Light Quality and Intensity Modulate Cold Acclimation in Arabidopsis
Abstract
1. Introduction
2. Results
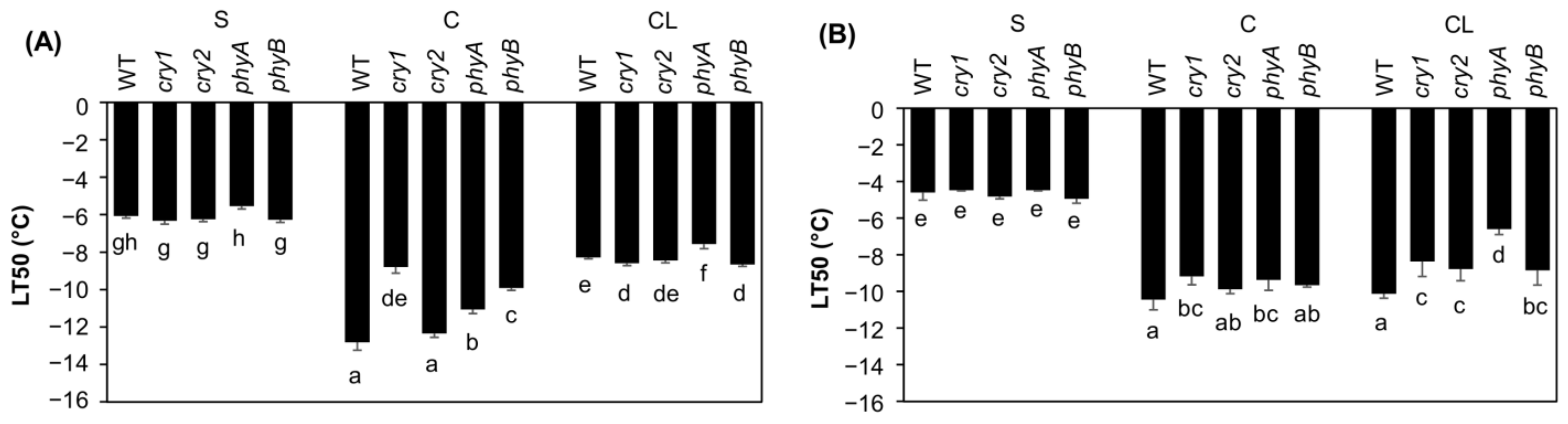
2.1. Freezing Tolerance at Optimal and Low Light Intensity
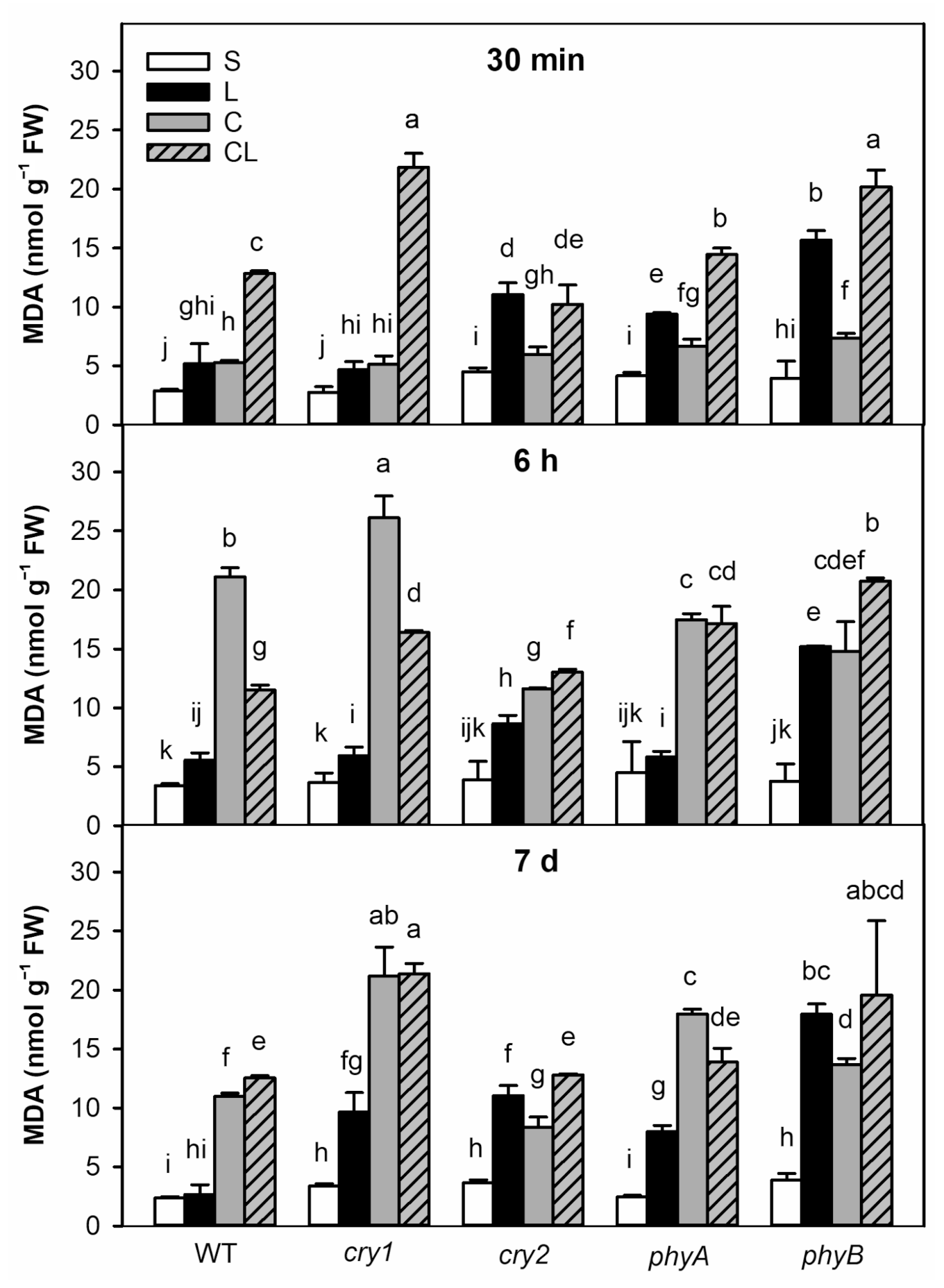
2.2. Lipid Peroxidation
2.3. Effects of Photoreceptor Mutations on Photosynthetic Parameters
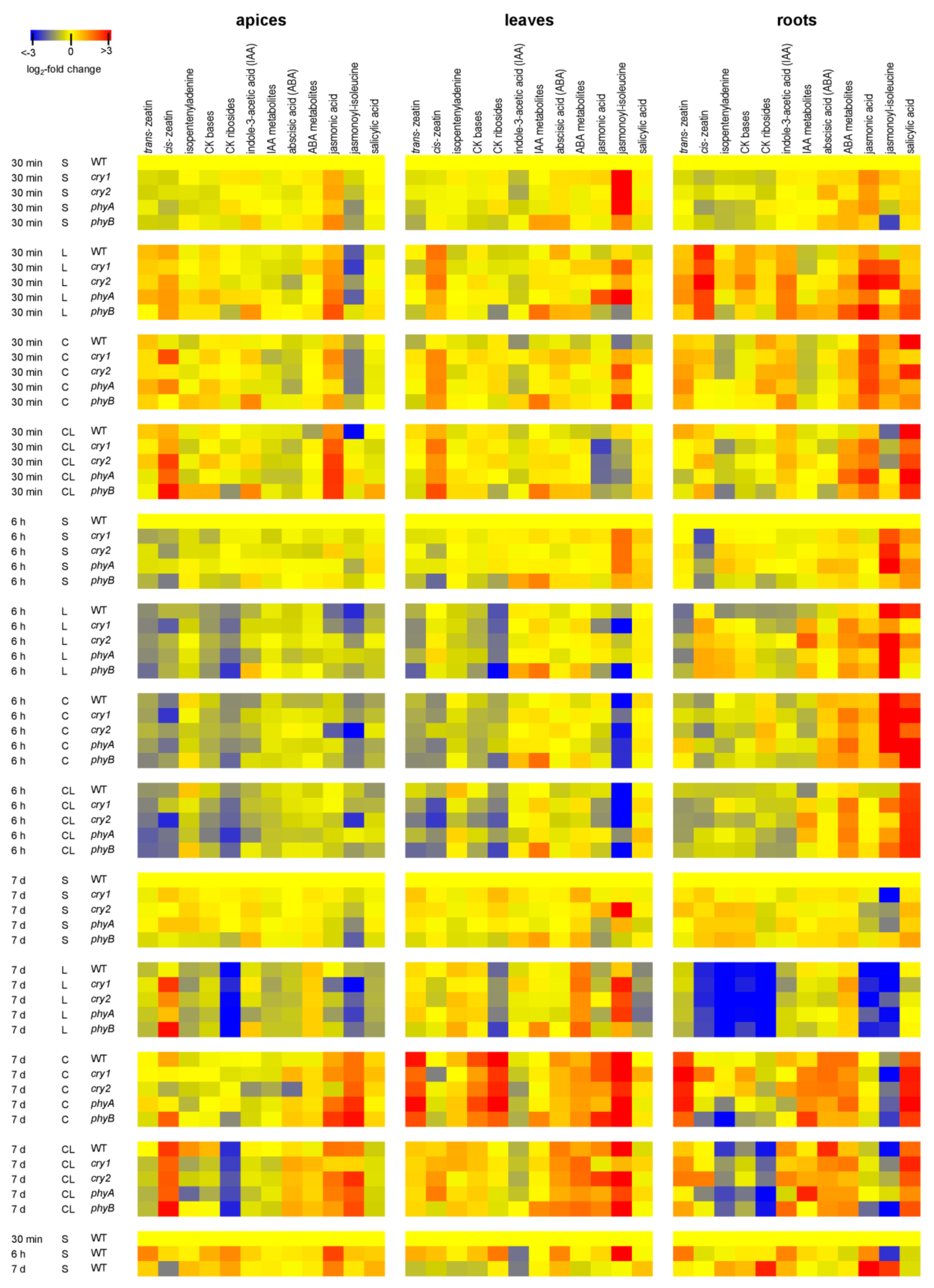
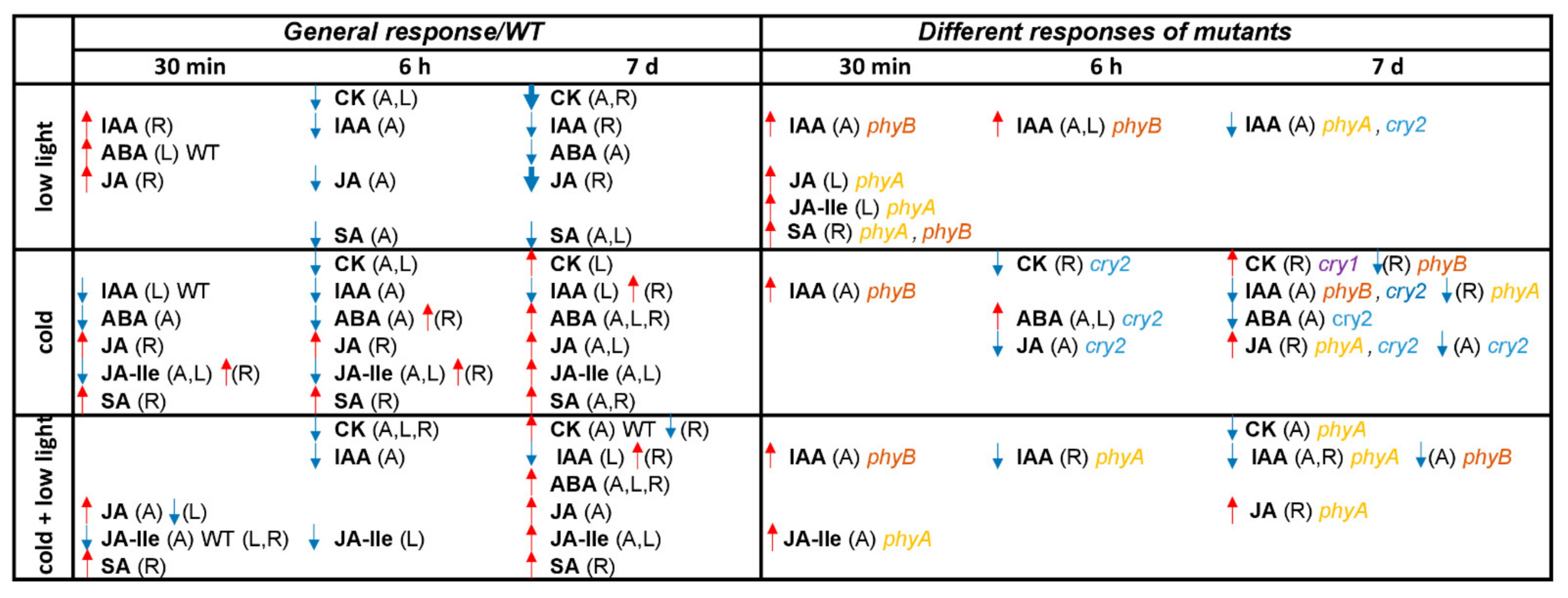
2.4. Effects of Cold and Low Light Intensity on Hormonal Pools in WT Plants and Photoreceptor Mutants
2.4.1. Cytokinins
2.4.2. Auxin
2.4.3. Abscisic Acid
2.4.4. Jasmonates
2.4.5. Salicylic Acid
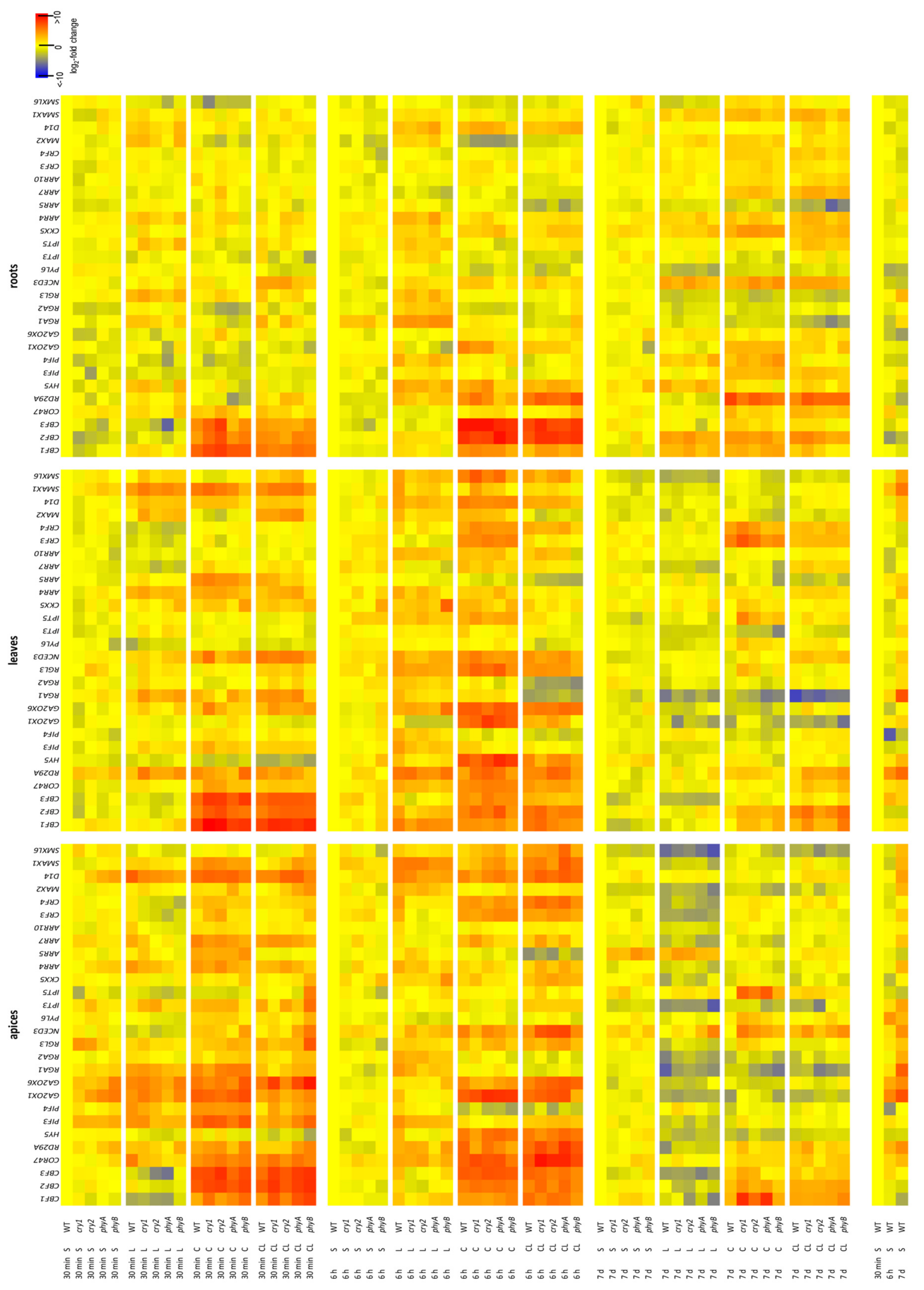
2.5. Transcription of Stress-Related Genes
2.5.1. CBFs
2.5.2. Other Stress-Inducible Genes
2.5.3. Hormone-Related Genes
3. Discussion
3.1. Low Light Intensity
3.2. Cold at Optimal Light Intensity
3.3. Cold at Low Light Intensity
3.4. Conclusion
4. Materials and Methods
4.1. Plant Material and Experimental Conditions
4.2. Freezing Tolerance
4.3. Lipid Peroxidation
4.4. Chlorophyll Fluorescence Measurement
4.5. Phytohormone Analysis
4.6. Gene Expression Quantification
4.7. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lau, O.S.; Deng, X.W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 2010, 13, 571–577. [Google Scholar] [CrossRef]
- Franklin, K.A. Light and temperature signal crosstalk in plant development. Curr. Opin. Plant Biol. 2009, 12, 63–68. [Google Scholar] [CrossRef]
- Huner, N.P.A.; Öquistm, G.; Sarhan, F. Energy balance and acclimation to light and cold. Trends Plant Sci. 1998, 3, 224–230. [Google Scholar] [CrossRef]
- Janda, T.; Majlath, I.; Szalai, G. Interaction of temperature and light in the development of freezing tolerance in plants. J. Plant Growth Regul. 2014, 33, 460–469. [Google Scholar] [CrossRef]
- Szalai, G.; Pap, M.; Janda, T. Light-induced frost tolerance differs in winter and spring wheat plants. J. Plant Physiol. 2009, 166, 1826–1831. [Google Scholar] [CrossRef]
- Ballaré, C.L.; Pierik, R. The shade-avoidance syndrome: Multiple signals and ecological consequences. Plant Cell Environ. 2017, 40, 2530–2543. [Google Scholar] [CrossRef]
- Debrieux, D.; Fankhauser, C. Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol. Biol. 2010, 73, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Shinomura, T.; Nagatani, A.; Hanzawa, H.; Kubota, M.; Watanabe, M.; Furuya, M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1996, 93, 8129–8133. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010, 61, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, L.; Chen, X.; Wu, X.; Xiang, X.; Zhou, J.; Xia, X.; Shi, K.; Yu, J.; Foyer, C.H.; et al. SlHY5 integrates temperature, light, and hormone signaling to balance plant growth and cold tolerance. Plant Physiol. 2019, 179, 749–760. [Google Scholar] [CrossRef]
- Wang, F.; Guo, Z.; Li, H.; Wang, M.; Onac, E.; Zhou, J.; Xia, X.; Shi, K.; Yu, J.; Zhou, Y. Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol. 2016, 170, 459–471. [Google Scholar] [CrossRef]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, J.; Gong, Z.; Thomashow, M.F.; Yang, S. Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Sharabi-Schwager, M.; Samach, A.; Porat, R. Overexpression of the CBF2 transcriptional activator in Arabidopsis counteracts hormone activation of leaf senescence. Plant Signal. Behav. 2010, 5, 296–299. [Google Scholar] [CrossRef]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef]
- Li, J.; Li, G.; Gao, S.; Martinez, C.; He, G.; Zhou, Z.; Huang, X.; Lee, J.H.; Zhang, H.; Shen, Y.; et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 2010, 22, 3634–3649. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of CBF expression–c-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Pluhařová, K.; Leontovyčová, H.; Stoudková, V.; Pospíchalová, R.; Maršík, P.; Klouček, P.; Starodubtseva, A.; Iakovenko, O.; Krčková, Z.; Valentová, O.; et al. “Salicylic Acid Mutant Collection” as a tool to explore the role of salicylic acid in regulation of plant growth under a changing environment. Int. J. Mol. Sci. 2019, 20, 6365. [Google Scholar] [CrossRef] [PubMed]
- Pál, M.; Janda, T.; Majláth, I.; Szalai, G. Involvement of salicylic acid and other phenolic compounds in light-dependent cold acclimation in maize. Int. J. Mol. Sci. 2020, 21, 1942. [Google Scholar] [CrossRef]
- Tajti, J.; Hamow, K.Á.; Majláth, I.; Gierczik, K.; Németh, E.; Janda, T.; Pál, M. Polyamine-induced hormonal changes in eds5 and sid2 mutant Arabidopsis plants. Int. J. Mol. Sci. 2019, 20, 5746. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef]
- Gray, W.M.; Östin, A.; Sandberg, G.; Romano, C.P.; Estelle, M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7197–7202. [Google Scholar] [CrossRef]
- Fankhauser, C. Light perception in plants: Cytokinins and red light join forces to keep phytochrome B active. Trends Plant Sci. 2002, 7, 143–145. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novák, O.; Ku, S.J.; Cho, C.; Lee, D.J.; Lee, E.J.; Strnad, M.; et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010, 285, 23371–23386. [Google Scholar] [CrossRef]
- Kosová, K.; Prášil, I.T.; Vítámvás, P.; Dobrev, P.; Motyka, V.; Floková, K.; Novák, O.; Turečková, V.; Rolčik, J.; Pešek, B.; et al. Complex phytohormone responses during the cold acclimation of two wheat cultivars differing in cold tolerance, winter Samanta and spring Sandra. J. Plant Physiol. 2012, 169, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Prerostova, S.; Černý, M.; Dobrev, P.I.; Motyka, V.; Hluskova, L.; Zupkova, B.; Gaudinova, A.; Knirsch, V.; Janda, T.; Brzobohatý, B.; et al. Light regulates the cytokinin-dependent cold stress responses in Arabidopsis. Front. Plant Sci. 2021, 11, 608711. [Google Scholar] [CrossRef]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.V.; Leyva-González, M.A.; Osakabe, Y.; Tran, U.T.; Nishiyama, R.; Watanabe, Y.; Tanaka, M.; Seki, M.; Yamaguchi, S.; Van Dong, N.; et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA 2014, 111, 851–856. [Google Scholar] [CrossRef]
- Ballaré, C.L. Light regulation of plant defense. Annu. Rev. Plant Biol. 2014, 65, 335–363. [Google Scholar] [CrossRef]
- De Wit, M.; Spoel, S.H.; Sanchez-Perez, G.F.; Gommers, C.M.; Pieterse, C.M.; Voesenek, L.A.; Pierik, R. Perception of low red:far-red ratio compromises both salicylic acid-and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 2013, 75, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Rahman, M.A.; Song, Y.; Ji, H.C.; Choi, G.J.; Kim, K.Y.; Lee, S.H. Cold stress-induced regulation of differentially expressed genes in barley (Hordeum vulgare L.) leaves. J. Anim. Plant Sci. 2019, 29, 1673–1679. [Google Scholar]
- Franklin, K.A.; Whitelam, G.C. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 2007, 39, 1410–1413. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wu, N.; Zhang, L.; Ahammed, G.J.; Chen, X.; Xiang, X.; Zhou, J.; Xia, X.; Shi, K.; Yu, J.; et al. Light signaling-dependent regulation of photoinhibition and photoprotection in tomato. Plant Physiol. 2018, 176, 1311–1326. [Google Scholar] [CrossRef]
- Imai, H.; Kawamura, Y.; Nagatani, A.; Uemura, M. Effects of the blue light–cryptochrome system on the early process of cold acclimation of Arabidopsis thaliana. Environ. Exp. Bot. 2020, 183, 104340. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, Y.K.; Park, J.Y.; Kim, J. Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 2002, 29, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.C.; Shi, Y.T.; Zhang, X.Y.; Xin, X.Y.; Qi, L.J.; Guo, H.W.; Li, J.G.; Yang, S.H. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef]
- Lin, L.; Liu, X.; Yin, R. PIF3 integrates light and low temperature signaling. Trends Plant Sci. 2018, 23, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Hahm, J.; Kim, K.; Qiu, Y.; Chen, M. Increasing ambient temperature progressively disassembles Arabidopsis phytochrome B from individual photobodies with distinct thermostabilities. Nat. Commun. 2020, 11, 1660. [Google Scholar] [CrossRef]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T. Wheat and barley dehydrins under cold, drought and salinity - what can LEA-II proteins tell us about plant stress response? Front. Plant Sci. 2014, 5, 343. [Google Scholar] [CrossRef] [PubMed]
- Hannah, M.A.; Heyer, A.G.; Hincha, D.K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005, 1, e26. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Janda, T.; Szalai, G.; Tari, I.; Paldi, E. Hydroponic treatment with salicylic acid decreases the effects of chilling injury in maize (Zea mays L.) plants. Planta 1999, 208, 175–180. [Google Scholar] [CrossRef]
- Janda, T.; Gondor, O.K.; Yordanova, R.; Szalai, G.; Pál, M. Salicylic acid and photosynthesis: Signalling and effects. Acta Physiol. Plant. 2014, 36, 2537–2546. [Google Scholar] [CrossRef]
- Dong, C.J.; Li, L.; Shang, Q.M.; Liu, X.Y.; Zhang, Z.G. Endogenous salicylic acid accumulation is required for chilling tolerance in cucumber (Cucumis sativus L.) seedlings. Planta 2014, 240, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, S.; Gilmour, S.J.; Thomashow, M.F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant. J. 2013, 75, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Cho, C.; Lee, M.R.; Binh, N.V.; Kim, J. CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in Arabidopsis. Plant. Cell 2016, 28, 1828–1843. [Google Scholar] [CrossRef]
- Zwack, P.J.; Compton, M.A.; Adams, C.I.; Rashotte, A.M. Cytokinin response factor 4 (CRF4) is induced by cold and involved in freezing tolerance. Plant. Cell Rep. 2016, 35, 573–584. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J. Arabidopsis response regulator1 and arabidopsis histidine phosphotransfer protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant. Physiol. 2013, 161, 408–424. [Google Scholar] [CrossRef]
- Dobrá, J.; Černý, M.; Štorchová, H.; Dobrev, P.; Skalák, J.; Jedelský, P.L.; Lukšanová, H.; Gaudinová, A.; Pešek, B.; Malbeck, J.; et al. The impact of heat stress targeting on the hormonal and transcriptomic response in Arabidopsis. Plant. Sci. 2015, 231, 52–61. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, K.X.; Wang, W.S.; Gong, W.; Liu, W.C.; Chen, H.G.; Xu, H.H.; Lu, Y.T. Low temperature inhibits root growth by reducing auxin accumulation via ARR1/12. Plant. Cell Physiol. 2015, 56, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, F.; Habricot, Y.; Condiff, A.S.; Maldiney, R.; Straeten, D.V.D.; Ahmad, M. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant. J. 2007, 49, 428–441. [Google Scholar] [CrossRef]
- Van der Schoot, C.; Rinne, P.L. Dormancy cycling at the shoot apical meristem: Transitioning between self-organization and self-arrest. Plant Sci. 2011, 180, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Janda, T.; Szalai, G.; Kissimon, J.; Páldi, E.; Márton, L.; Szigeti, Z. Role of irradiance in the chilling injury of young maize plants studied by chlorophyll fluorescence induction measurements. Photosynthetica 1994, 30, 293–299. [Google Scholar]
- Prášil, I.; Zámečník, J. The use of a conductivity measurement method for assessing freezing injury. I. Influence of leakage time, segment number, size and shape in a sample on evaluation of the degree of injury. Environ. Exp. Bot. 1998, 40, 1–10. [Google Scholar] [CrossRef]
- Janáček, J.; Prášil, I. Quantification of plant frost injury by nonlinear fitting of an s-shaped function. Cryo Lett. 1991, 12, 47–52. [Google Scholar]
- Hodges, D.M.; DeLong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Landi, M. Commentary to: “Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds” by Hodges et al., Planta (1999) 207: 604–611. Planta 2017, 245, 1067. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.M.; Baker, N.R. The relationship between quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Horton, P.; Ruban, A.V. Regulation of photosystem-II. Photosynth. Res. 1992, 34, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| S | L | C | CL | |||
|---|---|---|---|---|---|---|
| Fv/Fm | 6 h | WT | 0.838 ± 0.007 | 0.853 ± 0.004 | 0.783 ± 0.005 | 0.847 ± 0.005 |
| cry1 | 0.822 ± 0.019 | 0.844 ± 0.007 | 0.802 ± 0.012 | 0.847 ± 0.007 | ||
| cry2 | 0.835 ± 0.008 | 0.850 ± 0.001 | 0.810 ± 0.008 | 0.850 ± 0.006 | ||
| phyA | 0.835 ± 0.005 | 0.856 ± 0.006 | 0.821 ± 0.016 | 0.850 ± 0.006 | ||
| phyB | 0.837 ± 0.011 | 0.859 ± 0.003 | 0.827 ± 0.005 | 0.852 ± 0.009 | ||
| 7 d | WT | 0.846 ± 0.005 | 0.799 ± 0.014 | 0.810 ± 0.001 | 0.848 ± 0.004 | |
| cry1 | 0.840 ± 0.005 | 0.809 ± 0.014 | 0.797 ± 0.005 | 0.858 ± 0.004 | ||
| cry2 | 0.849 ± 0.003 | 0.800 ± 0.007 | 0.817 ± 0.005 | 0.852 ± 0.004 | ||
| phyA | 0.841 ± 0.010 | 0.802 ± 0.009 | 0.809 ± 0.015 | 0.850 ± 0.006 | ||
| phyB | 0.851 ± 0.003 | 0.828 ± 0.009 | 0.814 ± 0.007 | 0.856 ± 0.005 | ||
| QY_Lss | 6 h | WT | 0.540 ± 0.027 | 0.544 ± 0.024 | 0.547 ± 0.017 | 0.555 ± 0.050 |
| cry1 | 0.517 ± 0.019 | 0.523 ± 0.032 | 0.554 ± 0.014 | 0.597 ± 0.007 | ||
| cry2 | 0.543 ± 0.022 | 0.548 ± 0.013 | 0.573 ± 0.014 | 0.610 ± 0.001 | ||
| phyA | 0.575 ± 0.017 | 0.565 ± 0.015 | 0.583 ± 0.023 | 0.612 ± 0.007 | ||
| phyB | 0.512 ± 0.033 | 0.561 ± 0.041 | 0.577 ± 0.012 | 0.652 ± 0.057 | ||
| 7 d | WT | 0.576 ± 0.017 | 0.405 ± 0.023 | 0.520 ± 0.008 | 0.633 ± 0.005 | |
| cry1 | 0.544 ± 0.010 | 0.381 ± 0.021 | 0.530 ± 0.008 | 0.637 ± 0.005 | ||
| cry2 | 0.558 ± 0.016 | 0.424 ± 0.052 | 0.537 ± 0.033 | 0.642 ± 0.007 | ||
| phyA | 0.577 ± 0.024 | 0.482 ± 0.013 | 0.573 ± 0.022 | 0.630 ± 0.001 | ||
| phyB | 0.589 ± 0.020 | 0.498 ± 0.035 | 0.526 ± 0.026 | 0.610 ± 0.028 | ||
| NPQ_Lss | 6 h | WT | 1.093 ± 0.071 | 1.118 ± 0.090 | 0.310 ± 0.028 | 0.807 ± 0.112 |
| cry1 | 0.950 ± 0.114 | 1.007 ± 0.120 | 0.528 ± 0.028 | 0.619 ± 0.051 | ||
| cry2 | 0.955 ± 0.101 | 1.020 ± 0.060 | 0.477 ± 0.038 | 0.630 ± 0.050 | ||
| phyA | 0.882 ± 0.070 | 1.040 ± 0.102 | 0.575 ± 0.118 | 0.653 ± 0.023 | ||
| phyB | 1.118 ± 0.117 | 1.120 ± 0.128 | 0.743 ± 0.057 | 0.618 ± 0.153 | ||
| 7 d | WT | 1.002 ± 0.056 | 1.298 ± 0.106 | 0.697 ± 0.039 | 0.687 ± 0.039 | |
| cry1 | 1.173 ± 0.144 | 1.417 ± 0.114 | 0.547 ± 0.069 | 0.663 ± 0.039 | ||
| cry2 | 1.036 ± 0.094 | 1.196 ± 0.196 | 0.684 ± 0.113 | 0.660 ± 0.026 | ||
| phyA | 0.916 ± 0.139 | 0.880 ± 0.240 | 0.653 ± 0.078 | 0.777 ± 0.086 | ||
| phyB | 0.967 ± 0.090 | 0.957 ± 0.158 | 0.641 ± 0.069 | 0.784 ± 0.102 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prerostova, S.; Dobrev, P.I.; Knirsch, V.; Jarosova, J.; Gaudinova, A.; Zupkova, B.; Prášil, I.T.; Janda, T.; Brzobohatý, B.; Skalák, J.; et al. Light Quality and Intensity Modulate Cold Acclimation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 2736. https://doi.org/10.3390/ijms22052736
Prerostova S, Dobrev PI, Knirsch V, Jarosova J, Gaudinova A, Zupkova B, Prášil IT, Janda T, Brzobohatý B, Skalák J, et al. Light Quality and Intensity Modulate Cold Acclimation in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(5):2736. https://doi.org/10.3390/ijms22052736
Chicago/Turabian StylePrerostova, Sylva, Petre I. Dobrev, Vojtech Knirsch, Jana Jarosova, Alena Gaudinova, Barbara Zupkova, Ilja T. Prášil, Tibor Janda, Břetislav Brzobohatý, Jan Skalák, and et al. 2021. "Light Quality and Intensity Modulate Cold Acclimation in Arabidopsis" International Journal of Molecular Sciences 22, no. 5: 2736. https://doi.org/10.3390/ijms22052736
APA StylePrerostova, S., Dobrev, P. I., Knirsch, V., Jarosova, J., Gaudinova, A., Zupkova, B., Prášil, I. T., Janda, T., Brzobohatý, B., Skalák, J., & Vankova, R. (2021). Light Quality and Intensity Modulate Cold Acclimation in Arabidopsis. International Journal of Molecular Sciences, 22(5), 2736. https://doi.org/10.3390/ijms22052736







