Transcriptional Regulatory Networks Associate with Early Stages of Potato Virus X Infection of Solanum tuberosum
Abstract
1. Introduction
2. Results
2.1. Inoculation of Potato Plants with PVX-GFP and Evaluation of Virus Infection
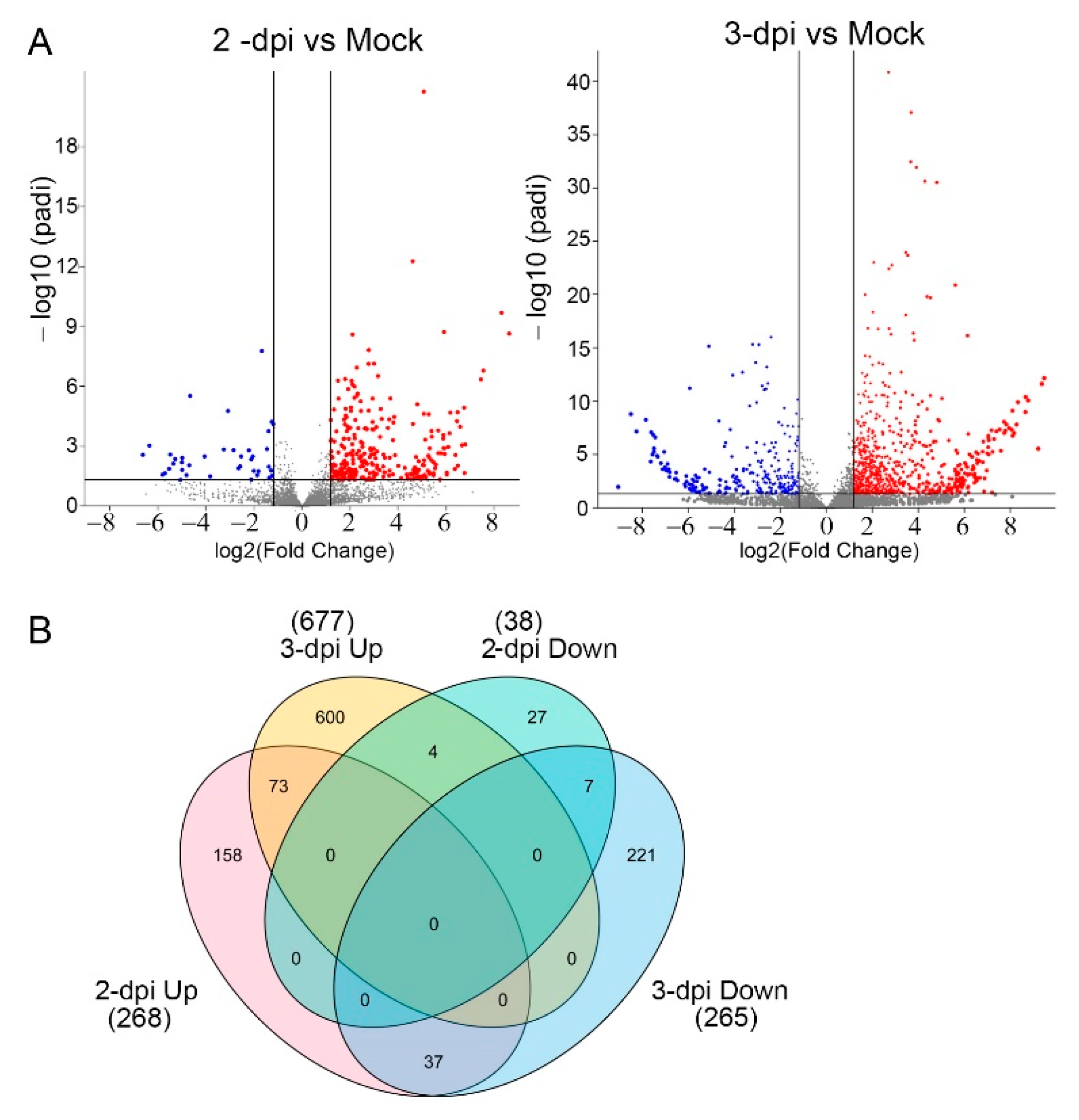
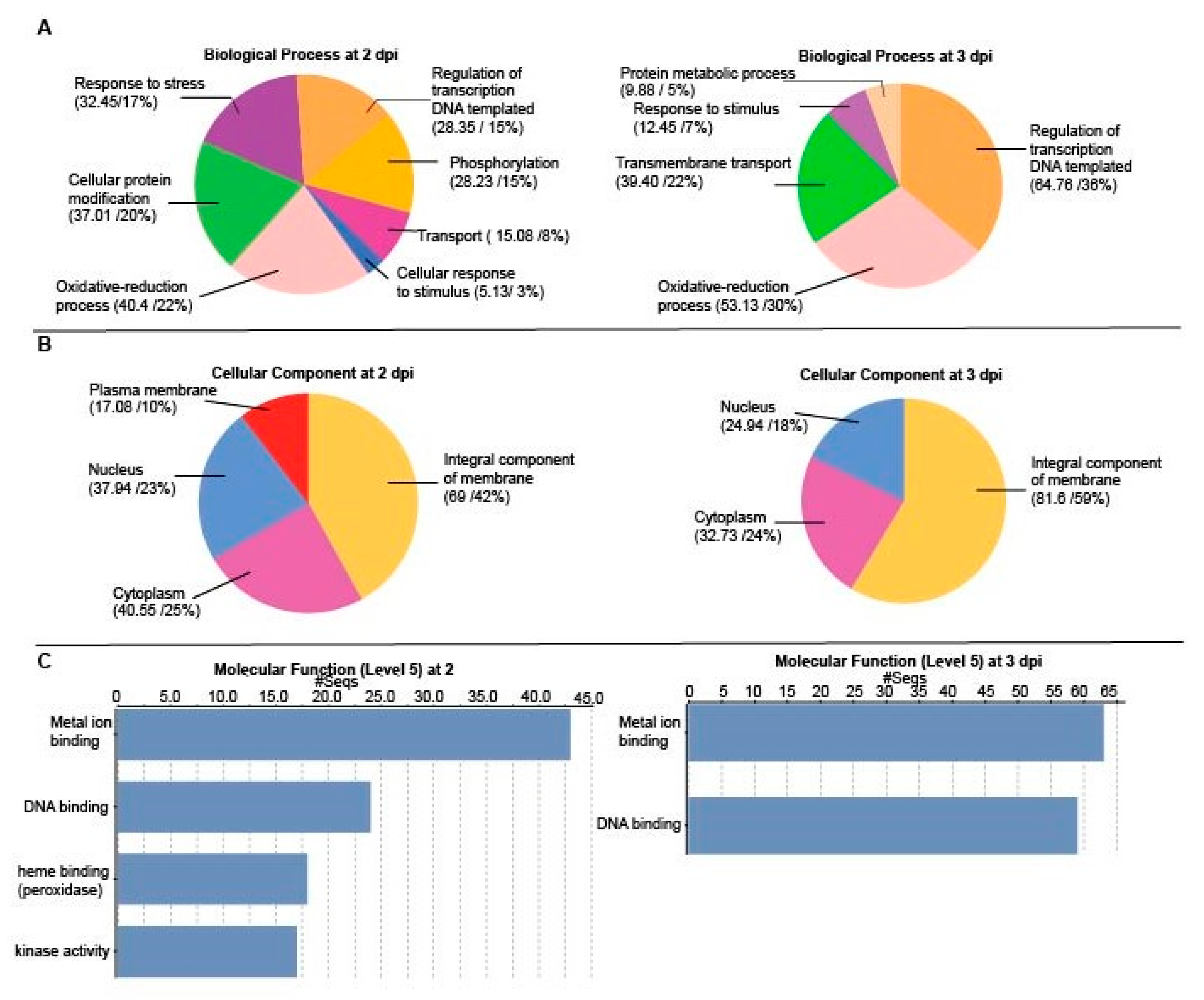
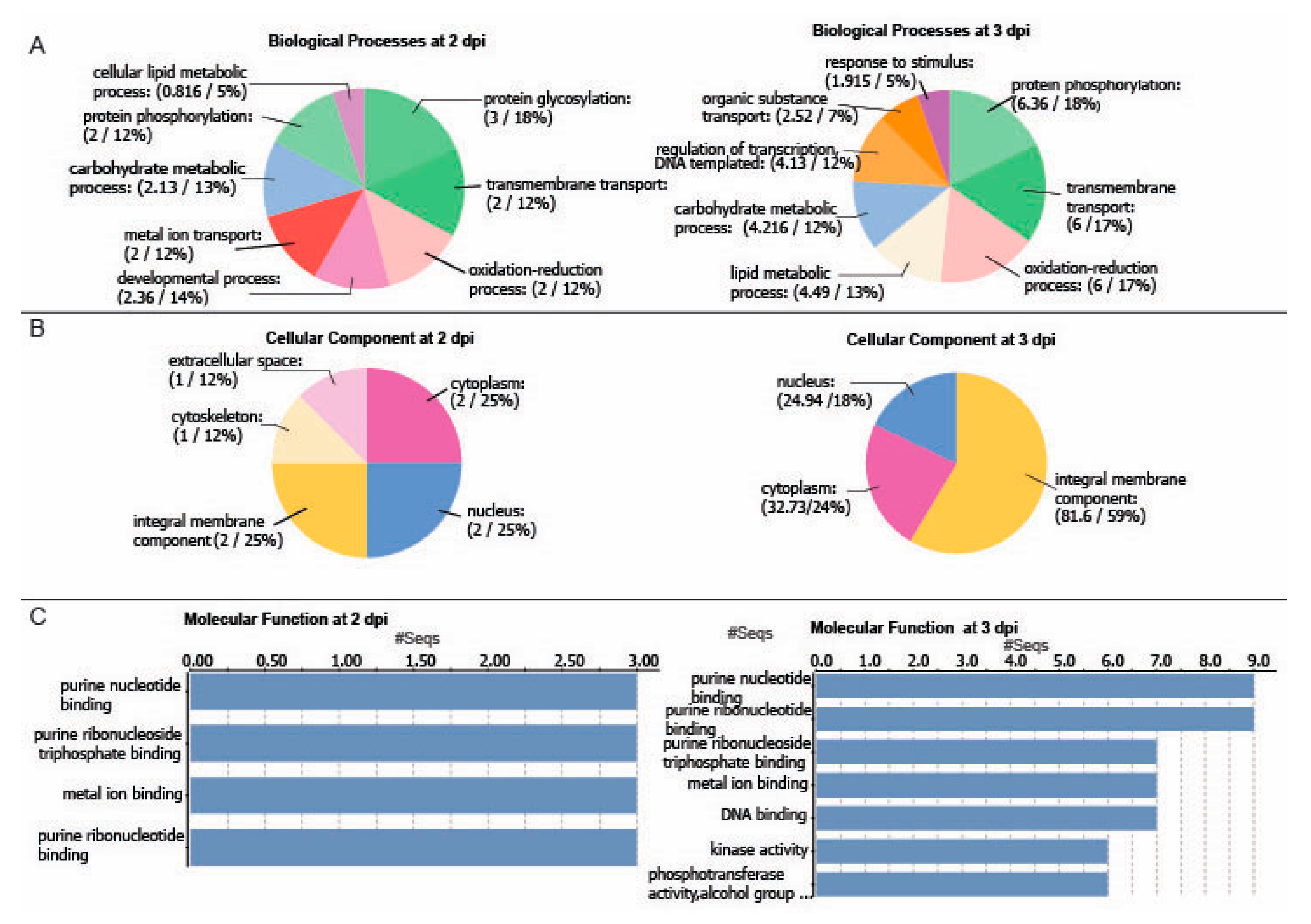
2.2. A Genome-Wide Analysis of Differentially Expressed Genes (DEGs) in Potato Inoculated with PVX-GFP
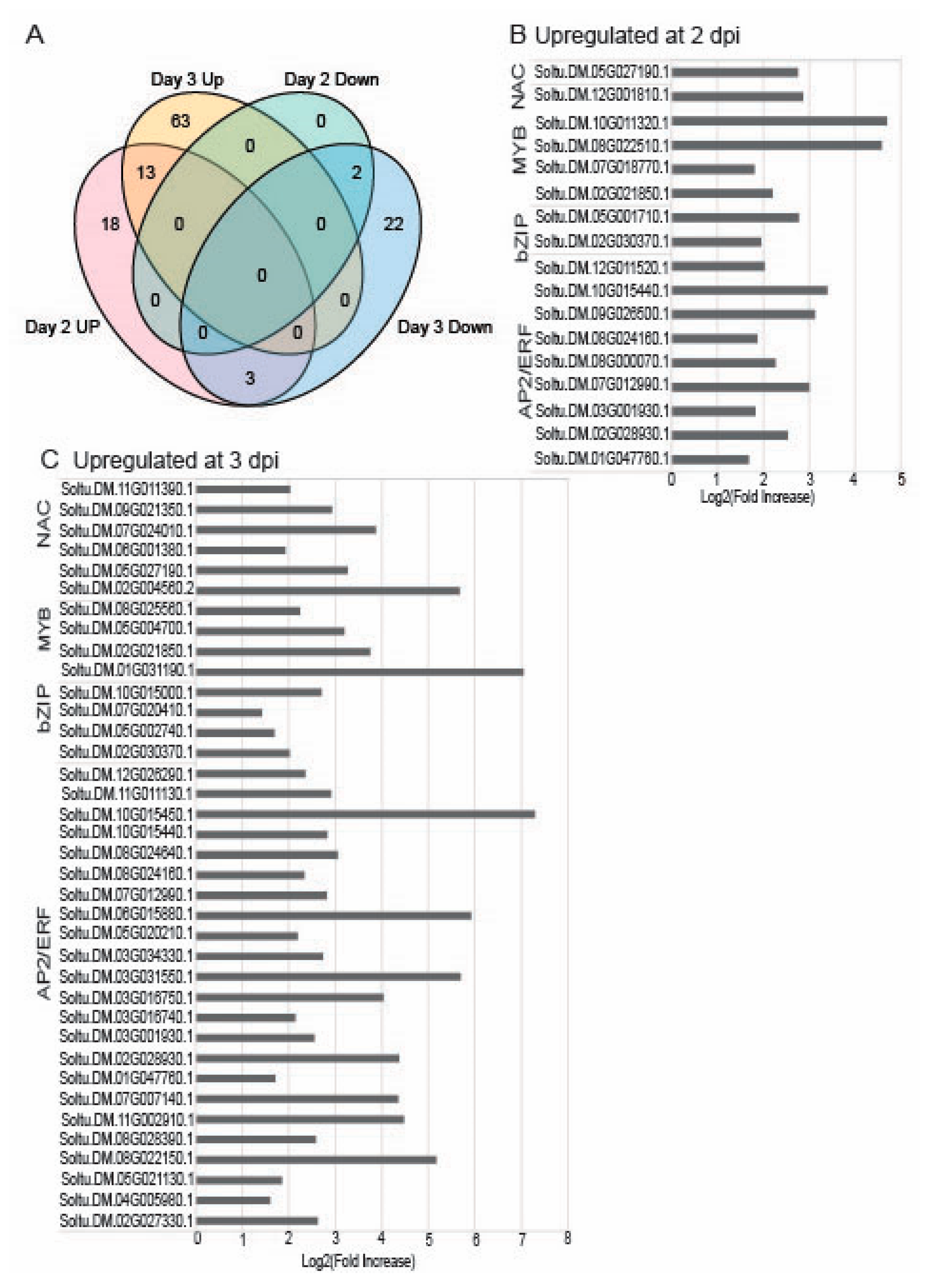
2.3. Differentially Regulated Transcription Factors (TFs) at 2 and 3 dpi
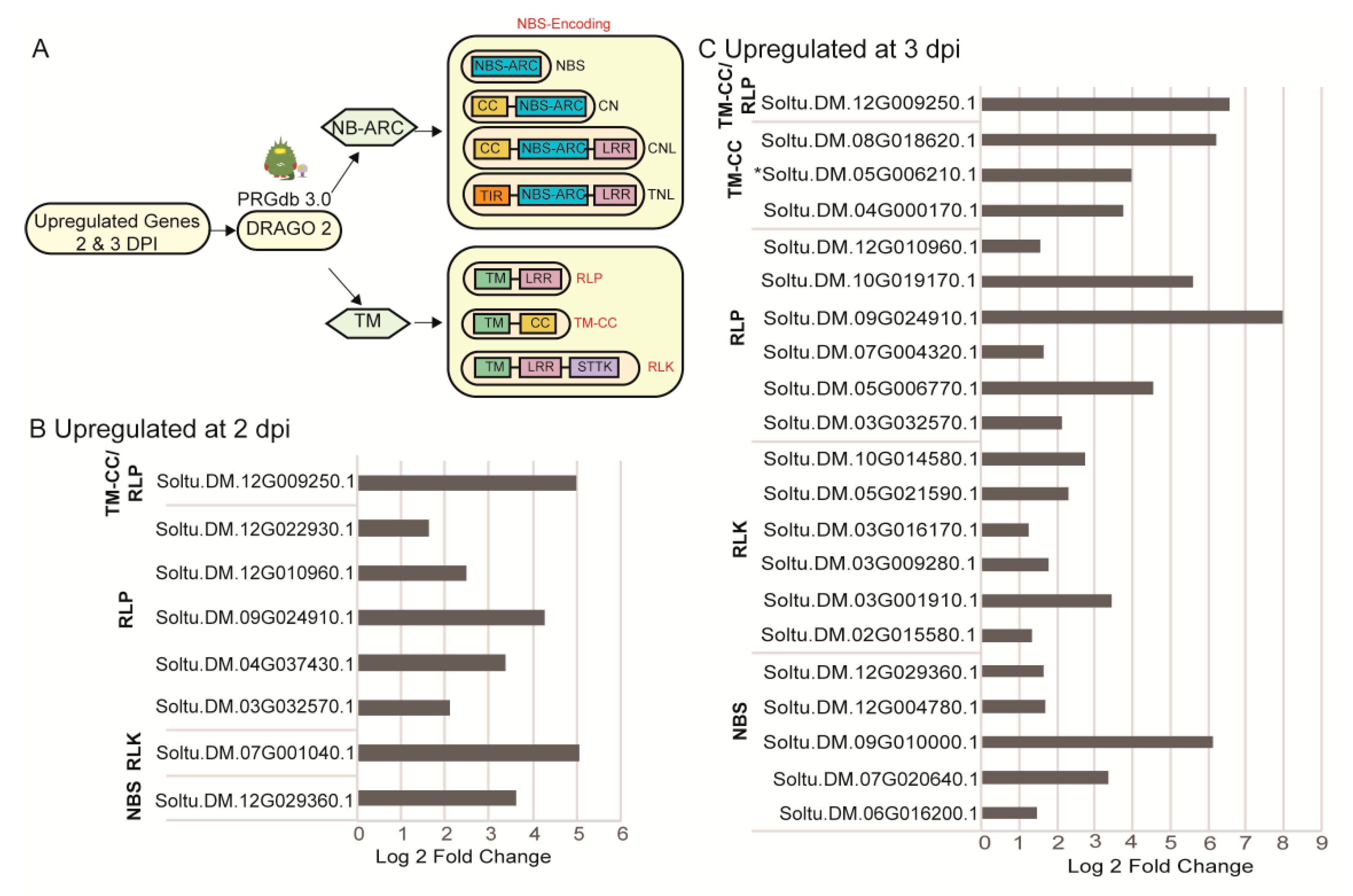
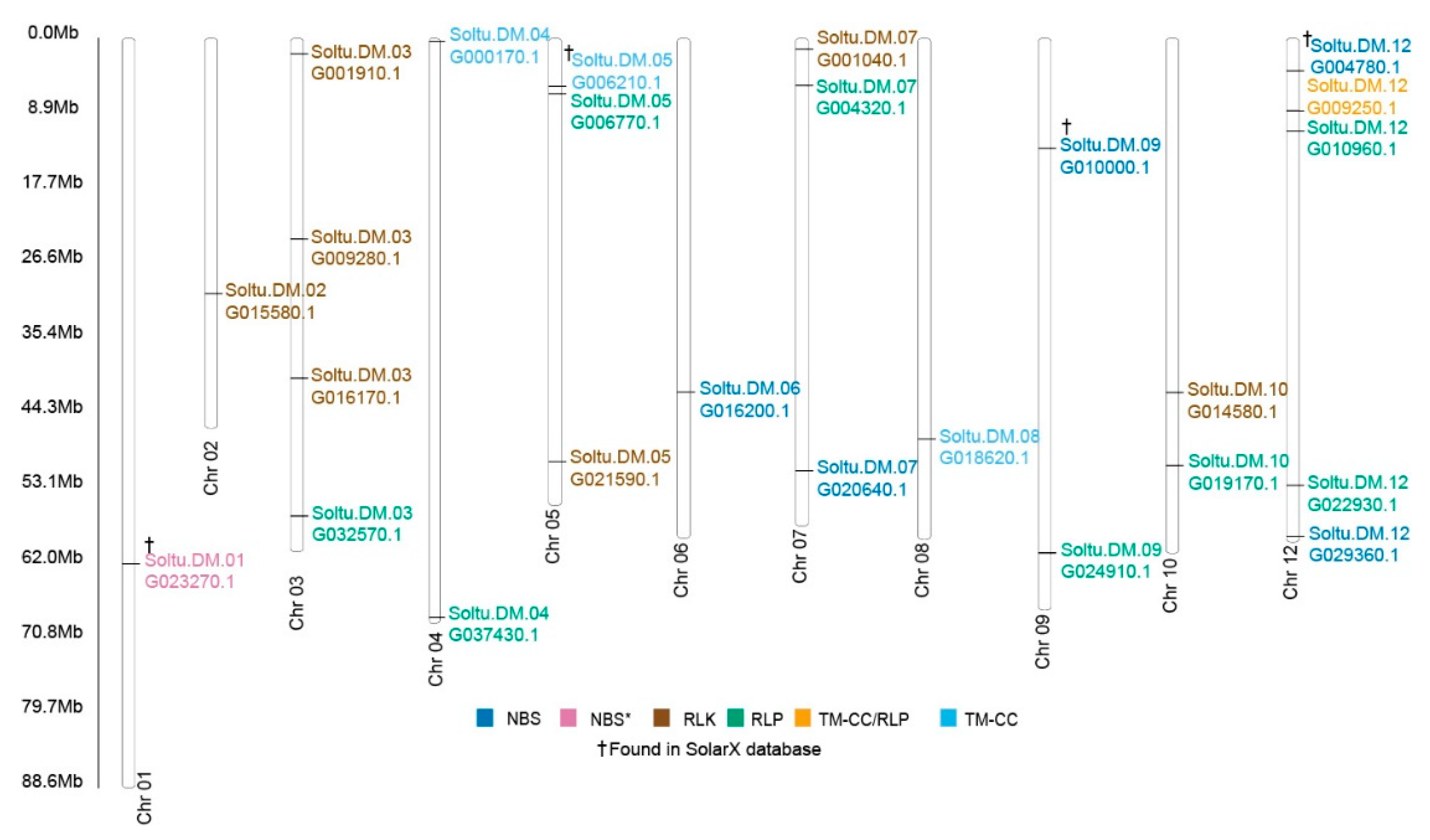
2.4. Identification of Upregulated Pathogen Resistance Gene Analogs (RGA)
2.5. DEGs Associated with the Unfolded Protein Response (UPR) that Are Common between Potato and Arabidopsis
3. Discussion
4. Materials and Methods
4.1. PVX Inoculation of Potato Leaves
4.2. RNA Extraction, cDNA Library Construction, Filtering Sequencing Reads
4.3. Genome Mapping and Gene Expression Analysis
4.4. Gene Ontology (GO)Enrichment Analysis
4.5. Functional Annotation of Transcription Factors and Resistance Gene Analogs (RGAs)
4.6. Comparative Analysis of PVX Induced DEGs in Potato and Published Tunicamycin Induced DEGs in Arabidopsis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spillane, C.; Verchot, J.; Kavanagh, T.A.; Baulcombe, D.C. Concurrent suppression of virus replication and rescue of movement-defective virus in transgenic plants expressing the coat protein of potato virus X. Virology 1997, 236. [Google Scholar] [CrossRef]
- Heppler, M.; Verchot-Lubicz, J. Cell-to-cell movement of potato virus X proteins in different plant species. Microsc. Microanal. 2004, 10. [Google Scholar] [CrossRef]
- Howard, A.R.; Heppler, M.L.; Ju, H.-J.; Krishnamurthy, K.; Payton, M.E.; Verchot-Lubicz, J. Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 2004, 328. [Google Scholar] [CrossRef]
- Heinlein, M. Plasmodesmata: Channels for viruses on the move. Methods Mol. Biol. 2015, 1217, 25–52. [Google Scholar] [CrossRef] [PubMed]
- Mekuria, T.; Bamunusinghe, D.; Payton, M.; Verchot-Lubicz, J. Phloem unloading of potato virus X movement protein is regulated by virus and host factors. Mol. Plant Microbe Interact. 2008, 21, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Oparka, K.J. Missing links?—The connection between replication and movement of plant RNA viruses. Curr. Opin. Virol. 2012, 2, 699–705. [Google Scholar] [CrossRef]
- Huang, X.; Stein, B.D.; Cheng, H.; Malyutin, A.; Tsvetkova, I.B.; Baxter, D.V.; Remmes, N.B.; Verchot, J.; Kao, C.; Bronstein, L.M.; et al. Magnetic virus-like nanoparticles in N. benthamiana plants: A new paradigm for environmental and agronomic biotechnological research. ACS Nano 2011, 5. [Google Scholar] [CrossRef]
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef]
- Tilsner, J.; Linnik, O.; Louveaux, M.; Roberts, I.M.; Chapman, S.N.; Oparka, K.J. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 2013, 201, 981–995. [Google Scholar] [CrossRef]
- Sasvari, Z.; Alatriste Gonzalez, P.; Nagy, P.D. Tombusvirus-yeast interactions identify conserved cell-intrinsic viral restriction factors. Front. Plant Sci. 2014, 5, 383. [Google Scholar] [CrossRef]
- Verchot, J. Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front. Plant Sci. 2012, 3, 275. [Google Scholar] [CrossRef]
- Verchot, J. Wrapping membranes around plant virus infection. Curr. Opin. Virol. 2011, 1. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Meier, N.; Hatch, C.; Nagalakshmi, U.; Dinesh-Kumar, S.P. Perspectives on intracellular perception of plant viruses. Mol. Plant Pathol. 2019, 20, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Gayral, M.; Arias Gaguancela, O.; Vasquez, E.; Herath, V.; Flores, F.J.; Dickman, M.B.; Verchot, J. Multiple ER-to-nucleus stress signaling pathways are activated during plantago asiatica mosaic virus and turnip mosaic virus infection in Arabidopsis thaliana. Plant J. 2020, 103, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Fahlgren, N.; Abbasi, A.; Berry, J.C.; Carrington, J.C. Antiviral ARGONAUTEs against turnip crinkle virus revealed by image-based trait analysis. Plant Physiol. 2019, 180, 1418–1435. [Google Scholar] [CrossRef] [PubMed]
- Gaguancela, O.; Zúñiga, L.P.; Arias, A.V.; Halterman, D.; Flores, F.J.; Johansen, I.E.; Wang, A.; Yamaji, Y.; Verchot, J. The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in Arabidopsis and Nicotiana benthamiana plants. Mol. Plant Microbe Interact. 2016, 29, 750–766. [Google Scholar] [CrossRef]
- Shukla, A.; López-González, S.; Hoffmann, G.; Hafrén, A. Diverse plant viruses: A toolbox for dissection of cellular pathways. J. Exp. Bot. 2019, 70, 3029–3034. [Google Scholar] [CrossRef]
- Hardigan, M.A.; Laimbeer, F.P.E.; Newton, L.; Crisovan, E.; Hamilton, J.P.; Vaillancourt, B.; Wiegert-Rininger, K.; Wood, J.C.; Douches, D.S.; Farré, E.M.; et al. Genome diversity of tuber-bearing solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc. Natl. Acad. Sci. USA 2017, 114, E9999–E10008. [Google Scholar] [CrossRef]
- Friedman, M. Potato glycoalkaloids and metabolites: Roles in the plant and in the diet. J. Agric. Food Chem. 2006, 54, 8655–8681. [Google Scholar] [CrossRef]
- Moyo, L.; Ramesh, S.V.; Kappagantu, M.; Mitter, N.; Sathuvalli, V.; Pappu, H.R. The effects of potato virus Y-derived virus small interfering RNAs of three biologically distinct strains on potato (Solanum tuberosum) transcriptome. Virol. J. 2017, 14, 1–17. [Google Scholar] [CrossRef]
- Stare, T.; Stare, K.; Weckwerth, W.; Wienkoop, S.; Gruden, K. Comparison between proteome and transcriptome response in potato (Solanum tuberosum L.) leaves following potato virus Y (PVY) infection. Proteomes 2017, 5, 14. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.; Ding, B.; Fei, Z. Comprehensive transcriptome analyses reveal that potato spindle tuber viroid triggers genome-wide changes in alternative splicing, inducible trans-acting activity of phased secondary small interfering RNAs, and immune responses. J. Virol. 2017, 91, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lukan, T.; Pompe-Novak, M.; Baebler, Š.; Tušek-Žnidarič, M.; Kladnik, A.; Križnik, M.; Blejec, A.; Zagorščak, M.; Stare, K.; Dušak, B.; et al. Precision transcriptomics of viral foci reveals the spatial regulation of immune-signaling genes and identifies RBOHD as an important player in the incompatible interaction between potato virus Y and potato. Plant J. 2020, 104, 645–661. [Google Scholar] [CrossRef] [PubMed]
- Pham, G.M.; Hamilton, J.P.; Wood, J.C.; Burke, J.T.; Zhao, H.; Vaillancourt, B.; Ou, S.; Jiang, J.; Buell, C.R. Construction of a chromosome-scale long-read reference genome assembly for potato. Gigascience 2020, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.-B.G. Making a Virus Visible: Francis, O. Holmes and a biological assay for tobacco mosaic virus. J. Hist. Biol. 2014, 47, 107–145. [Google Scholar] [CrossRef] [PubMed]
- Herath, V.; Verchot, J. Insight into the bzip gene family in Solanum tuberosum: Genome and transcriptome analysis to understand the roles of gene diversification in spatiotemporal gene expression and function. Int. J. Mol. Sci. 2021, 22, 253. [Google Scholar] [CrossRef]
- Rodrigo, G.; Zwart, M.P.; Elena, S.F. Onset of virus systemic infection in plants is determined by speed of cell-to-cell movement and number of primary infection foci. J. R. Soc. Interface 2014, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Rushton, P.J.; Bokowiec, M.T.; Han, S.; Zhang, H.; Brannock, J.F.; Chen, X.; Laudeman, T.W.; Timko, M.P. Tobacco transcription factors: Novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 2008, 147, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.; Kitazumi, A.; Cheung, C.Y.M.; Lakshmanan, M.; de los Reyes, B.G.; Jang, I.-C.; Lee, D.-Y. Identification of candidate network hubs involved in metabolic adjustments of rice under drought stress by integrating transcriptome data and genome-scale metabolic network. Plant Sci. 2016, 242, 224–239. [Google Scholar] [CrossRef]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef]
- Jha, P.; Kumar, V. BABY BOOM (BBM): A candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 2018. [Google Scholar] [CrossRef]
- Herath, V. Small family, big impact: In silico analysis of DREB2 transcription factor family in rice. Comput. Biol. Chem. 2016, 65, 128–139. [Google Scholar] [CrossRef]
- Herath, V.; Gayral, M.; Adhikari, N.; Miller, R.; Verchot, J. Genome-wide identification and characterization of Solanum tuberosum BiP genes reveal the role of the promoter architecture in BiP gene diversity. Sci. Rep. 2020, 10, 11327. [Google Scholar] [CrossRef]
- Yun, K.Y.; Park, M.R.; Mohanty, B.; Herath, V.; Xu, F.; Mauleon, R.; Wijaya, E.; Bajic, V.B.; Bruskiewich, R.; de los Reyes, B.G. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Jindrich, K.; Degnan, B.M. The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity Genome evolution and evolutionary systems biology. BMC Evol. Biol. 2016, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Benn, G.; Wang, C.Q.; Hicks, D.R.; Stein, J.; Guthrie, C.; Dehesh, K. A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J. 2014, 80, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Peng, J. Gibberellin and jasmonate crosstalk during stamen development. J. Integr. Plant Biol. 2009, 51, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Magyar, Z.; Bögre, L.; Ito, M. DREAMs make plant cells to cycle or to become quiescent. Curr. Opin. Plant Biol. 2016, 34, 100–106. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Collinge, M.; Boller, T. Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol. Biol. 2001, 46, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Jeong, R.D.; Chandra-Shekara, A.C.; Kachroo, A.; Klessig, D.F.; Kachroo, P. HRT-mediated hypersensitive response and resistance to turnip crinkle virus in Arabidopsis does not require the function of TIP, the presumed guardee protein. Mol. Plant Microbe Interact. 2008, 21, 1316–1324. [Google Scholar] [CrossRef]
- Carr, J.P.; Murphy, A.M.; Tungadi, T.; Yoon, J.Y. Plant defense signals: Players and pawns in plant-virus-vector interactions. Plant Sci. 2019, 279, 87–95. [Google Scholar] [CrossRef]
- Sekhwal, M.; Li, P.; Lam, I.; Wang, X.; Cloutier, S.; You, F. Disease resistance gene analogs (RGAs) in plants. Int. J. Mol. Sci. 2015, 16, 19248–19290. [Google Scholar] [CrossRef]
- Kourelis, J.; Kamoun, S. RefPlantNLR: A comprehensive collection of experimentally validated plant NLRs. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lee, W.-S.; Fu, S.-F.; Verchot-Lubicz, J.; Carr, J.P. Genetic modification of alternative respiration in Nicotiana benthamiana affects basal and salicylic acid-induced resistance to potato virus X. BMC Plant Biol. 2011, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.M.; Chivasa, S.; Singh, D.P.; Carr, J.P. Salicylic acid-induced resistance to viruses and other pathogens: A parting of the ways? Trends Plant Sci. 1999, 4, 155–160. [Google Scholar] [CrossRef]
- Brugmans, B.; Wouters, D.; Van Os, H.; Hutten, R.; Van Der Linden, G.; Visser, R.G.F.; Van Eck, H.J.; Van Der Vossen, E.A.G. Genetic mapping and transcription analyses of resistance gene loci in potato using NBS profiling. Theor. Appl. Genet. 2008, 117, 1379–1388. [Google Scholar] [CrossRef][Green Version]
- Louis, B.; Rey, C. Resistance gene analogs involved in tolerant cassava—Geminivirus interaction that shows a recovery phenotype. Virus Genes 2015, 51, 393–407. [Google Scholar] [CrossRef]
- Mushtaq, R.; Shahzad, K.; Mansoor, S.; Shah, Z.H.; Alsamadany, H.; Mujtaba, T.; Al-Zahrani, Y.; Alzahrani, H.A.S.; Ahmed, Z.; Bashir, A. Exploration of cotton leaf curl virus resistance genes and their screening in Gossypium arboreum by targeting resistance gene analogues. AoB Plants 2018, 10, 1–15. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, Z.; Jiao, F.; Yu, H.; Xiao, B.; Li, Y.; Lu, X. Cloning, structural features, and expression analysis of resistance gene analogs in tobacco. Mol. Biol. Rep. 2010, 37, 345–354. [Google Scholar] [CrossRef]
- Osuna-Cruz, C.M.; Paytuvi-Gallart, A.; Di Donato, A.; Sundesha, V.; Andolfo, G.; Cigliano, R.A.; Sanseverino, W.; Ercolano, M.R. PRGdb 3.0: A comprehensive platform for prediction and analysis of plant disease resistance genes. Nucleic Acids Res. 2018, 46, D1197–D1201. [Google Scholar] [CrossRef]
- Li, P.; Quan, X.; Jia, G.; Xiao, J.; Cloutier, S.; You, F.M. RGAugury: A pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genom. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Ye, C.; Dickman, M.B.; Whitham, S.A.; Payton, M.; Verchot, J. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 2011, 156, 741–755. [Google Scholar] [CrossRef]
- Song, Z.-T.; Sun, L.; Lu, S.-J.; Tian, Y.; Ding, Y.; Liu, J.-X. Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc. Natl. Acad. Sci. USA 2015, 112, 2900–2905. [Google Scholar] [CrossRef]
- Ozgur, R.; Uzilday, B.; Sekmen, A.H.; Turkan, I. The effects of induced production of reactive oxygen species in organelles on endoplasmic reticulum stress and on the unfolded protein response in arabidopsis. Ann. Bot. 2015. [Google Scholar] [CrossRef]
- Weis, C.; Pfeilmeier, S.; Glawischnig, E.; Isono, E.; Pachl, F.; Hahne, H.; Kuster, B.; Eichmann, R.; Hückelhoven, R. Co-immunoprecipitation-based identification of putative BAX INHIBITOR-1-interacting proteins involved in cell death regulation and plant-powdery mildew interactions. Mol. Plant Pathol. 2013, 14, 791–802. [Google Scholar] [CrossRef]
- Vitale, M.; Bakunts, A.; Orsi, A.; Lari, F.; Tadé, L.; Danieli, A.; Rato, C.; Valetti, C.; Sitia, R.; Raimondi, A.; et al. Inadequate BiP availability defines endoplasmic reticulum stress. Elife 2019, 8, 1–17. [Google Scholar] [CrossRef]
- Dorokhov, Y.L.; Sheshukova, E.V.; Komarova, T.V. Methanol in plant life. Front. Plant Sci. 2018, 871, 1–6. [Google Scholar] [CrossRef]
- Kogovšek, P.; Pompe-Novak, M.; Baebler, S.; Rotter, A.; Gow, L.; Gruden, K.; Foster, G.D.; Boonham, N.; Ravnikar, M. Aggressive and mild potato virus Y isolates trigger different specific responses in susceptible potato plants. Plant Pathol. 2010, 59, 1121–1132. [Google Scholar] [CrossRef]
- Park, M.-R.; Jeong, R.-D.; Kim, K.-H. Understanding the intracellular trafficking and intercellular transport of potexviruses in their host plants. Front. Plant Sci. 2014, 5, 60. [Google Scholar] [CrossRef]
- Kanneganti, V.; Gupta, A.K. Wall associated kinases from plants—An overview. Physiol. Mol. Biol. Plants 2008, 14, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Bernat-Silvestre, C.; Vieira, V.D.S.; Sanchez-Simarro, J.; Pastor-Cantizano, N.; Hawes, C.; Marcote, M.J.; Aniento, F. P24 Family proteins are involved in transport to the plasma membrane of gpi-anchored proteins in plants. Plant Physiol. 2020, 184, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, J.C.; Langhans, M.; Sturm, S.; Hillmer, S.; Aniento, F.; Robinson, D.G.; Marcote, M.J. Putative p24 complexes in Arabidopsis contain members of the delta and beta subfamilies and. J. Exp. Bot. 2013, 64, 3147–3167. [Google Scholar] [CrossRef]
- Chen, J.; Qi, X.; Zheng, H. Subclass-specific localization and trafficking of Arabidopsis p24 proteins in the ER-Golgi interface. Traffic 2012, 13, 400–415. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zhang, X.; Sun, S.; Wang, X. Factors affecting the accumulation of curcumin in microrhizomes of Curcuma aromatica Salisb. Biomed. Res. Int. 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Chen, Q.; Tian, Z.; Jiang, R.; Zheng, X.; Xie, C.; Liu, J. StPOTHR1, a NDR1/HIN1-like gene in Solanum tuberosum, enhances resistance against Phytophthora infestans. Biochem. Biophys. Res. Commun. 2018, 496, 1155–1161. [Google Scholar] [CrossRef]
- Trivedi, D.K.; Ansari, M.W.; Tuteja, N. Multiple abiotic stress responsive rice cyclophilin: (OsCYP-25) mediates a wide range of cellular responses. Commun. Integr. Biol. 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.Q.; Sun, Y.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Seebeck, T.; Schrenker, D.; Yu, O. CYP709B3, a cytochrome P450 monooxygenase gene involved in salt tolerance in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 1–13. [Google Scholar] [CrossRef]
- Poku, S.A.; Chukwurah, P.N.; Aung, H.H.; Nakamura, I. Over-expression of a melon Y3SK2-type LEA gene confers drought and salt tolerance in transgenic tobacco plants. Plants 2020, 9, 1749. [Google Scholar] [CrossRef]
- Barah, P.; Naika, B.N.M.; Jayavelu, N.D.; Sowdhamini, R.; Shameer, K.; Bones, A.M. Transcriptional regulatory networks in Arabidopsis thaliana during single and combined stresses. Nucleic Acids Res. 2015, 44, 3147–3164. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Baebler, Š.; Krečič-Stres, H.; Rotter, A.; KogovŠek, P.; Cankar, K.; Kok, E.J.; Gruden, K.; KovaČ, M.; Žel, J.; Pompe-Novak, M.; et al. PVYNTN elicits a diverse gene expression response in different potato genotypes in the first 12 h after inoculation. Mol. Plant Pathol. 2009, 10, 263–275. [Google Scholar] [CrossRef]
- Stare, T.; Ramšak, Ž.; Križnik, M.; Gruden, K. Multiomics analysis of tolerant interaction of potato with potato virus Y. Sci. Data 2019, 6, 1–11. [Google Scholar] [CrossRef]
- Whitham, S.A.; Quan, S.; Chang, H.S.; Cooper, B.; Estes, B.; Zhu, T.; Wang, X.; Hou, Y.M. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 2003, 33, 271–283. [Google Scholar] [CrossRef]
- Di Carli, M.; Benvenuto, E.; Donini, M. Recent insights into plant-virus interactions through proteomic analysis. J. Proteome Res. 2012, 11, 4765–4780. [Google Scholar] [CrossRef]
- Lee, W.S.; Fu, S.F.; Li, Z.; Murphy, A.M.; Dobson, E.A.; Garland, L.; Chaluvadi, S.R.; Lewsey, M.G.; Nelson, R.S.; Carr, J.P. Salicylic acid treatment and expression of an RNA-dependent RNA polymerase 1 transgene inhibit lethal symptoms and meristem invasion during tobacco mosaic virus infection in Nicotiana benthamiana. BMC Plant Biol. 2016. [Google Scholar] [CrossRef]
- Zhang, Y.; Pei, X.; Zhang, C.; Lu, Z.; Wang, Z.; Jia, S.; Li, W. De novo foliar transcriptome of Chenopodium amaranticolor and analysis of its gene expression during virus-induced hypersensitive response. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef][Green Version]
- Allie, F.; Pierce, E.J.; Okoniewski, M.J.; Rey, C. Transcriptional analysis of South African cassava mosaic virus-infected susceptible and tolerant landraces of cassava highlights differences in resistance, basal defense and cell wall associated genes during infection. BMC Genom. 2014, 15, 1006. [Google Scholar] [CrossRef]
- Takken, F.L.W.; Goverse, A. How to build a pathogen detector: Structural basis of NB-LRR function. Curr. Opin. Plant Biol. 2012, 15, 375–384. [Google Scholar] [CrossRef]
- Howell, S.H. When is the unfolded protein response not the unfolded protein response? Plant Sci. 2017, 260, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Afrin, T.; Diwan, D.; Sahawneh, K.; Pajerowska-Mukhtar, K. Multilevel regulation of endoplasmic reticulum stress responses in plants: Where old roads and new paths meet. J. Exp. Bot. 2020, 71, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, R.; Takahashi, M.; Suzuki, N. Coordination between bZIP28 and HSFA2 in the regulation of heat response signals in Arabidopsis. Plant Signal. Behav. 2017, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.Y.; Chen, M.X.; Ye, N.H.; Qiao, W.M.; Gao, B.; Law, W.K.; Tian, Y.; Zhang, D.; Zhang, D.; Liu, T.Y.; et al. Comparative performance of the BGISEQ-500 and Illumina HiSeq4000 sequencing platforms for transcriptome analysis in plants. Plant Methods 2018, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015. [Google Scholar] [CrossRef]
- Etherington, G.J.; Ramirez-Gonzalez, R.H.; MacLean, D. Bio-samtools 2: A package for analysis and visualization of sequence and alignment data with SAMtools in Ruby. Bioinformatics 2015. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Haynes, W. Benjamini—Hochberg Method. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; p. 78. ISBN 978-1-4419-9863-7. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools—An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2020, 49, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Törönen, P.; Medlar, A.; Holm, L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef] [PubMed]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herath, V.; Verchot, J. Transcriptional Regulatory Networks Associate with Early Stages of Potato Virus X Infection of Solanum tuberosum. Int. J. Mol. Sci. 2021, 22, 2837. https://doi.org/10.3390/ijms22062837
Herath V, Verchot J. Transcriptional Regulatory Networks Associate with Early Stages of Potato Virus X Infection of Solanum tuberosum. International Journal of Molecular Sciences. 2021; 22(6):2837. https://doi.org/10.3390/ijms22062837
Chicago/Turabian StyleHerath, Venura, and Jeanmarie Verchot. 2021. "Transcriptional Regulatory Networks Associate with Early Stages of Potato Virus X Infection of Solanum tuberosum" International Journal of Molecular Sciences 22, no. 6: 2837. https://doi.org/10.3390/ijms22062837
APA StyleHerath, V., & Verchot, J. (2021). Transcriptional Regulatory Networks Associate with Early Stages of Potato Virus X Infection of Solanum tuberosum. International Journal of Molecular Sciences, 22(6), 2837. https://doi.org/10.3390/ijms22062837







