CFTR Cooperative Cis-Regulatory Elements in Intestinal Cells
Abstract
1. Introduction
2. Results
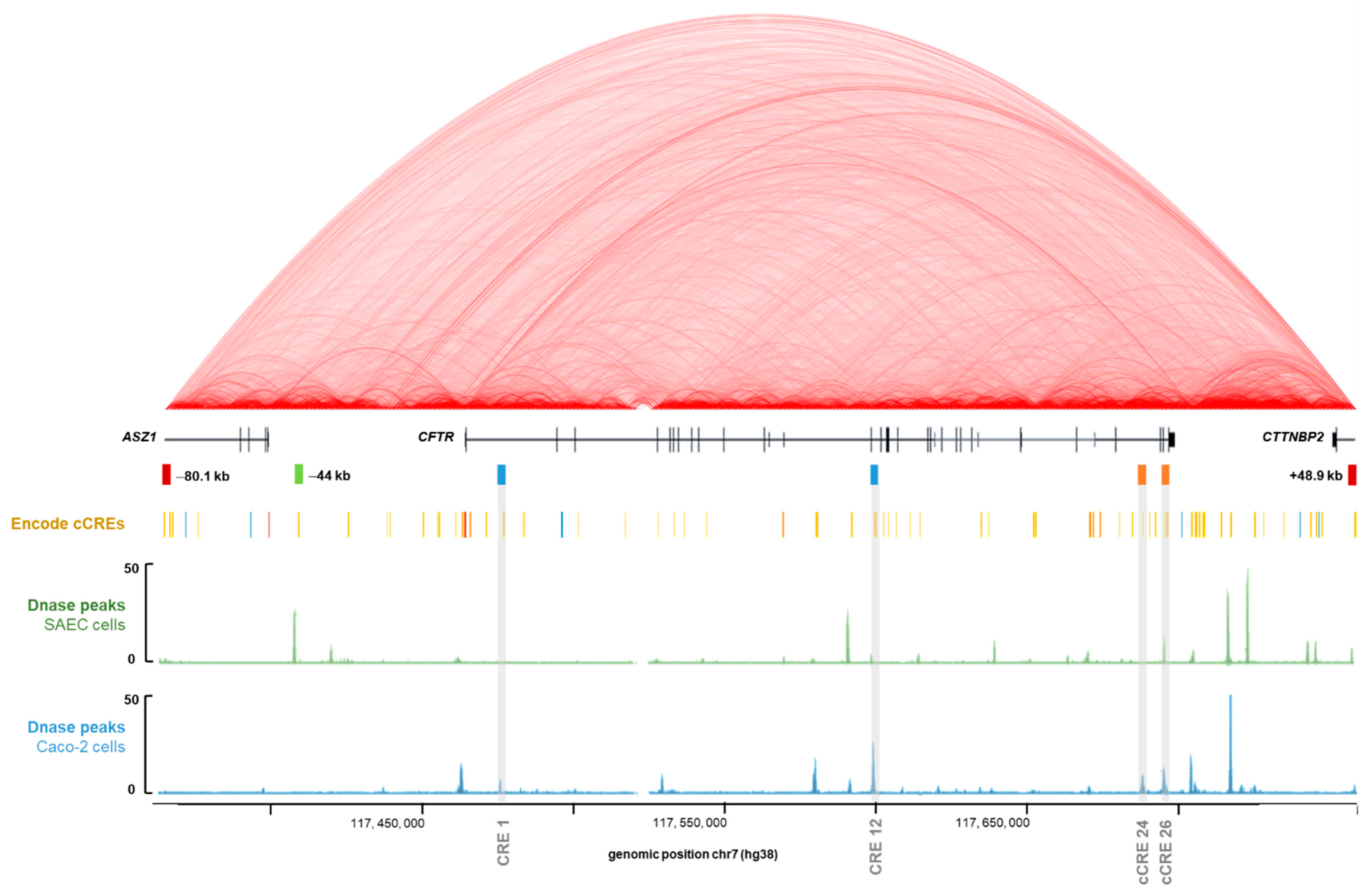
2.1. GWAS3D Score Predicts Four Regions as Candidate Cis-Regulatory Elements of the CFTR Gene
2.2. Intestinal Specific Activity of Intron 24 CRE
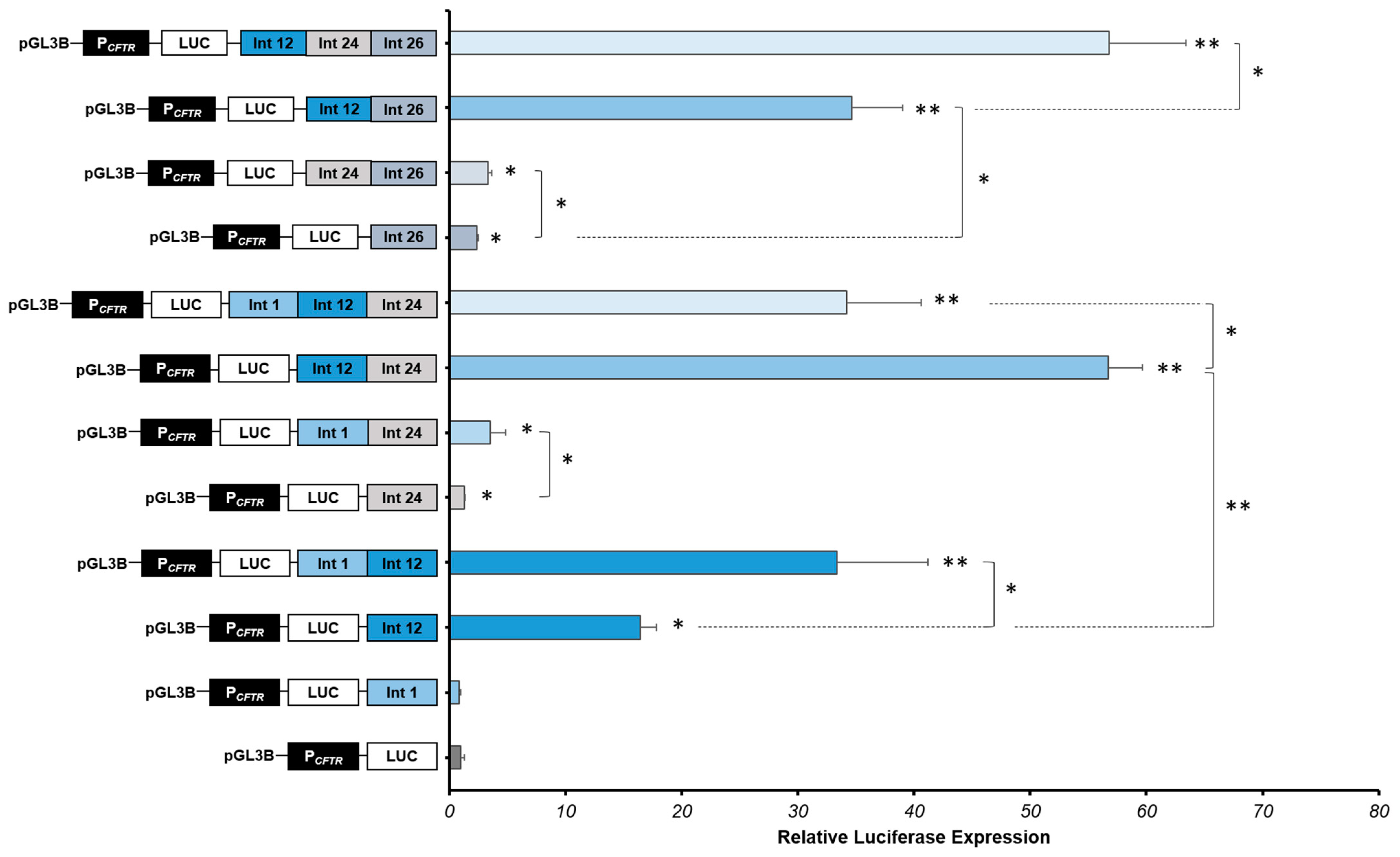
2.3. Strong Cooperative Effects between Relevant Introns CREs in Intestinal Cells
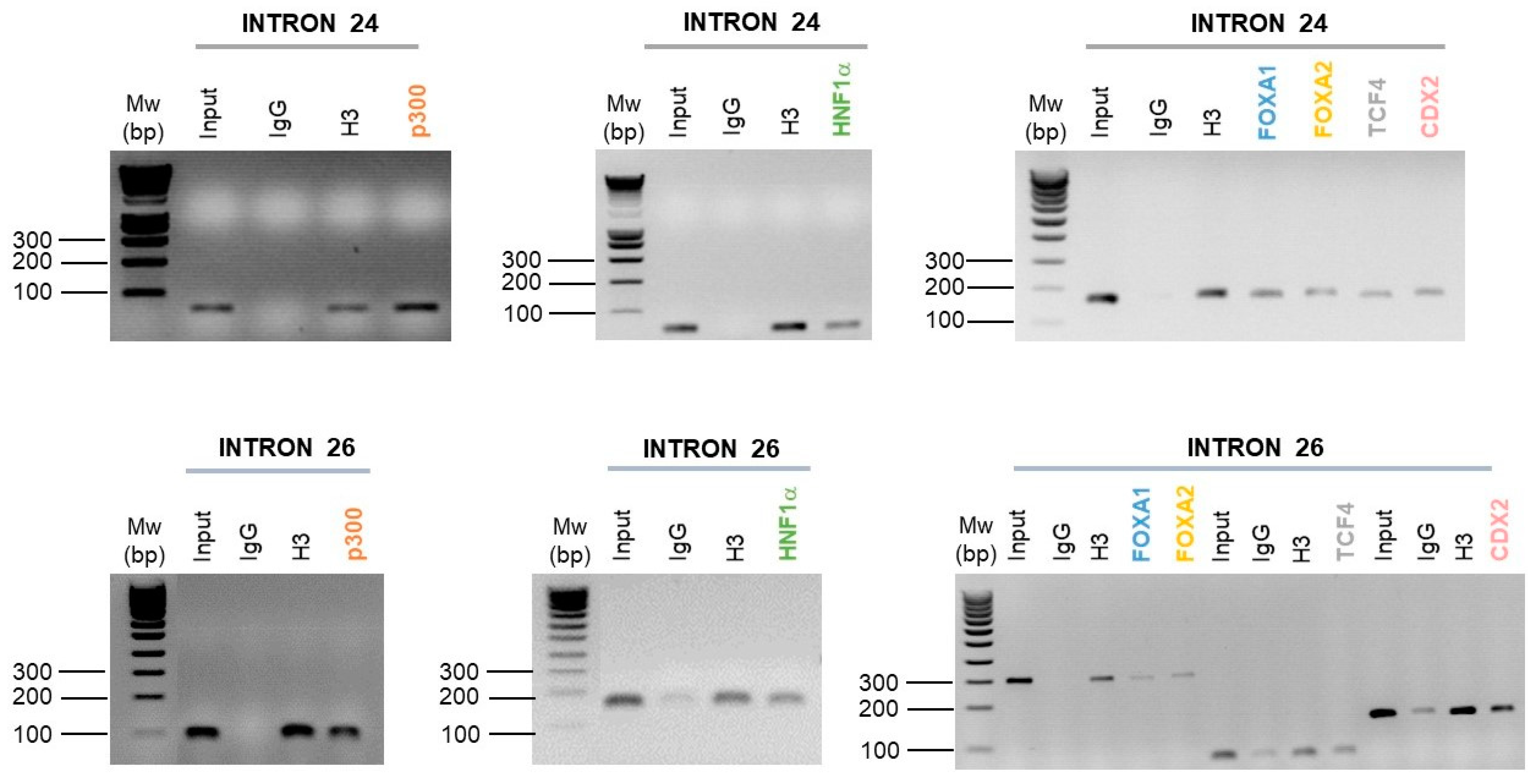
2.4. Intestinal Cell-Specific Transcription Factors Bind on Introns 24 and 26 CREs
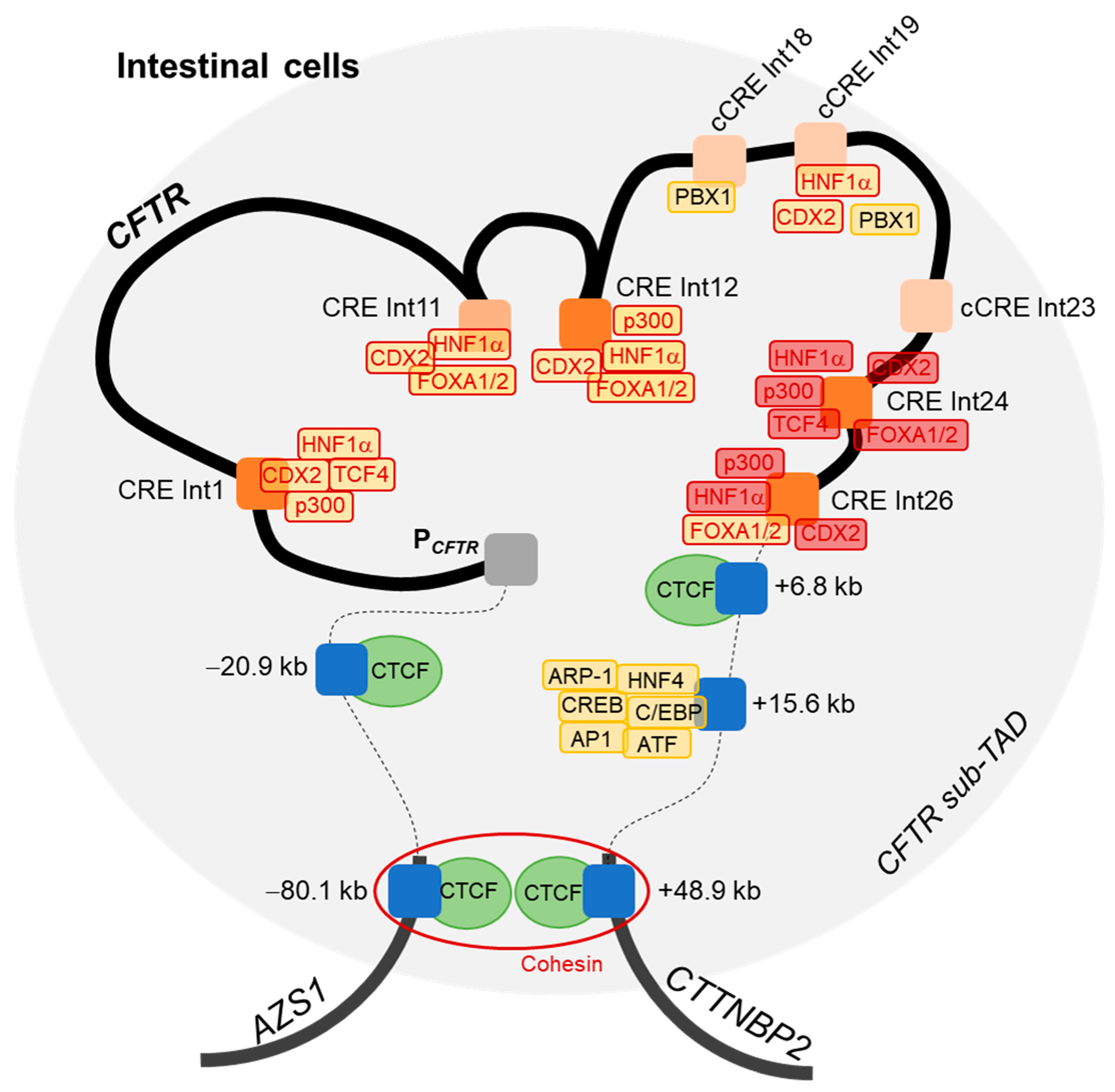
3. Discussion
4. Materials and Methods
4.1. GWAS3D Score
4.2. Databases and URLs
4.3. Culture Cells
4.4. Plasmid Construction
4.5. Luciferase Assays
4.6. Chromatin Immunoprecipitation and Polymerase Chain Reaction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Regions Name | Coordinates (hg38) | Size (pb) |
|---|---|---|
| Promoter CFTR | chr7: 117, 479, 274–117, 480, 060 | 787 |
| −44 kb | chr7: 117, 434, 807–117, 436, 588 | 1782 |
| Intron 1 | chr7: 117, 489, 621–117, 490, 501 | 881 |
| Intron 12 | chr7: 117, 587, 777–117, 589, 290 | 1514 |
| Intron 24 | chr7: 117, 659, 283–117, 660, 272 | 990 |
| Intron 26 | chr7: 117, 665, 734–117, 666, 846 | 1113 |
| Primers Name | Primers Sequence |
|---|---|
| pGL3B_BamHI_F | TTCAGGTTCAGGGGGAGGTG |
| pGL3B_BamHI_R | GTGCGGCGACGATAGTCATG |
| IF_BamHI_PCFTR_F | CGATCTAAGTAAGCTTGGAGTTCACTCACCTAAACCTGA |
| IF_BamHI_PCFTR_R | CCGGAATGCCAAGCTTTCTGGGCTCAAGCTCCTAATG |
| IF_BamHI_Int24_F | AAATCGATAAGGATCCGCATTTCTCACTCTGGCTGG |
| IF_BamHI_Int24_R | ATCGGTCGACGGATCCCCTGTCCAAACTAAGAAGACTTC |
| IF_BamHI_Int12_F | AAATCGATAAGGATCCTGGAGAAGGTGGAATCACACTG |
| IF_BamHI_Int12_R | ATCGGTCGACGGATCCGAAGACAGTATGCAAGAGCTACAT |
| IF_Int12-24_F | CTTGCATACTGTCTTCGCATTTCTCACTCTGGCTGG |
| IF_Int12-24_R | CCAGAGTGAGAAATGCGAAGACAGTATGCAAGAGCTACAT |
| IF_BamHI_Int1_F | AAATCGATAAGGATCCGTAGTGACATGCTCACTAATGC |
| IF_BamHI_Int1_R | ATCGGTCGACGGATCCCAGAGAAGGACGAAATTC |
| IF_Int1-Int24_F | ATTTCGTCCTTCTCTGGCATTTCTCACTCTGGCTGG |
| IF_Int1-Int24_R | CCAGAGTGAGAAATGCCAGAGAAGGACGAAATTCC |
| IF_Int1-Int12_F | ATTTCGTCCTTCTCTGTGGAGAAGGTGGAATCACACTG |
| IF_Int1-Int12_R | GATTCCACCTTCTCCACAGAGAAGGACGAAATTCC |
| IF_BamHI_Int26_F | AAATCGATAAGGATCCATCTGAGCCATGTGGTGAGGTTGA |
| IF_BamHI_Int26_R | ATCGGTCGACGGATCCGAAACTGGCACAGGCTCAAAGGAA |
| IF_Int12-26_F | CATACTGTCTTCATCTGAGCCATGTGGTGAGGTTGA |
| IF_Int12-26_R | CATGGCTCAGATGAAGACAGTATGCAAGAGCTACAT |
| IF_Int24-26_F | AGTTTGGACAGGATCTGAGCCATGTGGTGAGGTTGA |
| IF_Int24-26_R | CATGGCTCAGATCCTGTCCAAACTAAGAAGACTTC |
| Primers Name | Primers Sequence | Transcription Factors Specificity |
|---|---|---|
| ChIP_Int24_F1 | GCACATGTGCTTTCTGTTCTGC | HNF1, p300 |
| ChIP_Int24_R1 | GATGGTAGAAGGAGGCAGTG | HNF1, p300 |
| ChIP_Int26_F1 | CCCTATGGTTTAGTCACAAGGAAGTT | HNF1, p300 |
| ChIP_Int26_R1 | GGCTCAAAAGCCTGAACAGAA | HNF1, p300 |
| ChIP_Int24_F2 | CAGCAGTTTGGGAAGTACATAC | FOXA1/2, CDX2, TCF4 |
| ChIP_Int24_R2 | CGTCCTGCCCCCAATATAATC | FOXA1/2, CDX2, TCF4 |
| ChIP_Int26_F2 | TCTAGCCATGATTTGGGTTC | FOXA1/2 |
| ChIP_Int26_R2 | GGCTCAAAAGCCTGAACAGAA | FOXA1/2 |
| ChIP_Int26_F3 | CATACAGCCATCCAGCTTTAC | TCF4 |
| ChIP_Int26_R3 | TCAACCTCACCACATGGCTC | TCF4 |
| ChIP_Int26_F4 | GAAGCAATGCTGGAATGCCAAC | CDX2 |
| ChIP_Int26_R4 | GTAAAGCTGGATGGCTGTATG | CDX2 |
References
- Gasperini, M.; Tome, J.M.; Shendure, J. Towards a Comprehensive Catalogue of Validated and Target-Linked Human Enhancers. Nat. Rev. Genet. 2020. [Google Scholar] [CrossRef] [PubMed]
- Epstein, D.J. Cis-Regulatory Mutations in Human Disease. Brief. Funct. Genom. Proteomic 2009, 8, 310–316. [Google Scholar] [CrossRef]
- Kleinjan, D.A.; van Heyningen, V. Long-Range Control of Gene Expression: Emerging Mechanisms and Disruption in Disease. Am. J. Hum. Genet. 2005, 76, 8–32. [Google Scholar] [CrossRef]
- Loots, G.G.; Kneissel, M.; Keller, H.; Baptist, M.; Chang, J.; Collette, N.M.; Ovcharenko, D.; Plajzer-Frick, I.; Rubin, E.M. Genomic Deletion of a Long-Range Bone Enhancer Misregulates Sclerostin in Van Buchem Disease. Genome Res. 2005, 15, 928–935. [Google Scholar] [CrossRef]
- Kleinjan, D.A.; Seawright, A.; Schedl, A.; Quinlan, R.A.; Danes, S.; van Heyningen, V. Aniridia-Associated Translocations, DNase Hypersensitivity, Sequence Comparison and Transgenic Analysis Redefine the Functional Domain of PAX6. Hum. Mol. Genet. 2001, 10, 2049–2059. [Google Scholar] [CrossRef] [PubMed]
- Benko, S.; Fantes, J.A.; Amiel, J.; Kleinjan, D.-J.; Thomas, S.; Ramsay, J.; Jamshidi, N.; Essafi, A.; Heaney, S.; Gordon, C.T.; et al. Highly Conserved Non-Coding Elements on Either Side of SOX9 Associated with Pierre Robin Sequence. Nat. Genet. 2009, 41, 359–364. [Google Scholar] [CrossRef]
- Bhatia, S.; Bengani, H.; Fish, M.; Brown, A.; Divizia, M.T.; de Marco, R.; Damante, G.; Grainger, R.; van Heyningen, V.; Kleinjan, D.A. Disruption of Autoregulatory Feedback by a Mutation in a Remote, Ultraconserved PAX6 Enhancer Causes Aniridia. Am. J. Hum. Genet. 2013, 93, 1126–1134. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Ma, F.; Zeng, X.; Liu, Q.; Zhao, X.-L.; Wu, F.-X.; Wu, G.-P.; Zhang, Z.-F.; Gu, B.; Zhao, Y.-F.; et al. Triphalangeal Thumb–Polysyndactyly Syndrome and Syndactyly Type IV Are Caused by Genomic Duplications Involving the Long Range, Limb-Specific SHH Enhancer. J. Med. Genet. 2008, 45, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef]
- Kleinjan, D.-J.; Coutinho, P. Cis-Ruption Mechanisms: Disruption of Cis-Regulatory Control as a Cause of Human Genetic Disease. Brief. Funct. Genom. Proteomic 2009, 8, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Rommens, J.M.; Iannuzzi, M.C.; Kerem, B.; Drumm, M.L.; Melmer, G.; Dean, M.; Rozmahel, R.; Cole, J.L.; Kennedy, D.; Hidaka, N.; et al. Identification of the Cystic Fibrosis Gene: Chromosome Walking and Jumping. Science 1989, 245, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Cystic Fibrosis Genotype-Phenotype Consortium. Correlation between Genotype and Phenotype in Patients with Cystic Fibrosis. N. Engl. J. Med. 1993, 329, 1308–1313. [Google Scholar] [CrossRef] [PubMed]
- Bombieri, C.; Claustres, M.; De Boeck, K.; Derichs, N.; Dodge, J.; Girodon, E.; Sermet, I.; Schwarz, M.; Tzetis, M.; Wilschanski, M.; et al. Recommendations for the Classification of Diseases as CFTR-Related Disorders. J. Cyst. Fibros. 2011, 10, S86–S102. [Google Scholar] [CrossRef]
- Bombieri, C.; Seia, M.; Castellani, C. Genotypes and Phenotypes in Cystic Fibrosis and Cystic Fibrosis Transmembrane Regulator-Related Disorders. Semin. Respir. Crit. Care Med. 2015, 36, 180–193. [Google Scholar] [CrossRef]
- Kerem, E.; Corey, M.; Kerem, B.S.; Rommens, J.; Markiewicz, D.; Levison, H.; Tsui, L.C.; Durie, P. The Relation between Genotype and Phenotype in Cystic Fibrosis—Analysis of the Most Common Mutation (Delta F508). N. Engl. J. Med. 1990, 323, 1517–1522. [Google Scholar] [CrossRef]
- Drumm, M.L.; Konstan, M.W.; Schluchter, M.D.; Handler, A.; Pace, R.; Zou, F.; Zariwala, M.; Fargo, D.; Xu, A.; Dunn, J.M.; et al. Genetic Modifiers of Lung Disease in Cystic Fibrosis. N. Engl. J. Med. 2005, 353, 1443–1453. [Google Scholar] [CrossRef]
- Corvol, H.; Blackman, S.M.; Boëlle, P.-Y.; Gallins, P.J.; Pace, R.G.; Stonebraker, J.R.; Accurso, F.J.; Clement, A.; Collaco, J.M.; Dang, H.; et al. Genome-Wide Association Meta-Analysis Identifies Five Modifier Loci of Lung Disease Severity in Cystic Fibrosis. Nat. Commun. 2015, 6, 8382. [Google Scholar] [CrossRef]
- Broackes-Carter, F.C.; Mouchel, N.; Gill, D.; Hyde, S.; Bassett, J.; Harris, A. Temporal Regulation of CFTR Expression during Ovine Lung Development: Implications for CF Gene Therapy. Hum. Mol. Genet. 2002, 11, 125–131. [Google Scholar] [CrossRef]
- Crawford, I.; Maloney, P.C.; Zeitlin, P.L.; Guggino, W.B.; Hyde, S.C.; Turley, H.; Gatter, K.C.; Harris, A.; Higgins, C.F. Immunocytochemical Localization of the Cystic Fibrosis Gene Product CFTR. Proc. Natl. Acad. Sci. USA 1991, 88, 9262–9266. [Google Scholar] [CrossRef]
- Gosalia, N.; Harris, A. Chromatin Dynamics in the Regulation of CFTR Expression. Genes 2015, 6, 543–558. [Google Scholar] [CrossRef]
- Trezise, A.E.O.; Chambers, J.A.; Wardle, C.J.; Gould, S.; Harris, A. Expression of the Cystic Fibrosis Gene in Human Foetal Tissues. Hum. Mol. Genet. 1993, 2, 213–218. [Google Scholar] [CrossRef]
- Nuthall, H.N.; Moulin, D.S.; Huxley, C.; Harris, A. Analysis of DNase-I-Hypersensitive Sites at the 3’ End of the Cystic Fibrosis Transmembrane Conductance Regulator Gene (CFTR). Biochem. J. 1999, 341, 601–611. [Google Scholar] [CrossRef]
- Rowntree, R.K.; Vassaux, G.; McDowell, T.L.; Howe, S.; McGuigan, A.; Phylactides, M.; Huxley, C.; Harris, A. An Element in Intron 1 of the CFTR Gene Augments Intestinal Expression in Vivo. Hum. Mol. Genet. 2001, 10, 1455–1464. [Google Scholar] [CrossRef]
- Smith, A.N.; Barth, M.L.; McDowell, T.L.; Moulin, D.S.; Nuthall, H.N.; Hollingsworth, M.A.; Harris, A. A Regulatory Element in Intron 1 of the Cystic Fibrosis Transmembrane Conductance Regulator Gene. J. Biol. Chem. 1996, 271, 9947–9954. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Nuthall, H.N.; Majetti, M.E.; Harris, A. Multiple Potential Intragenic Regulatory Elements in the CFTR Gene. Genomics 2000, 64, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing Chromosome Conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Splinter, E.; de Wit, E.; van de Werken, H.J.G.; Klous, P.; de Laat, W. Determining Long-Range Chromatin Interactions for Selected Genomic Sites Using 4C-Seq Technology: From Fixation to Computation. Methods 2012, 58, 221–230. [Google Scholar] [CrossRef]
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome Conformation Capture Carbon Copy (5C): A Massively Parallel Solution for Mapping Interactions between Genomic Elements. Genome Res. 2006, 16, 1299–1309. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Ott, C.J.; Gillen, A.E.; Harris, A. An Insulator Element 3′ to the CFTR Gene Binds CTCF and Reveals an Active Chromatin Hub in Primary Cells. Nucleic Acids Res. 2009, 37, 1086–1094. [Google Scholar] [CrossRef]
- Ott, C.J.; Blackledge, N.P.; Kerschner, J.L.; Leir, S.-H.; Crawford, G.E.; Cotton, C.U.; Harris, A. Intronic Enhancers Coordinate Epithelial-Specific Looping of the Active CFTR Locus. Proc. Natl. Acad. Sci. USA 2009, 106, 19934–19939. [Google Scholar] [CrossRef]
- Ott, C.J.; Suszko, M.; Blackledge, N.P.; Wright, J.E.; Crawford, G.E.; Harris, A. A Complex Intronic Enhancer Regulates Expression of the CFTR Gene by Direct Interaction with the Promoter. J. Cell. Mol. Med. 2009, 13, 680–692. [Google Scholar] [CrossRef]
- Gheldof, N.; Smith, E.M.; Tabuchi, T.M.; Koch, C.M.; Dunham, I.; Stamatoyannopoulos, J.A.; Dekker, J. Cell-Type-Specific Long-Range Looping Interactions Identify Distant Regulatory Elements of the CFTR Gene. Nucleic Acids Res. 2010, 38, 4325–4336. [Google Scholar] [CrossRef] [PubMed]
- Moisan, S.; Berlivet, S.; Ka, C.; Gac, G.L.; Dostie, J.; Férec, C. Analysis of Long-Range Interactions in Primary Human Cells Identifies Cooperative CFTR Regulatory Elements. Nucleic Acids Res. 2016, 44, 2564–2576. [Google Scholar] [CrossRef]
- Gosalia, N.; Neems, D.; Kerschner, J.L.; Kosak, S.T.; Harris, A. Architectural Proteins CTCF and Cohesin Have Distinct Roles in Modulating the Higher Order Structure and Expression of the CFTR Locus. Nucleic Acids Res. 2014, 42, 9612–9622. [Google Scholar] [CrossRef]
- Smith, E.M.; Lajoie, B.R.; Jain, G.; Dekker, J. Invariant TAD Boundaries Constrain Cell-Type-Specific Looping Interactions between Promoters and Distal Elements around the CFTR Locus. Am. J. Hum. Genet. 2016, 98, 185–201. [Google Scholar] [CrossRef]
- Yang, R.; Kerschner, J.L.; Gosalia, N.; Neems, D.; Gorsic, L.K.; Safi, A.; Crawford, G.E.; Kosak, S.T.; Leir, S.-H.; Harris, A. Differential Contribution of Cis-Regulatory Elements to Higher Order Chromatin Structure and Expression of the CFTR Locus. Nucleic Acids Res. 2016, 44, 3082–3094. [Google Scholar] [CrossRef]
- Li, M.J.; Wang, L.Y.; Xia, Z.; Sham, P.C.; Wang, J. GWAS3D: Detecting Human Regulatory Variants by Integrative Analysis of Genome-Wide Associations, Chromosome Interactions and Histone Modifications. Nucleic Acids Res. 2013, 41, W150–W158. [Google Scholar] [CrossRef]
- Nishizaki, S.S.; Boyle, A.P. Mining the Unknown: Assigning Function to Noncoding Single Nucleotide Polymorphisms. Trends Genet. 2017, 33, 34–45. [Google Scholar] [CrossRef]
- Zhang, S.; He, Y.; Liu, H.; Zhai, H.; Huang, D.; Yi, X.; Dong, X.; Wang, Z.; Zhao, K.; Zhou, Y.; et al. RegBase: Whole Genome Base-Wise Aggregation and Functional Prediction for Human Non-Coding Regulatory Variants. Nucleic Acids Res. 2019, 47, e134. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- The ENCODE Project Consortium. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Weintraub, H.; Groudine, M. Chromosomal Subunits in Active Genes Have an Altered Conformation. Science 1976, 193, 848–856. [Google Scholar] [CrossRef]
- Gross, D.S.; Garrard, W.T. Nuclease Hypersensitive Sites in Chromatin. Annu. Rev. Biochem. 1988, 57, 159–197. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium; Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; et al. Expanded Encyclopaedias of DNA Elements in the Human and Mouse Genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Thurman, R.E.; Rynes, E.; Humbert, R.; Vierstra, J.; Maurano, M.T.; Haugen, E.; Sheffield, N.C.; Stergachis, A.B.; Wang, H.; Vernot, B.; et al. The Accessible Chromatin Landscape of the Human Genome. Nature 2012, 489, 75–82. [Google Scholar] [CrossRef]
- Miga, K.H.; Eisenhart, C.; Kent, W.J. Utilizing Mapping Targets of Sequences Underrepresented in the Reference Assembly to Reduce False Positive Alignments. Nucleic Acids Res. 2015, 43, e133. [Google Scholar] [CrossRef] [PubMed]
- Yigit, E.; Bischof, J.M.; Zhang, Z.; Ott, C.J.; Kerschner, J.L.; Leir, S.-H.; Buitrago-Delgado, E.; Zhang, Q.; Wang, J.-P.Z.; Widom, J.; et al. Nucleosome Mapping across the CFTR Locus Identifies Novel Regulatory Factors. Nucleic Acids Res. 2013, 41, 2857–2868. [Google Scholar] [CrossRef]
- Paul, T.; Li, S.; Khurana, S.; Leleiko, N.S.; Walsh, M.J. The Epigenetic Signature of CFTR Expression Is Co-Ordinated via Chromatin Acetylation through a Complex Intronic Element. Biochem. J. 2007, 408, 317–326. [Google Scholar] [CrossRef]
- Kerschner, J.L.; Harris, A. Transcriptional Networks Driving Enhancer Function in the CFTR Gene. Biochem. J. 2012, 446, 203–212. [Google Scholar] [CrossRef]
- Kerschner, J.L.; Gosalia, N.; Leir, S.-H.; Harris, A. Chromatin Remodeling Mediated by the FOXA1/A2 Transcription Factors Activates CFTR Expression in Intestinal Epithelial Cells. Epigenetics 2014, 9, 557–565. [Google Scholar] [CrossRef]
- Barker, N.; Huls, G.; Korinek, V.; Clevers, H. Restricted High Level Expression of Tcf-4 Protein in Intestinal and Mammary Gland Epithelium. Am. J. Pathol. 1999, 154, 29–35. [Google Scholar] [CrossRef]
- Zhang, Z.; Ott, C.J.; Lewandowska, M.A.; Leir, S.-H.; Harris, A. Molecular Mechanisms Controlling CFTR Gene Expression in the Airway. J. Cell. Mol. Med. 2012, 16, 1321–1330. [Google Scholar] [CrossRef]
- Phylactides, M.; Rowntree, R.; Nuthall, H.; Ussery, D.; Wheeler, A.; Harris, A. Evaluation of Potential Regulatory Elements Identified as DNase I Hypersensitive Sites in the CFTR Gene. Eur. J. Biochem. 2002, 269, 553–559. [Google Scholar] [CrossRef]
- Swahn, H.; Harris, A. Cell-Selective Regulation of CFTR Gene Expression: Relevance to Gene Editing Therapeutics. Genes 2019, 10, 235. [Google Scholar] [CrossRef]
- Trezíse, A.E.O.; Buchwald, M. In Vivo Cell-Specific Expression of the Cystic Fibrosis Transmembrane Conductance Regulator. Nature 1991, 353, 434–437. [Google Scholar] [CrossRef]
- Zhang, Z.; Leir, S.-H.; Harris, A. Oxidative Stress Regulates CFTR Gene Expression in Human Airway Epithelial Cells through a Distal Antioxidant Response Element. Am. J. Respir. Cell Mol. Biol. 2015, 52, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Kerschner, J.L.; Harris, A. Hepatocyte Nuclear Factor 1 Coordinates Multiple Processes in a Model of Intestinal Epithelial Cell Function. Biochim. Biophys. Acta 2016, 1859, 591–598. [Google Scholar] [CrossRef][Green Version]
- Kreft, Ł.; Soete, A.; Hulpiau, P.; Botzki, A.; Saeys, Y.; De Bleser, P. ConTra v3: A Tool to Identify Transcription Factor Binding Sites across Species, Update 2017. Nucleic Acids Res. 2017, 45, W490–W494. [Google Scholar] [CrossRef] [PubMed]
- Lopez, E.; Viart, V.; Guittard, C.; Templin, C.; Georges, M.D.; Claustres, M.; Romey-chatelain, M.; Taulan, M. Variants in CFTR Untranslated Regions Are Associated with Congenital Bilateral Absence of the Vas Deferens. J. Med. Genet. 2011, 48, 152–159. [Google Scholar] [CrossRef]
- Lukowski, S.W.; Bombieri, C.; Trezise, A.E.O. Disrupted Posttranscriptional Regulation of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) by a 5′UTR Mutation Is Associated with a Cftr-Related Disease. Hum. Mutat. 2011, 32, E2266–E2282. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A Three-Dimensional Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Robson, M.I.; Ringel, A.R.; Mundlos, S. Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D. Mol. Cell 2019, 74, 1110–1122. [Google Scholar] [CrossRef]
- Vecchio-Pagán, B.; Blackman, S.M.; Lee, M.; Atalar, M.; Pellicore, M.J.; Pace, R.G.; Franca, A.L.; Raraigh, K.S.; Sharma, N.; Knowles, M.R.; et al. Deep Resequencing of CFTR in 762 F508del Homozygotes Reveals Clusters of Non-Coding Variants Associated with Cystic Fibrosis Disease Traits. Hum. Genome Var. 2016, 3, 16038. [Google Scholar] [CrossRef]
- Hsieh, T.-H.S.; Fudenberg, G.; Goloborodko, A.; Rando, O.J. Micro-C XL: Assaying Chromosome Conformation from the Nucleosome to the Entire Genome. Nat. Methods 2016, 13, 1009–1011. [Google Scholar] [CrossRef] [PubMed]
- Krietenstein, N.; Abraham, S.; Venev, S.V.; Abdennur, N.; Gibcus, J.; Hsieh, T.-H.S.; Parsi, K.M.; Yang, L.; Maehr, R.; Mirny, L.A.; et al. Ultrastructural Details of Mammalian Chromosome Architecture. Mol. Cell 2020, 78, 554–565.e7. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Collobert, M.; Bocher, O.; Le Nabec, A.; Génin, E.; Férec, C.; Moisan, S. CFTR Cooperative Cis-Regulatory Elements in Intestinal Cells. Int. J. Mol. Sci. 2021, 22, 2599. https://doi.org/10.3390/ijms22052599
Collobert M, Bocher O, Le Nabec A, Génin E, Férec C, Moisan S. CFTR Cooperative Cis-Regulatory Elements in Intestinal Cells. International Journal of Molecular Sciences. 2021; 22(5):2599. https://doi.org/10.3390/ijms22052599
Chicago/Turabian StyleCollobert, Mégane, Ozvan Bocher, Anaïs Le Nabec, Emmanuelle Génin, Claude Férec, and Stéphanie Moisan. 2021. "CFTR Cooperative Cis-Regulatory Elements in Intestinal Cells" International Journal of Molecular Sciences 22, no. 5: 2599. https://doi.org/10.3390/ijms22052599
APA StyleCollobert, M., Bocher, O., Le Nabec, A., Génin, E., Férec, C., & Moisan, S. (2021). CFTR Cooperative Cis-Regulatory Elements in Intestinal Cells. International Journal of Molecular Sciences, 22(5), 2599. https://doi.org/10.3390/ijms22052599








