, I.; Tarquis-Medina, M.; Salinno, C.; Schirge, S.; Beckenbauer, J.; Bakhti, M.; Lickert, H. Generation of a Novel Nkx6-1 Venus Fusion Reporter Mouse Line. Int. J. Mol. Sci. 2021, 22, 3434. https://doi.org/10.3390/ijms22073434
Burtscher I, Tarquis-Medina M, Salinno C, Schirge S, Beckenbauer J, Bakhti M, Lickert H. Generation of a Novel Nkx6-1 Venus Fusion Reporter Mouse Line. International Journal of Molecular Sciences. 2021; 22(7):3434. https://doi.org/10.3390/ijms22073434
Chicago/Turabian StyleBurtscher, Ingo, Marta Tarquis-Medina, Ciro Salinno, Silvia Schirge, Julia Beckenbauer, Mostafa Bakhti, and Heiko Lickert. 2021. "Generation of a Novel Nkx6-1 Venus Fusion Reporter Mouse Line" International Journal of Molecular Sciences 22, no. 7: 3434. https://doi.org/10.3390/ijms22073434
APA StyleBurtscher, I., Tarquis-Medina, M., Salinno, C., Schirge, S., Beckenbauer, J., Bakhti, M., & Lickert, H. (2021). Generation of a Novel Nkx6-1 Venus Fusion Reporter Mouse Line. International Journal of Molecular Sciences, 22(7), 3434. https://doi.org/10.3390/ijms22073434





