A High-Throughput Screening System Based on Fluorescence-Activated Cell Sorting for the Directed Evolution of Chitinase A
Abstract
1. Introduction
2. Results and Discussion
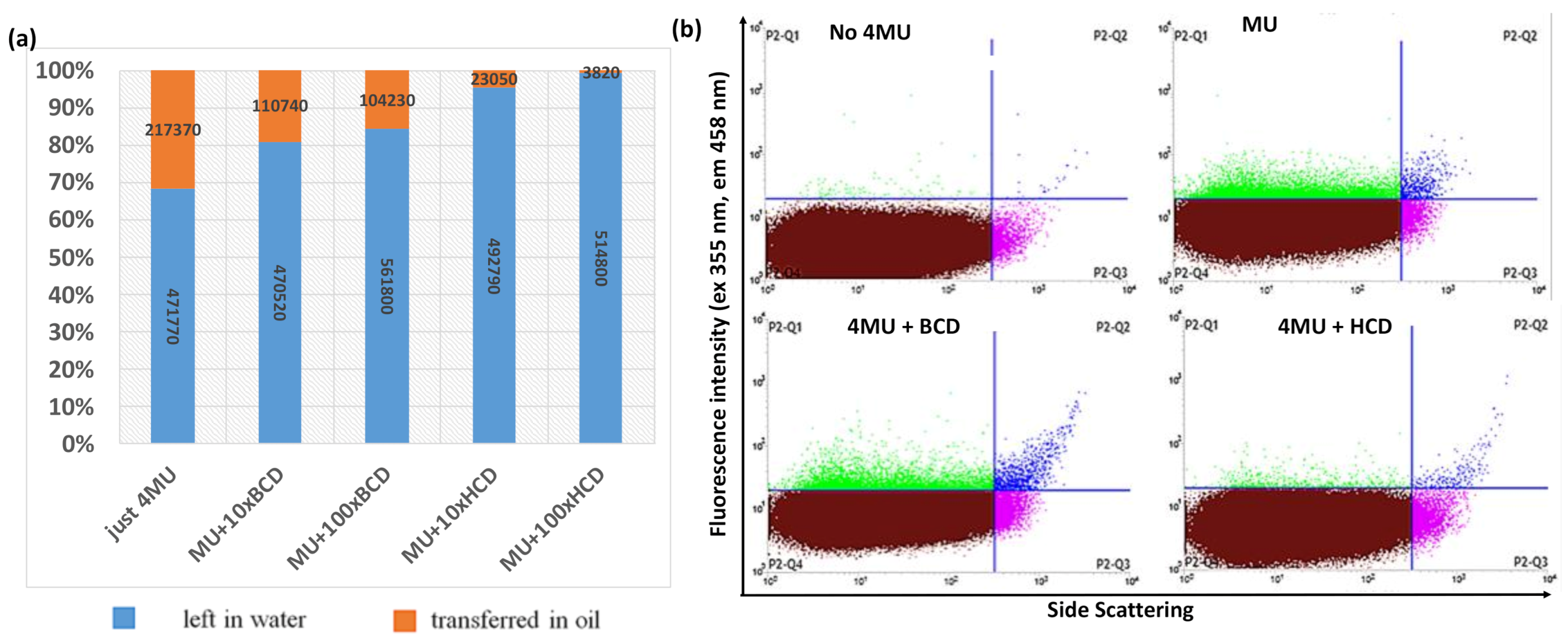
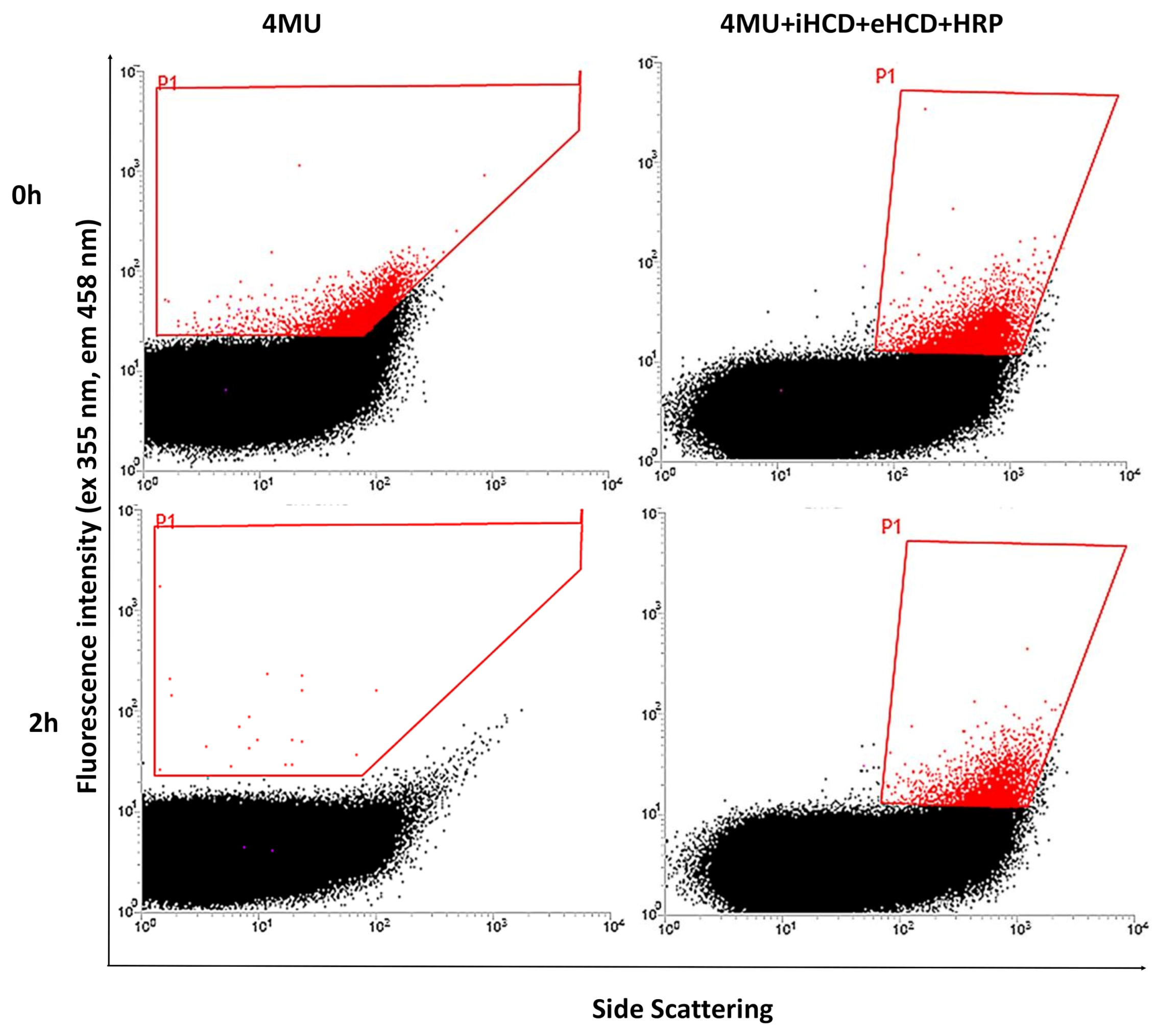
2.1. Development of the FACS Assay
2.2. Sorting of Reference and Mutant ChiA Gene Libraries
2.3. Screening of Mutants Using the MTP-Based Fluorescence Assay
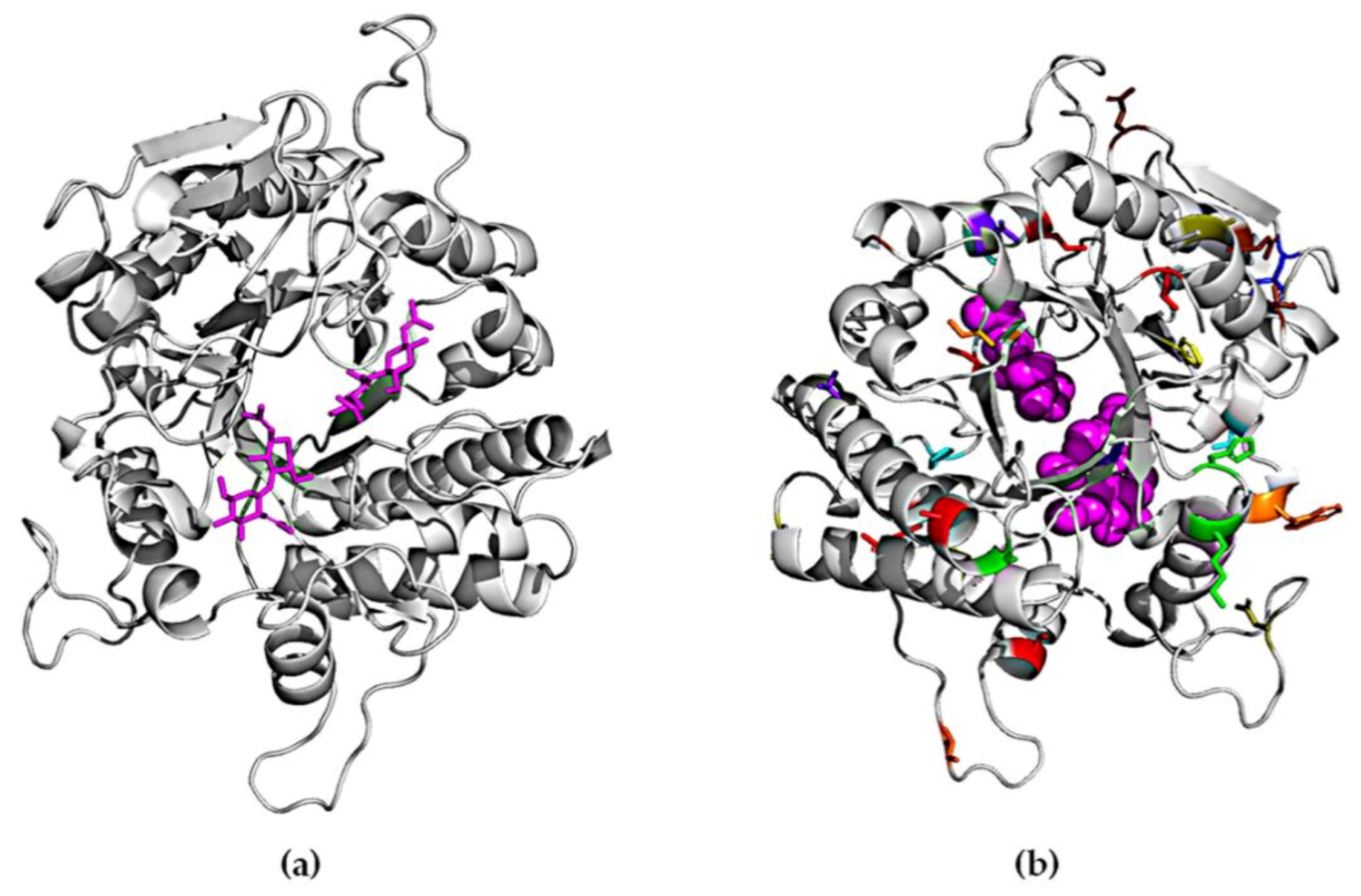
2.4. Homology Modeling of ChiA
3. Materials and Methods
3.1. Chemicals and Enzymes
3.2. Directed Evolution of ChiA and Library Preparation
3.3. MTP Assay for ChiA Activity
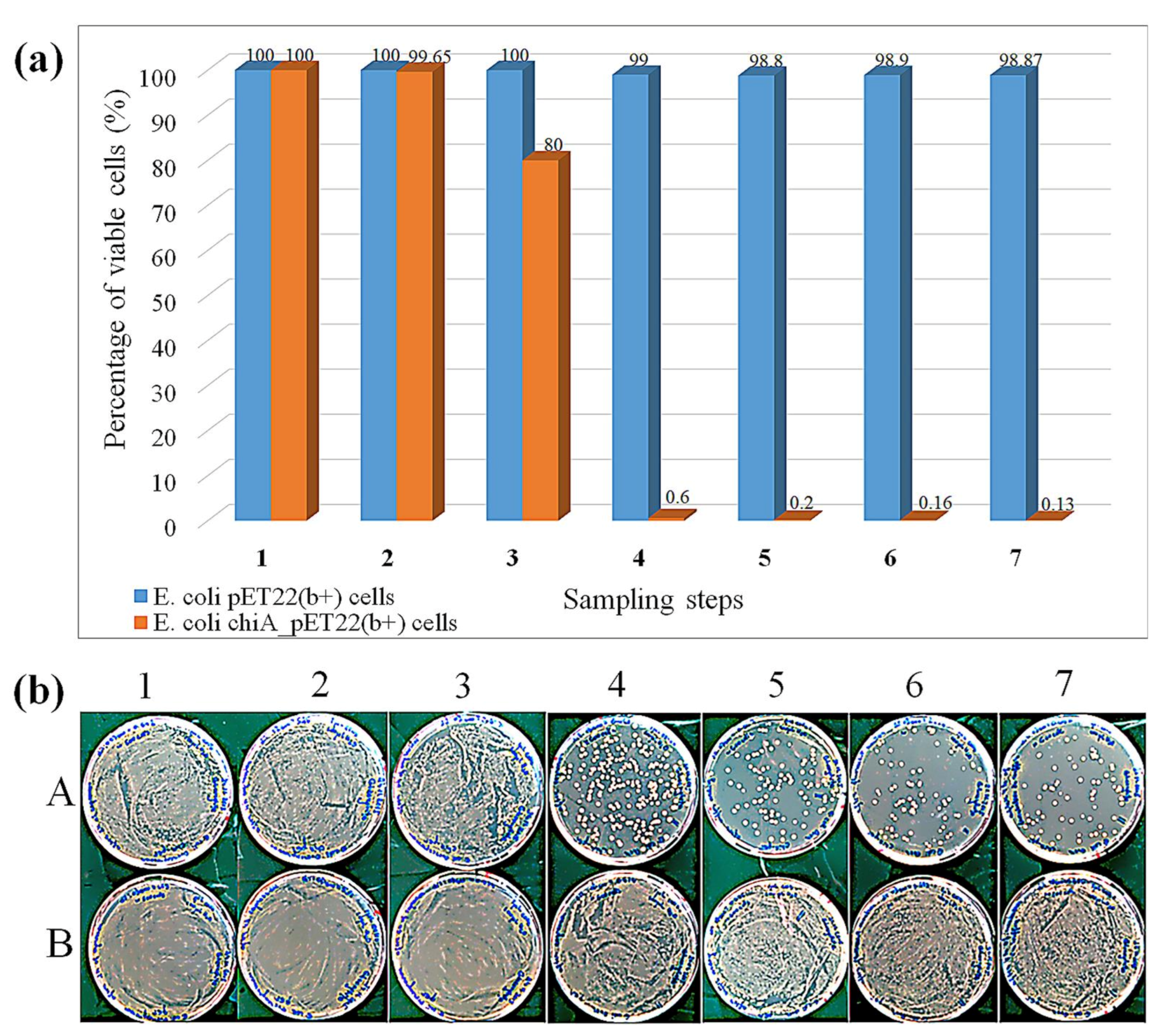
3.4. Cell Viability Assay
3.5. FACS Assay
3.6. Testing of Cyclodextrins and Different Oils
3.7. Sorting and Selection of ChiA Mutants
3.8. MTP Fluorescence Assay
3.9. Homology Modeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lenardon, M.D.; Munro, C.A.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef]
- Yadav, M.; Goswami, P.; Paritosh, K.; Kumar, M.; Pareek, N.; Vivekanand, V. Seafood waste: A source for preparation of commercially employable chitin/chitosan materials. Bioresour. Bioprocess. 2019, 6, 1–20. [Google Scholar] [CrossRef]
- Santos, V.P.; Marques, N.S.S.; Maia, P.C.S.V.; De Lima, M.A.B.; Franco, L.D.O.; De Campos-Takaki, G.M. Seafood waste as attractive source of chitin and chitosan production and their applications. Int. J. Mol. Sci. 2020, 21, 4290. [Google Scholar] [CrossRef]
- Songsiriritthigul, C.; Lapboonrueng, S.; Pechsrichuang, P.; Pesatcha, P.; Yamabhai, M. Expression and characterization of Bacillus licheniformis chitinase (ChiA), suitable for bioconversion of chitin waste. Bioresour. Technol. 2010, 101, 4096–4103. [Google Scholar] [CrossRef] [PubMed]
- Muzzarelli, R.A.; Mattioli-Belmonte, M.; Pugnaloni, A.; Biagini, G. Biochemistry, histology and clinical uses of chitins and chitosans in wound healing. EXS 1999, 87, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Matica, M.A.; Aachmann, F.L.; Tøndervik, A.; Sletta, H.; Ostafe, V. Chitosan as a wound dressing starting material: Antimicrobial properties and mode of action. Int. J. Mol. Sci. 2019, 20, 5889. [Google Scholar] [CrossRef]
- Mincea, M.; Negrulescu, A.; Ostafe, V. Preparation, modification, and applications of chitin nanowhiskers: A review. Rev. Adv. Mater. Sci. 2012, 30, 225–242. [Google Scholar]
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, limitations, and trends in engineering for suitable applications. Biosci. Rep. 2018, 38, 2018032300. [Google Scholar] [CrossRef] [PubMed]
- Ostafe, R.; Prodanovic, R.; Nazor, J.; Fischer, R. Ultra-high-throughput screening method for the directed evolution of glucose oxidase. Chem. Biol. 2014, 21, 414–421. [Google Scholar] [CrossRef]
- Prodanovic, R.; Ostafe, R.; Scacioc, A.; Schwaneberg, U. Ultrahigh throughput screening system for directed glucose oxidase evolution in yeast cells. Comb. Chem. High Throughput Screen. 2010, 14, 55–60. [Google Scholar] [CrossRef]
- Farinas, E.; Schwaneberg, U.; Glieder, A.; Arnold, F. Directed evolution of a cytochrome P450 monooxygenase for alkane oxidation. Adv. Synth. Catal. 2001, 343. [Google Scholar] [CrossRef]
- Goedegebuur, F.; Dankmeyer, L.; Gualfetti, P.; Karkehabadi, S.; Hansson, H.; Jana, S.; Huynh, V.; Kelemen, B.R.; Kruithof, P.; Larenas, E.A.; et al. Improving the thermal stability of cellobiohydrolase Cel7A from Hypocrea jecorina by directed evolution. J. Biol. Chem. 2017, 292, 17418–17430. [Google Scholar] [CrossRef]
- Đurđić, I.K.; Ece, S.; Ostafe, R.; Vogel, S.; Schillberg, S.; Fischer, R.; Prodanović, R. Improvement in oxidative stability of versatile peroxidase by flow cytometry-based high-throughput screening system. Biochem. Eng. J. 2020, 157, 107555. [Google Scholar] [CrossRef]
- Blažić, M.; Balaž, A.M.; Tadić, V.; Draganić, B.; Ostafe, R.; Fischer, R.; Prodanović, R. Protein engineering of cellobiose dehydrogenase from Phanerochaete chrysosporium in yeast Saccharomyces cerevisiae Invsc1 for increased activity and stability. Biochem. Eng. J. 2019, 146, 179–185. [Google Scholar] [CrossRef]
- Weiß, M.S.; Pavlidis, I.V.; Spurr, P.; Hanlon, S.P.; Wirz, B.; Iding, H.; Bornscheuer, U.T. Protein-engineering of an amine transaminase for the stereoselective synthesis of a pharmaceutically relevant bicyclic amine. Org. Biomol. Chem. 2016, 14, 10249–10254. [Google Scholar] [CrossRef] [PubMed]
- Giver, L.; Gershenson, A.; Freskgard, P.-O.; Arnold, F.H. Directed evolution of a thermostable esterase. Proc. Natl. Acad. Sci. USA 1998, 95, 12809–12813. [Google Scholar] [CrossRef]
- Liebeton, K.; Zonta, A.; Schimossek, K.; Nardini, M.; Lang, D.; Dijkstra, B.W.; Reetz, M.T.; Jaeger, K.E. Directed evolution of an enantioselective lipase. Chem. Biol. 2000, 7, 709–718. [Google Scholar] [CrossRef]
- Yoo, T.H.; Pogson, M.; Iverson, B.L.; Georgiou, G. Directed evolution of highly selective proteases by using a novel FACS-based screen that capitalizes on the p53 regulator MDM2. ChemBioChem 2012, 13, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Fang, W.; Xiao, Y.; Yang, X.; Zhang, Y.; Bidochka, M.J.; Pei, Y. Directed evolution for increased chitinase activity. Appl. Microbiol. Biotechnol. 2007, 76, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Songsiriritthigul, C.; Pesatcha, P.; Eijsink, V.G.; Yamabhai, M. Directed evolution of a Bacillus chitinase. Biotechnol. J. 2009, 4, 501–509. [Google Scholar] [CrossRef]
- Wang, S.; Fu, G.; Li, J.; Wei, X.; Fang, H.; Huang, D.; Lin, J.; Zhang, D. High-efficiency secretion and directed evolution of chitinase bcchia1 in Bacillus subtilis for the conversion of chitinaceous wastes into chitooligosaccharides. Front. Bioeng. Biotechnol. 2020, 8, 432. [Google Scholar] [CrossRef] [PubMed]
- Neylon, C. Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: Library construction methods for directed evolution. Nucleic Acids Res. 2004, 32, 1448–1459. [Google Scholar] [CrossRef]
- Cirino, P.C.; Mayer, K.M.; Umeno, D.; Arnold, F.H.; Georgiou, G. Generating mutant libraries using error-prone PCR. In Directed Evolution Library Creation; Humana Press: Totowa, NJ, USA, 2003; pp. 3–9. [Google Scholar] [CrossRef]
- Fujii, R.; Kitaoka, M.; Hayashi, K. Error-prone rolling circle amplification: The simplest random mutagenesis protocol. Nat. Protoc. 2006, 1, 2493. [Google Scholar] [CrossRef]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef]
- Xiao, H.; Bao, Z.; Zhao, H. High throughput screening and selection methods for directed enzyme evolution. Ind. Eng. Chem. Res. 2015, 54, 4011–4020. [Google Scholar] [CrossRef]
- Yang, J.; Liu, X.; Sun, S.; Liu, X.; Yang, L. Screening platform based on robolid microplate for immobilized enzyme-based assays. ACS Omega 2017, 2, 5079–5086. [Google Scholar] [CrossRef]
- Griffiths, A.D.; Tawfik, D.S. Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 2003, 22, 24–35. [Google Scholar] [CrossRef]
- Mastrobattista, E.; Taly, V.; Chanudet, E.; Treacy, P.; Kelly, B.T.; Griffiths, A.D. High-throughput screening of enzyme libraries: In vitro evolution of a Β-galactosidase by fluorescence-activated sorting of double emulsions. Chem. Biol. 2005, 12, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, D.S.; Griffiths, A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998, 16, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Rich, J.R.; Gilbert, M.; Wakarchuk, W.W.; Feng, Y.; Withers, S.G. Fluorescence activated cell sorting as a general ultra-high-throughput screening method for directed evolution of glycosyltransferases. J. Am. Chem. Soc. 2010, 132, 10570–10577. [Google Scholar] [CrossRef] [PubMed]
- Ruff, A.J.; Dennig, A.; Wirtz, G.; Blanusa, M.; Schwaneberg, U. Flow cytometer-based high-throughput screening system for accelerated directed evolution of P450 monooxygenases. ACS Catal. 2012, 2, 2724–2728. [Google Scholar] [CrossRef]
- Körfer, G.D.J.; Pitzler, C.; Vojcic, L.; Martinez, R.; Schwaneberg, U. In vitro flow cytometry-based screening platform for cellulase engineering. Sci. Rep. 2016, 6, 26128. [Google Scholar] [CrossRef]
- Markel, U.; Essani, K.D.; Besirlioglu, V.; Schiffels, J.; Streit, W.R.; Schwaneberg, U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 2020, 49, 233–262. [Google Scholar] [CrossRef]
- Arnold, F.H. The nature of chemical innovation: New enzymes by evolution. Q. Rev. Biophys. 2015, 48, 404–410. [Google Scholar] [CrossRef]
- Kaczmarek, B.M.; Struszczyk-Swita, K.; Li, X.; Szczęsna-Antczak, M.; Daroch, M. Enzymatic modifications of chitin, chitosan, and chitooligosaccharides. Front. Bioeng. Biotechnol. 2019, 7, 243. [Google Scholar] [CrossRef] [PubMed]
- Menghiu, G.; Ostafe, V.; Prodanovic, R.; Fischer, R.; Ostafe, R. Biochemical characterization of chitinase a from Bacillus licheniformis Dsm8785 expressed in Pichia pastoris KM71H. Protein Expr. Purif. 2019, 154, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Raunio, H.; Pentikäinen, O.; Juvonen, R.O. Coumarin-based profluorescent and fluorescent substrates for determining xenobiotic-metabolizing enzyme activities in vitro. Int. J. Mol. Sci. 2020, 21, 4708. [Google Scholar] [CrossRef] [PubMed]
- Gee, R.K.; Sun, W.C.; Bhalgat, M.K.; Upson, R.H.; Klaubert, D.H.; Latham, K.A.; Haugland, R.P. Fluorogenic substrates based on fluorinated umbelliferones for continuous assays of phosphatases and Β-galactosidases. Anal. Biochem. 1999, 273, 41–48. [Google Scholar] [CrossRef]
- Pritsch, K.; Raidl, S.; Marksteiner, E.; Blaschke, H.; Agerer, R.; Schloter, M.; Hartmann, A. A Rapid and highly sensitive method for measuring enzyme activities in single mycorrhizal tips using 4-methylumbelliferone-labelled fluorogenic substrates in a microplate system. J. Microbiol. Methods 2004, 58, 233–241. [Google Scholar] [CrossRef]
- Blažić, M.; Balaž, A.M.; Prodanović, O.; Popović, N.; Ostafe, R.; Fischer, R.; Prodanović, R. Directed evolution of cellobiose dehydrogenase on the surface of yeast cells using resazurin-based fluorescent assay. Appl. Sci. 2019, 9, 1413. [Google Scholar] [CrossRef]
- Tiwari, G.; Tiwari, R.; Rai, A.K. Cyclodextrins in delivery systems: Applications. J. Pharm. Bioallied Sci. 2010, 2, 72–79. [Google Scholar] [CrossRef]
- Winger, T. Enzymatic Assays Using Umbelliferone Substrates with Cyclodextrins in Droplets in Oil. U.S. Patent 8,093,062, 10 January 2012. [Google Scholar]
- Peng, A.C.; Hsu, Y.C. Perfluorocarbon-Soluble Compounds. U.S. Patent WO/2005/107764, 17 November 2005. [Google Scholar]
- Fontana, M.; Costa, M.; Mosca, L.; Rosei, M.A. A specific assay for discriminating between peroxidase and lipoxy-genase activities. Biochem. Mol. Biol. Int. 1997, 42, 163–168. [Google Scholar] [CrossRef]
- Rathore, A.S.; Gupta, R.D. Chitinases from bacteria to human: Properties, applications, and future perspectives. Enzym. Res. 2015, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lobo, M.D.P.; Silva, F.D.A.; Landim, P.G.D.C.; Da Cruz, P.R.; De Brito, T.L.; De Medeiros, S.C.; Oliveira, J.T.A.; Vasconcelos, I.M.; Pereira, H.D.; Grangeiro, T.B. Expression and efficient secretion of a functional chitinase from Chromobacterium violaceum in Escherichia coli. BMC Biotechnol. 2013, 13, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Huang, Y.; Zhang, R.; Diao, G.; Fan, H.; Wang, Z. Chitinase genes Lbchi31 and Lbchi32 from Limonium bicolor were successfully expressed in Escherichia coli and exhibit recombinant chitinase activities. Sci. World J. 2013, 648382. [Google Scholar] [CrossRef]
- Okazaki, K.; Yamashita, Y.; Noda, M.; Sueyoshi, N.; Kameshita, I.; Hayakawa, S. Molecular cloning and expression of the gene encoding family 19 chitinase from Streptomyces sp. J-13-3. Biosci. Biotechnol. Biochem. 2004, 68, 341–351. [Google Scholar] [CrossRef]
- Pettersen, F.E.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [PubMed]
- Ashenberg, O.; Gong, L.I.; Bloom, J.D. Mutational effects on stability are largely conserved during protein evolution. Proc. Natl. Acad. Sci. USA 2013, 110, 21071–21076. [Google Scholar] [CrossRef]
- Zhou, H.-X.; Pang, X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef]
- Lee, H.S.; Ryu, E.J.; Kang, M.J.; Wang, E.S.; Piao, Z.; Choi, Y.J.; Jung, K.H.; Jeon, J.Y.J.; Shin, Y.C. A new approach to directed gene evolution by recombined extension on truncated templates (RETT). J. Mol. Catal. B Enzym. 2003, 26, 119–129. [Google Scholar] [CrossRef]
- Arnold, H.F.; Georgiou, G. Directed Enzyme Evolution. Screening and Selection Methods; Humana Press: Totowa, NJ, USA, 2003. [Google Scholar]
- Ostafe, R.; Prodanovic, R.; Commandeur, U.; Fischer, R. Flow cytometry-based ultra-high-throughput screening assay for cellulase activity. Anal. Biochem. 2013, 435, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]


| No | Mutants | Mutation Points Number | Amino Acids Replaced (Mutations) |
|---|---|---|---|
| 1 | ChiA (wild-type) | - | - |
| 2 | AB01 | 5 | D154E, F315L, E389D, S530P, G568S |
| 3 | AE01 | 8 | V46I, P190A, N192K, T295S, N446I, G516R, T537M, R574Q |
| 4 | AE02 | 6 | K16I, A17V, W42R, I87N, I252V, V474L |
| 5 | AD03 | 2 | R448C, A471E |
| 6 | AH03 | 2 | V484I, A506V |
| 8 | AH09 | 7 | K24N, S79L, R147L, A218V, S230C, I292F, S441P |
| 7 | AF06 | 1 | Y362S |
| 9 | BA09 | 4 | L7S, N53S, V296L, V304D |
| 10 | CA02 | 4 | D110N, P436T, A443V, N469D |
| 11 | CA10 | 2 | A165T, D427N |
| 12 | GM | 5 | M3R, G303D, Q369L, E389D, T419M |
| 13 | DH08 | 3 | K128E, H130N, D220Y |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menghiu, G.; Ostafe, V.; Prodanović, R.; Fischer, R.; Ostafe, R. A High-Throughput Screening System Based on Fluorescence-Activated Cell Sorting for the Directed Evolution of Chitinase A. Int. J. Mol. Sci. 2021, 22, 3041. https://doi.org/10.3390/ijms22063041
Menghiu G, Ostafe V, Prodanović R, Fischer R, Ostafe R. A High-Throughput Screening System Based on Fluorescence-Activated Cell Sorting for the Directed Evolution of Chitinase A. International Journal of Molecular Sciences. 2021; 22(6):3041. https://doi.org/10.3390/ijms22063041
Chicago/Turabian StyleMenghiu, Gheorghita, Vasile Ostafe, Radivoje Prodanović, Rainer Fischer, and Raluca Ostafe. 2021. "A High-Throughput Screening System Based on Fluorescence-Activated Cell Sorting for the Directed Evolution of Chitinase A" International Journal of Molecular Sciences 22, no. 6: 3041. https://doi.org/10.3390/ijms22063041
APA StyleMenghiu, G., Ostafe, V., Prodanović, R., Fischer, R., & Ostafe, R. (2021). A High-Throughput Screening System Based on Fluorescence-Activated Cell Sorting for the Directed Evolution of Chitinase A. International Journal of Molecular Sciences, 22(6), 3041. https://doi.org/10.3390/ijms22063041







