Plant Growth Promotion Function of Bacillus sp. Strains Isolated from Salt-Pan Rhizosphere and Their Biocontrol Potential against Macrophomina phaseolina
Abstract
1. Introduction
2. Results
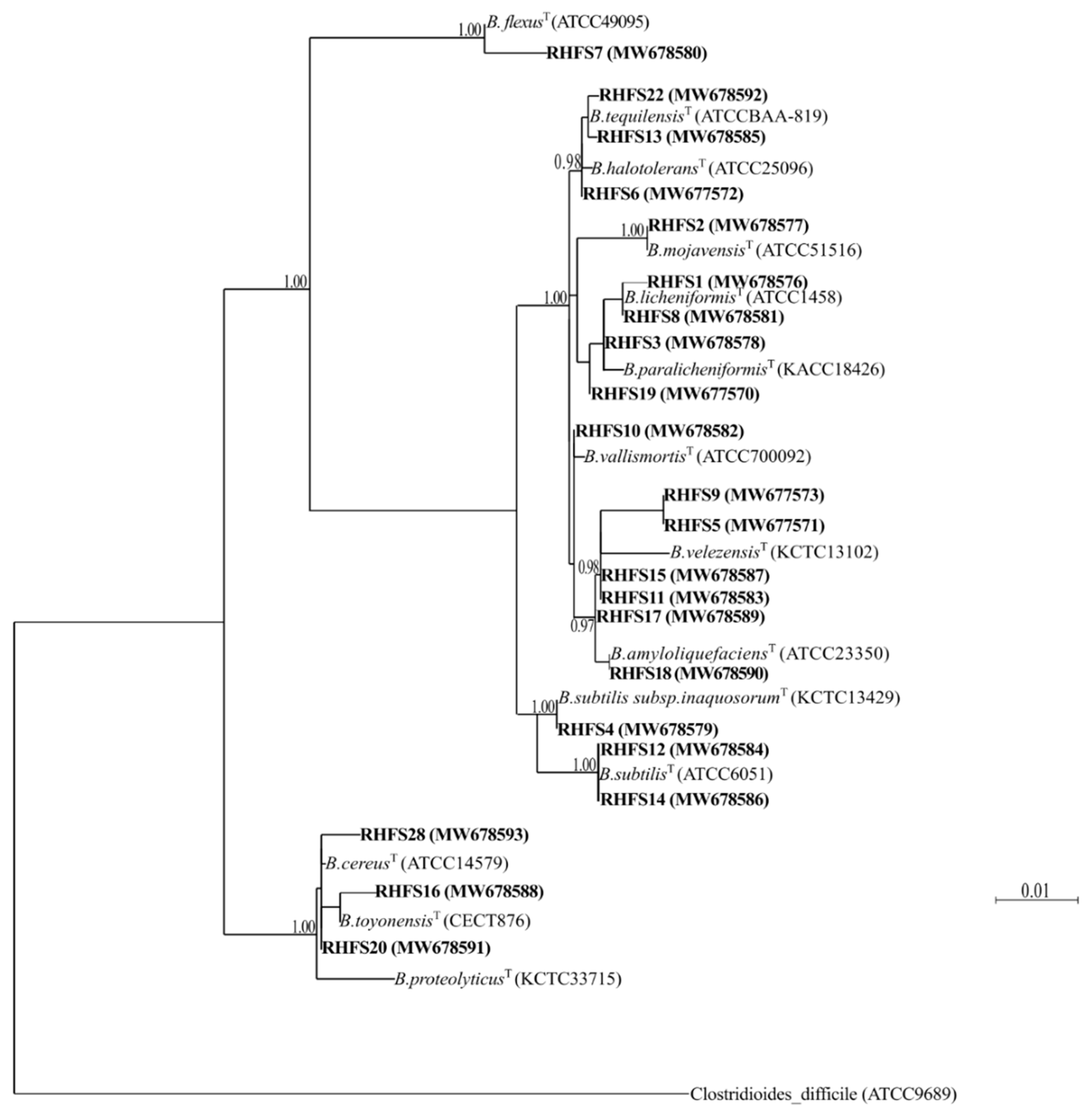
2.1. Isolation and Screening of Plant Growth-Promoting Spore-Forming Rhizobacteria
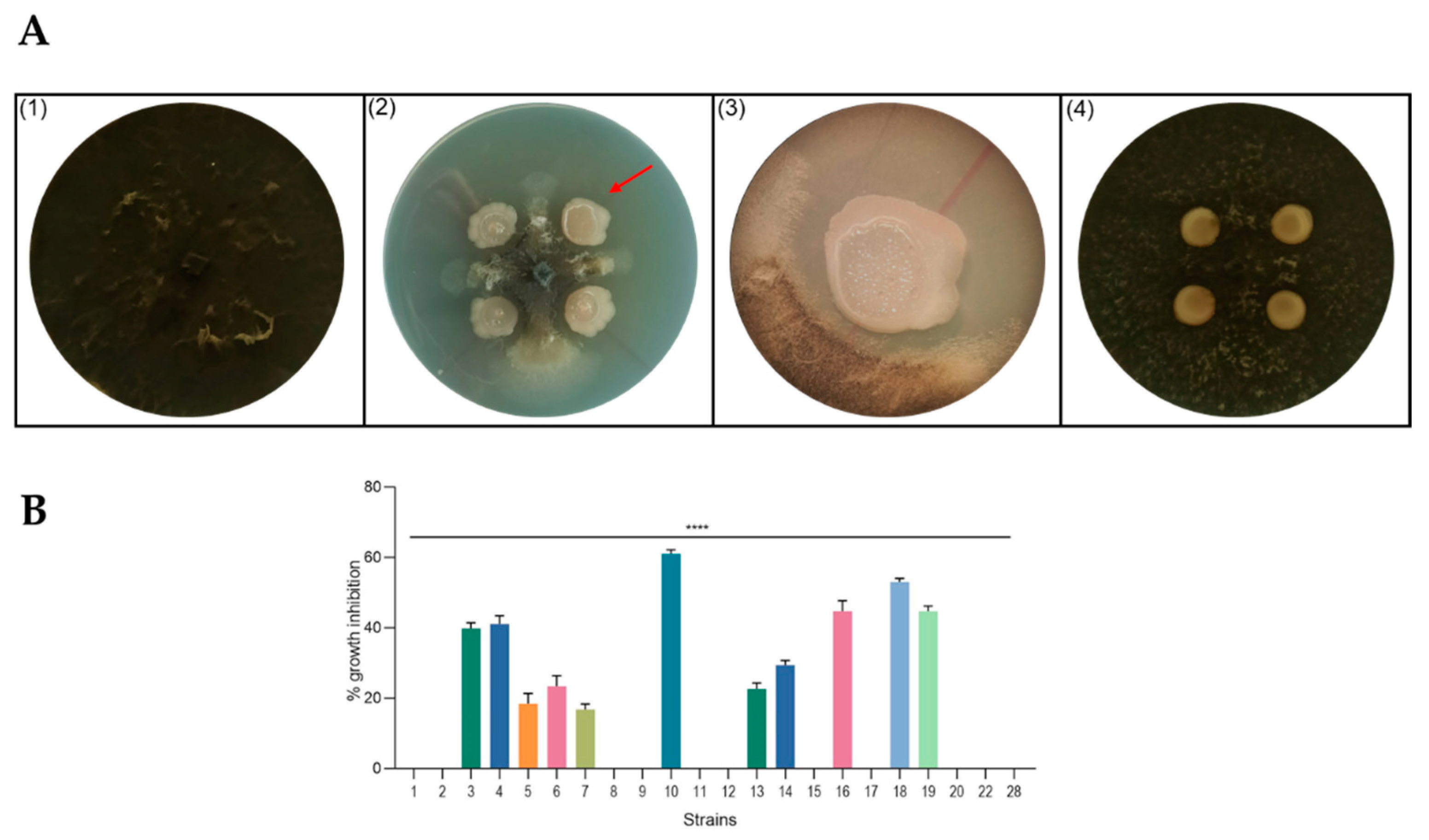
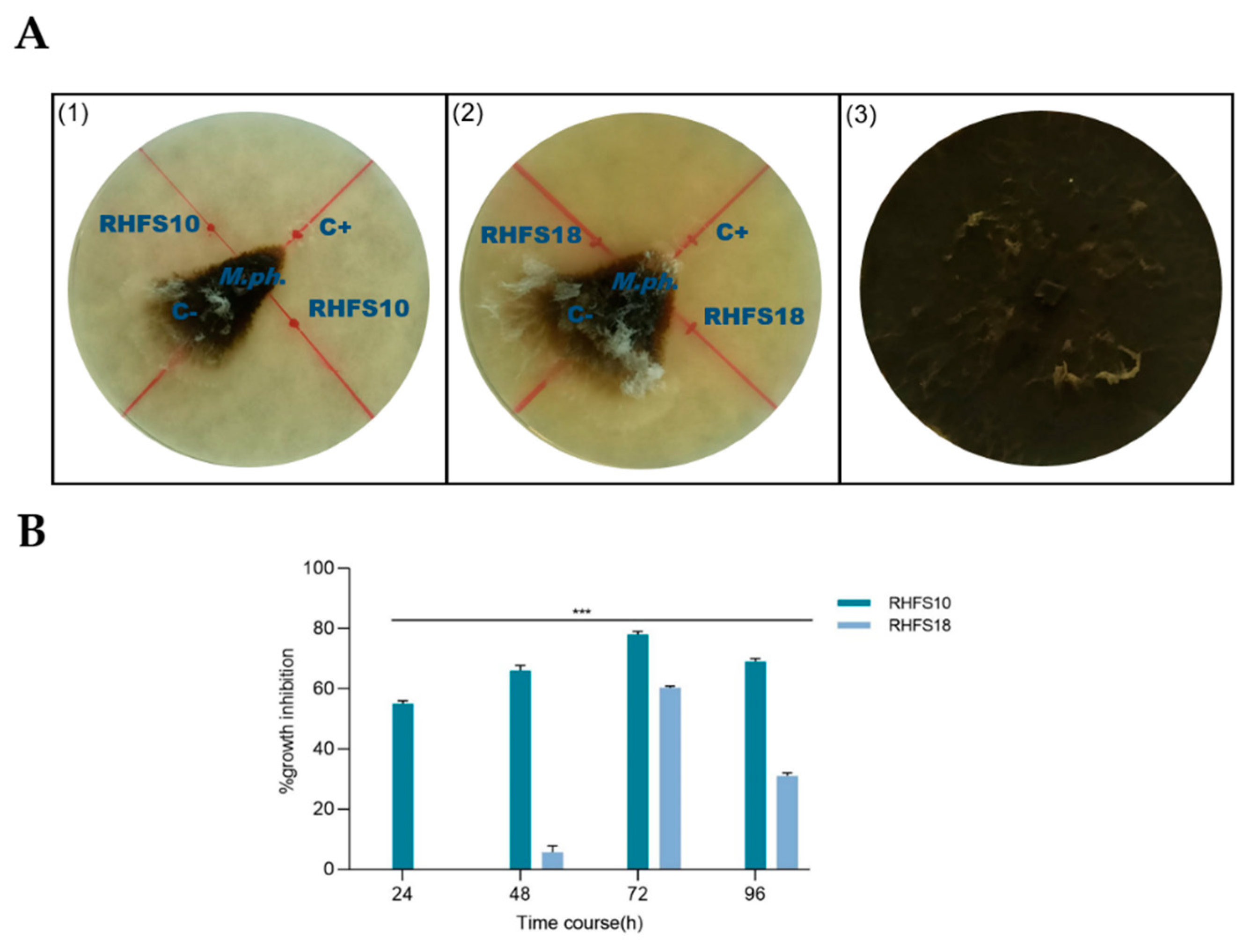
2.2. Antagonistic Activity of Spore-Forming Isolates against Fungal Plant Pathogen
2.3. Characterization of Antifungal Metabolites
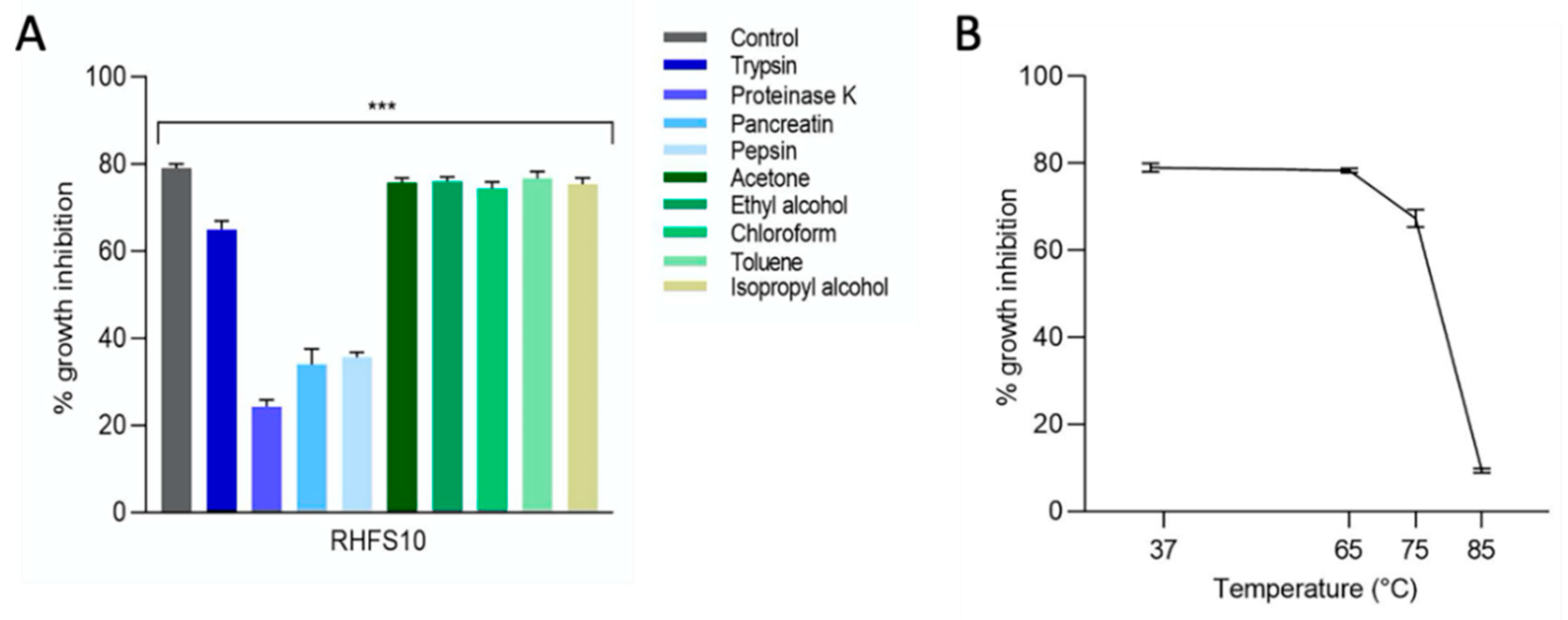
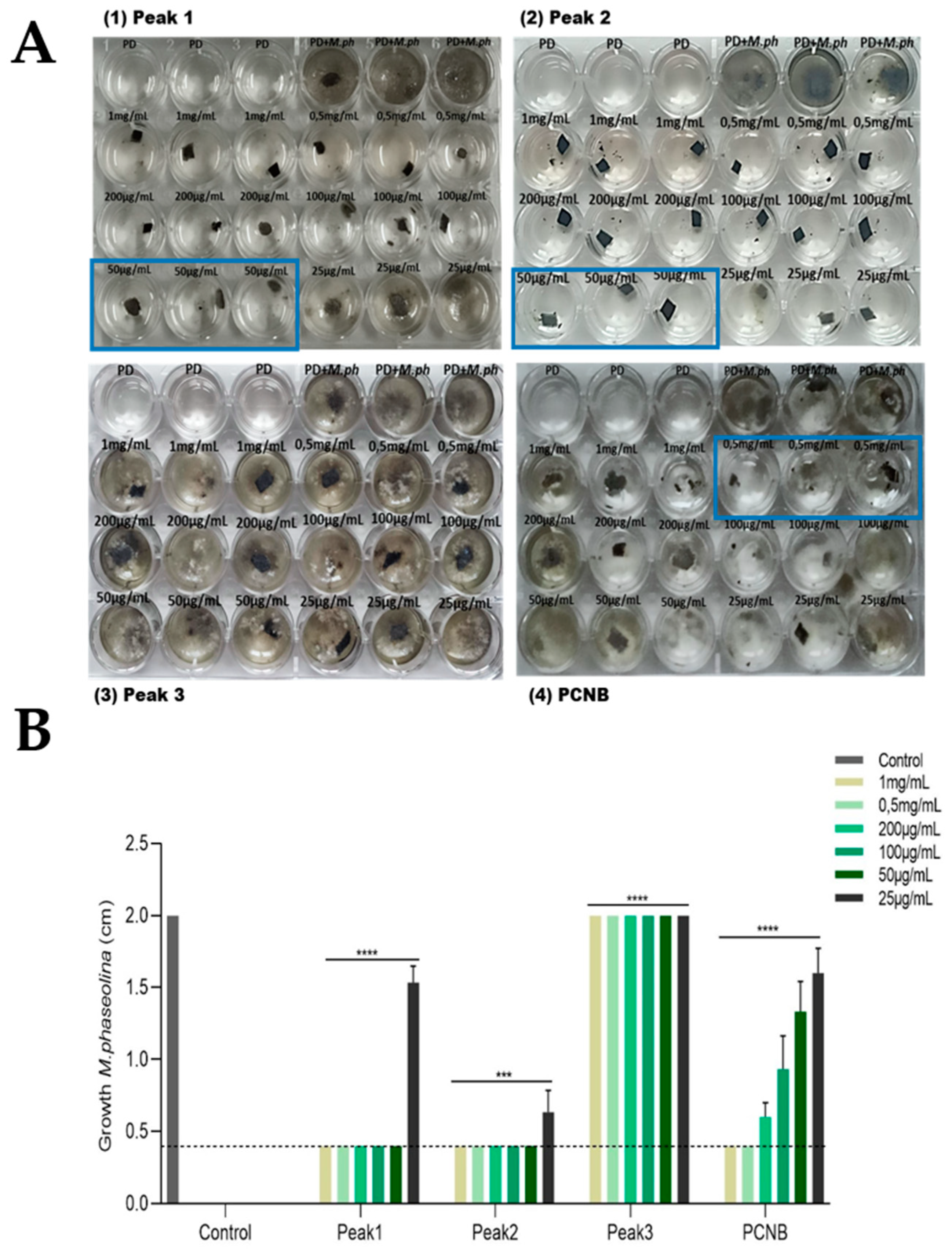
2.4. Purification of Antifungal Metabolites
2.4.1. Minimum Inhibitory Concentration
2.4.2. Preliminary Identification of Bioactive Compounds
3. Discussion
4. Materials and Methods
4.1. Isolation of Bacteria
4.2. Growth Conditions
4.3. Isolates Identification by PCR Amplification of 16S rRNA
4.4. In Vitro Screening for Plant Growth-Promoting (PGP) Traits
4.4.1. Phosphate Solubilization
4.4.2. Siderophore Production
4.4.3. Indole Acetic Acid Detection
4.4.4. Biosurfactant Production
4.4.5. Swarming Motility
4.4.6. Biofilm Production
4.5. Evaluation of Potential Biocontrol Features
4.5.1. Screening for Hydrolytic Enzymatic Activity
4.5.2. Dual-Culture Assay
4.5.3. Antifungal Assay of Cell-Free Supernatants (CFSs)
4.6. Extraction of Secondary Metabolites
4.7. Stability of Antifungal Metabolites at Different Enzymes, Temperatures and Organic Solvent Conditions
4.8. Size-Fractionated Supernatants Tested for Antifungal Activity
4.9. LC–MS/MS Analyses
4.10. Detection of Antifungal Metabolites
4.11. Minimum Inhibitory Concentrations
4.12. Whole-Genome Sequencing
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nicolopoulou-Stamati, P.; Maipas, S.; Kotampasi, C.; Stamatis, P.; Hens, L. Chemical Pesticides and Human Health: The Urgent Need for a New Concept in Agriculture. Front. Public Health 2006, 4, 148. [Google Scholar] [CrossRef]
- Syed Ab Rahman, S.F.; Singh, E.; Pieterse, C.M.J.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef]
- Cimmino, A.; Masi, M.; Evidente, M.; Superchi, S.; Evidente, A. Fungal phytotoxins with potential herbicidal activity: Chemical and biological characterization. Nat. Prod. Rep. 2015, 32, 1629–1653. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecick, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.A.; Kumar, V.; Bhat, M.A.; Wani, I.A.; Dar, F.L.; Farooq, I.; Bhatti, F.; Koser, R.; Rahman, S.; Jan, A.T. Mechanistic Insights of the Interaction of Plant Growth-Promoting Rhizobacteria (PGPR) With Plant Roots Toward Enhancing Plant Productivity by Alleviating Salinity Stress. Front. Microbiol. 2020, 11, 1952. [Google Scholar] [CrossRef] [PubMed]
- Pal, K.K.; McSpadden Gardener, B. Biological Control of Plant Pathogens. Plant Health Instr. 2006, 36, 37–45. [Google Scholar] [CrossRef]
- Miljaković, D.; Marinković, J.; Balešević-Tubić, S. The Significance of Bacillus spp. in Disease Suppression and Growth Promotion of Field and Vegetable Crops. Microorganisms 2020, 7, 1037. [Google Scholar] [CrossRef]
- Meena, M.; Swapnil, P.; Divyanshu, K.; Kumar, S.; Harish; Tripathi, Y.N.; Zehra, A.; Marwal, A.; Upadhyay, R.S. PGPR-mediated induction of systemic resistance and physiochemical alterations in plants against the pathogens: Current perspectives. J. Basic Microbiol. 2020, 60, 828–861. [Google Scholar] [CrossRef]
- Gamez, R.; Montes, M.; Ramirez, S.; Schnell, S.; Rodriguez, F. Screening, plant growth promotion and root colonization pattern of two rhizobacteria (Pseudomonas fluorescens Ps006 and Bacillus amyloliquefaciens Bs006) on banana cv. Williams (Musa acuminata Colla). Microbiol. Res. 2019, 220, 12–20. [Google Scholar] [CrossRef]
- Tamošiūnė, I.; Stanienė, G.; Haimi, P.; Stanys, V.; Rugienius, R.; Baniulis, D. Endophytic Bacillus and Pseudomonas spp. Modulate Apple Shoot Growth, Cellular Redox Balance, and Protein Expression Under in Vitro Conditions. Front. Plant Sci. 2018, 9, 889. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.H. Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [PubMed]
- Pesce, G.; Rusciano, G.; Sasso, A.; Isticato, R.; Sirec, T.; Ricca, E. Surface charge and hydrodynamic coefficient measurements of Bacillus subtilis spore by optical tweezers. Colloids Surf. B 2014, 116, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Gardner, B.B.M. Ecology of Bacillus and Paenibacillus spp. in Agricultural System. Phytopathology 2004, 94, 1252–1258. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [PubMed]
- Rodrìgue-Echeverrìa, S. Influence of soil microbiota in nurse plant systems. Funct. Ecol. 2015, 30, 30–40. [Google Scholar] [CrossRef]
- Kaur, S.; Dhillon, G.S.; Brar, S.K.; Vallad, G.E.; Chand, R.; Chauhan, V.B. Emerging phytopathogen Macrophomina phaseolina: Biology, economic importance and current diagnostic trends. CR Microbiol. 2012, 38, 136–151. [Google Scholar] [CrossRef]
- Pagano, M.C.; Miransari, M. The importance of soybean production worldwide. Soybean Prod. 2016, 1, 1–26. [Google Scholar] [CrossRef]
- Simonetti, E.; Viso, N.P.; Montecchia, M.; Zilli, C.; Balestrasse, K.; Carmona, M. Evaluation of native bacteria and manganese phosphite for alternative control of charcoal root rot of soybean. Microbiol. Res. 2015, 180, 40–48. [Google Scholar] [CrossRef]
- Bacher, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Road map to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Jadhav, H.; Shaikh, S.S.; Sayyed, R. Role of Hydrolytic Enzymes of Rhizoflora in Biocontrol of Fungal Phytopathogens: An Overview. Rhizotrophs Plant Growth Promot. Bioremed. 2017, 978, 183–203. [Google Scholar] [CrossRef]
- Hong, T.-T.; Meng, M. Biochemical characterization and antifungal activity of an endo-1,3-beta-glucanase of Paenibacillus sp. isolated from garden soil. Appl. Microbiol. Biotechnol. 2003, 61, 472–478. [Google Scholar] [CrossRef]
- Tariq, M.; Noman, M.; Ahmed, T.; Hameed, A.; Manzoor, N.; Zafar, M. Antagonistic features displayed by Plant Growth Promoting Rhizobacteria (PGPR): A Review. J. Plant Sci. Phytopathol. 2017, 1, 38–43. [Google Scholar] [CrossRef]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.L. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 48, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Masi, M.; Sautua, F.; Zatout, R.; Castaldi, S.; Arrico, L.; Isticato, R.; Pecitelli, G.; Carmona, M.A.; Evidente, A. Phaseocyclopentenones A and B, Phytotoxic Penta-and Tetrasubstituted Cyclopentenones Produced by Macrophomina phaseolina, the Causal Agent of Charcoal Rot of Soybean in Argentina. J. Nat. Prod. 2021, 84, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Martìnez-Hidaldo, P.; Ice, T.A.; Maymon, M.; Humm, E.A.; Nejat, N.; Sanders, E.R.; Hirsch, A.M. Antifungal Activity of Bacillus Species Against Fusarium and Analysis of the Potential Mechanisms Used in Biocontrol. Front. Microbiol. 2018, 320, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Cazorla, F.M.; Romeo, D.; Pérez-García, A.; Lugtenberg, B.J.J.; De Vincente, A.; Bloemberg, G. Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J. Appl. Microbiol. 2007, 103, 1950–1959. [Google Scholar] [CrossRef]
- Taniguchi, T.; Usuki, H.; Kikuchi, J.; Hirobe, M.; Miki, N.; Fukuda, K.; Zhang, G.; Wang, L.; Yoshikawa, K.; Yamanaka, N. Colonization and community structure of root-associated microorganisms of Sabina vulgaris with soil depth in a semiarid desert ecosystem with shallow groundwater. Mycorrhiza 2012, 22, 419–428. [Google Scholar] [CrossRef]
- Corvey, C.; Stein, T.; Düsterhus, S.; Karas, M.; Entian, K.-D. Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem. Biophys. Res. Commun. 2003, 304, 48–54. [Google Scholar] [CrossRef]
- Yan, L.; Qian, Y. Cloning and heterologous expression of SS10, a subtilisin-like protease displaying antifungal activity from Trichoderma harzianum. FEMS Microbiol. Lett. 2009, 290, 54–61. [Google Scholar] [CrossRef]
- Schönbichler, A.; Díaz-Moreno, S.M.; Srivastava, V.; McKee, S.L. Exploring the Potential for Fungal Antagonism and Cell Wall Attack by Bacillus subtilis natto. Front. Microbiol. 2020, 31, 11–521. [Google Scholar] [CrossRef]
- Cangiano, G.; Sirec, T.; Panarella, C.; Isticato, R.; Baccigalupi, L.; De Felice, M.; Ricca, E. The sps Gene Products Affect the Germination, Hydrophobicity, and Protein Adsorption of Bacillus subtilis Spores. Appl. Environ. Microbiol. 2014, 80, 7293–7302. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, C.; Castaldi, S.; Lanzilli, M.; Saggese, A.; Donadio, G.; Baccigalupi, L.; Ricca, E.; Isticato, R. The temperature of growth and sporulation modulates the efficiency of spore-display in Bacillus subtilis. Microb. Cell Factories 2020, 19, 185. [Google Scholar] [CrossRef] [PubMed]
- Gronemeyer, J.L.; Burbano Roa, C.S.; Hurek, T.; Reinhold-Hurek, B. Isolation and characterization of root-associated bacteria from agricultural crops in the Kavango region of Namibia for Plant Growth Promoting Characteristics. Plant Soil 2012, 71, 566–571. [Google Scholar] [CrossRef]
- Schoebitz, M.; Ceballos, C.; Ciampl, L. Effect of immobilized phosphate solubilizing bacteria on wheat growth and phosphate uptake. J. Soil Sci. Plant Nutr. 2013, 12, 1–10. [Google Scholar] [CrossRef]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a fast and universal method for siderophore detection. J. Microb. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef]
- Damodaran, T.; Sah, V.; Nishra, V.K.; Jha, S.K.; Kannan, R. Isolation of salt tolerant endophytic and rhizospheric bacteria by natural selection and screening for promising plant growth-promoting rhizobacteria (PGPR) and growth vigour in tomato under sodic environment. Afr. J. Microb. Res. 2013, 7, 5082–5089. [Google Scholar] [CrossRef]
- Sarwar, S.; Brader, G.; Corretto, E.; Aleti, G.; Abaidullah, M.; Sessitsch, A.; Hafeez, F.Y. Qualitative analysis of biosurfactants from bacillus species exhibiting antifungal activity. PLoS ONE 2018, 13, e0198107. [Google Scholar] [CrossRef]
- Haney, E.F.; Brito-Sánchez, Y.; Trimble, M.J.; Mansour, S.C.; Cherkasov, A.; Hancock, R.E.W. Computer-aided discovery of peptides that Effect of phosphate solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere specifically attack bacterial biofilms. Sci. Rep. 2018, 8, 1871. [Google Scholar] [CrossRef]
- Morris, L.S.; Evans, J.; Marchesi, R. A robust plate assay for detection of extracellular microbial protease activity in metagenomic screens and pure cultures. J. Microbiol. Methods 2012, 91, 144–146. [Google Scholar] [CrossRef]
- Parsad, M.P.; Manjunath, K. Effect of media and process parameters in the enhancement of extracellular lipase production by bacterial isolates from industrial effluents. Int. J. Microbiol. Res. 2012, 4, 308–311. [Google Scholar] [CrossRef]
- Sethi, S.; Alariya, S.S.; Gupta, S.; Gupta, L.B. Amylase activity of a starch degrading bacteria isolated from soil. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 659–671. [Google Scholar] [CrossRef]
- Meddeb-Mouelhi, F.; Moisan, J.K.; Beauregard, M. A comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzym. Microb. Technol. 2014, 66, 16–19. [Google Scholar] [CrossRef]
- Hankin, L.; Anagnostakis, S. Solid media containing carboxy methyl cellulose to detect CM cellulase activity of microorganisms. Microbiology 1977, 98, 109–115. [Google Scholar] [CrossRef]
- Kuddus, S.M.; Ahmad, R.I.Z. Isolation of novel chitinolytic bacteria and production optimization of extracellular chitinase. J. Genet. Eng. Biotechnol. 2013, 11, 39–46. [Google Scholar] [CrossRef]
- Geetha, K.; Venkatesham, E.V.; Hindumathi, A.; Bhadraiah, B. Isolation, screening and characterization of plant growth promoting bacteria and their effect on Vigna Radia (L.) R. Wilczek. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 799–809. [Google Scholar] [CrossRef]
- Khamna, S.; Yokota, A.; Lumyong, S. Actinomycetes isolated from medicinal plant rhizospheric soils: Diversity and screening of antifungal compounds, indole-3-acetic acid and siderophore production. World J. Microbiol. Biotechnol. 2009, 25, 649–655. [Google Scholar] [CrossRef]
- Kumar, P.; Dubey, R.C.; Maheshwari, D.K. Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol. Res. 2012, 8, 493–499. [Google Scholar] [CrossRef]
- Lavermicocca, P.; Lonigro, S.L.; Evidente, A.; Andolfi, A. Bacteriocin production by Pseudomonas syringae pv.ciccaronei NCPPB2355. Isolation and partial characterization of the antimicrobial compound. J. Appl. Microbiol. 2001, 86, 257–265. [Google Scholar] [CrossRef]
- Agrillo, B.; Mirino, S.; Tatè, R.; Gratino, L.; Gogliettino, M.; Cocca, E.; Tabli, N.; Nabti, E.; Palmieri, G. An alternative biocontrol agent of soil-borne phytopathogens: A new antifungal compound produced by a plant growth promoting bacterium isolated from North Algeria. Microbiol. Res. 2019, 221, 60–69. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]

| Biofertilizer Activities | Biocontrol Activities | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains Code |
Siderophores
Production | PVK * | IAA * | Biofilm | Swarming |
Protease
Activity |
Amylase
Activity |
Lipase
Activity |
Xylanase
Activity |
Cellulase
Activity |
Chitinase
Activity |
Catalase
Activity |
| RHFS1 | + | - | - | - | - | +++ | +++ | + | ++ | ++ | + | +++ |
| RHFS2 | - | ++ | - | + | +++ | +++ | +++ | + | ++ | +++ | ++ | - |
| RHFS3 | - | + | + | - | − | +++ | +++ | ++ | + | +++ | - | - |
| RHFS4 | - | + | + | - | - | +++ | +++ | - | +++ | +++ | ++ | +++ |
| RHFS5 | + | - | + | - | - | +++ | +++ | - | + | + | ++ | ++ |
| RHFS6 | - | + | ++ | - | - | +++ | +++ | - | +++ | ++ | - | +++ |
| RHFS7 | - | - | ++ | - | - | + | - | + | +++ | +++ | ++ | +++ |
| RHFS8 | ++ | + | - | - | - | +++ | +++ | - | ++ | + | ++ | +++ |
| RHFS9 | + | - | - | + | +++ | +++ | +++ | ++ | - | +++ | ++ | ++ |
| RHFS10 | +++ | ++ | + | ++ | +++ | +++ | +++ | ++ | +++ | +++ | ++ | +++ |
| RHFS11 | + | + | + | - | - | +++ | +++ | + | +++ | - | ++ | ++ |
| RHFS12 | - | + | + | + | - | - | +++ | - | +++ | +++ | ++ | + |
| RHFS13 | - | - | ++ | - | - | +++ | - | - | ++ | - | ++ | ++ |
| RHFS14 | - | ++ | ++ | - | - | + | + | - | +++ | - | + | +++ |
| RHFS15 | + | + | +++ | - | - | +++ | +++ | + | ++ | +++ | ++ | - |
| RHFS16 | + | + | + | + | - | ++ | ++ | - | +++ | +++ | - | - |
| RHFS17 | + | + | +++ | - | - | +++ | ++ | - | + | + | ++ | +++ |
| RHFS18 | +++ | ++ | ++ | +++ | ++ | +++ | +++ | ++ | +++ | +++ | ++ | +++ |
| RHFS19 | ++ | ++ | + | +++ | ++ | +++ | +++ | ++ | +++ | +++ | + | +++ |
| RHFS20 | + | - | + | - | - | + | ++ | - | ++ | ++ | ++ | +++ |
| RHFS22 | + | + | + | - | ++ | +++ | - | - | + | ++ | ++ | - |
| RHFS28 | - | - | - | - | - | +++ | +++ | - | ++ | ++ | ++ | ++ |
| Fractions | Mass (Da) a | Swiss Prot AC | Significant Sequences | Score | Description |
|---|---|---|---|---|---|
| Peak 1 | 47.924 | XYNC_BACIU | 18 | 1776 | Glucuronoxylanase XynC OS = Bacillus subtilis |
| 39.483 | SUBN_BACNA | 5 | 1080 | Subtilisin NAT OS = Bacillus subtilis subsp. natto | |
| 27.42 | SUBN_BACNA | 5 | 865 | Subtilisin NAT OS = Bacillus subtilis subsp. natto | |
| 75.961 | SACC_BACSU | 1 | 795 | Levanase OS = Bacillus subtilis | |
| 38.141 | PEL2_BACIU | 3 | 566 | Pectin lyase OS = Bacillus subtilis | |
| Peak 2 | 27.365 | GUB_BACAM | 8 | 990 | Beta-glucanase OS = Bacillus amyloliquefaciens |
| 39.483 | SUBN_BACNA | 5 | 800 | Subtilisin NAT OS = Bacillus subtilis subsp. natto | |
| 27.42 | SUBN_BACNA | 5 | 637 | Subtilisin NAT OS = Bacillus subtilis subsp. natto | |
| Peak 3 | 72.39 | AMY_BACSU | 1 | 41 | Alpha-amylase OS = Bacillus subtilis |
| 34.106 | MPR_BACSU | 1 | 39 | Extracellular metalloprotease OS = Bacillus subtilis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castaldi, S.; Petrillo, C.; Donadio, G.; Piaz, F.D.; Cimmino, A.; Masi, M.; Evidente, A.; Isticato, R. Plant Growth Promotion Function of Bacillus sp. Strains Isolated from Salt-Pan Rhizosphere and Their Biocontrol Potential against Macrophomina phaseolina. Int. J. Mol. Sci. 2021, 22, 3324. https://doi.org/10.3390/ijms22073324
Castaldi S, Petrillo C, Donadio G, Piaz FD, Cimmino A, Masi M, Evidente A, Isticato R. Plant Growth Promotion Function of Bacillus sp. Strains Isolated from Salt-Pan Rhizosphere and Their Biocontrol Potential against Macrophomina phaseolina. International Journal of Molecular Sciences. 2021; 22(7):3324. https://doi.org/10.3390/ijms22073324
Chicago/Turabian StyleCastaldi, Stefany, Claudia Petrillo, Giuliana Donadio, Fabrizio Dal Piaz, Alessio Cimmino, Marco Masi, Antonio Evidente, and Rachele Isticato. 2021. "Plant Growth Promotion Function of Bacillus sp. Strains Isolated from Salt-Pan Rhizosphere and Their Biocontrol Potential against Macrophomina phaseolina" International Journal of Molecular Sciences 22, no. 7: 3324. https://doi.org/10.3390/ijms22073324
APA StyleCastaldi, S., Petrillo, C., Donadio, G., Piaz, F. D., Cimmino, A., Masi, M., Evidente, A., & Isticato, R. (2021). Plant Growth Promotion Function of Bacillus sp. Strains Isolated from Salt-Pan Rhizosphere and Their Biocontrol Potential against Macrophomina phaseolina. International Journal of Molecular Sciences, 22(7), 3324. https://doi.org/10.3390/ijms22073324











