Deciphering the Transcriptomic Heterogeneity of Duodenal Coeliac Disease Biopsies
Abstract
1. Introduction
2. Results
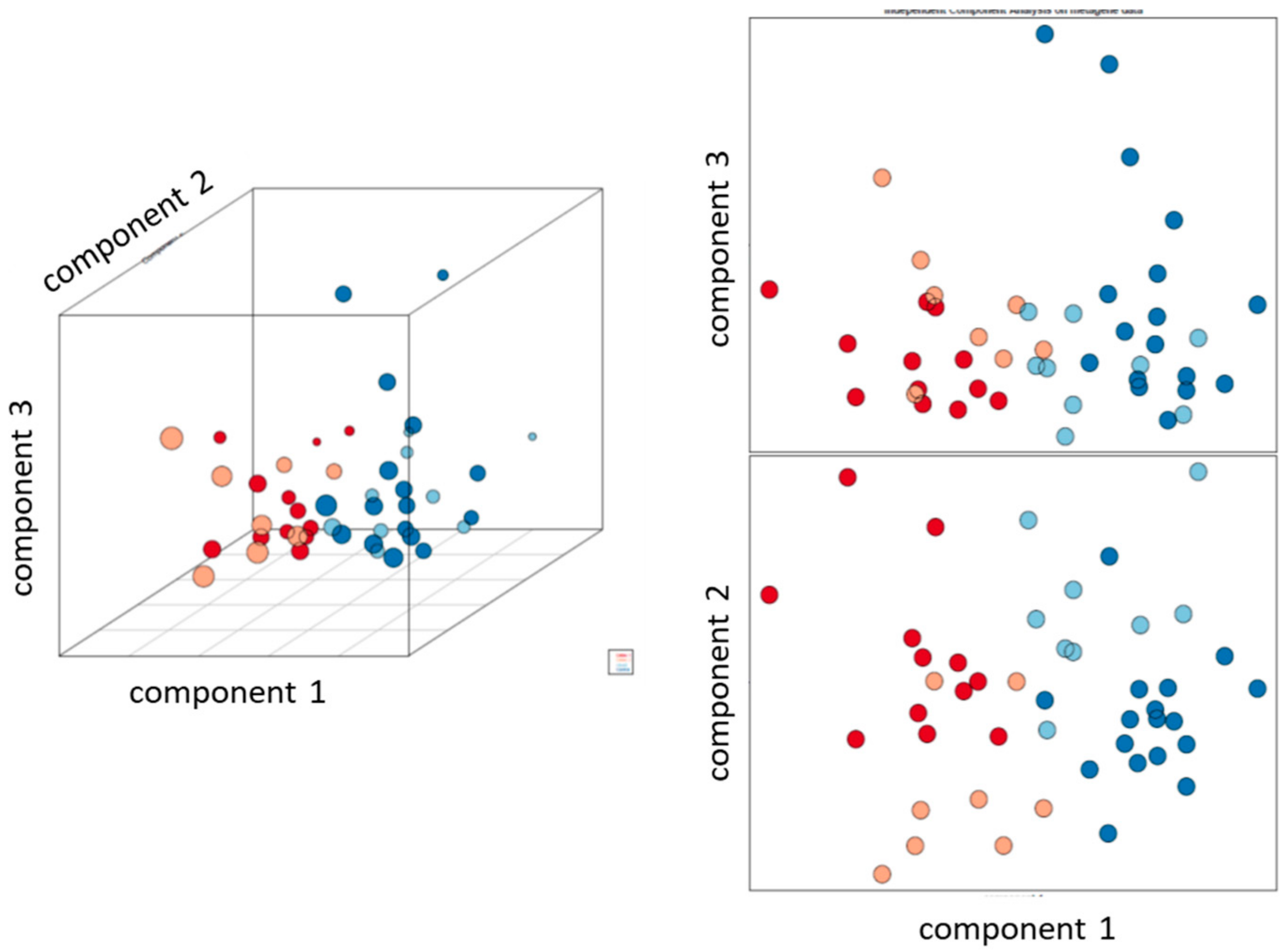
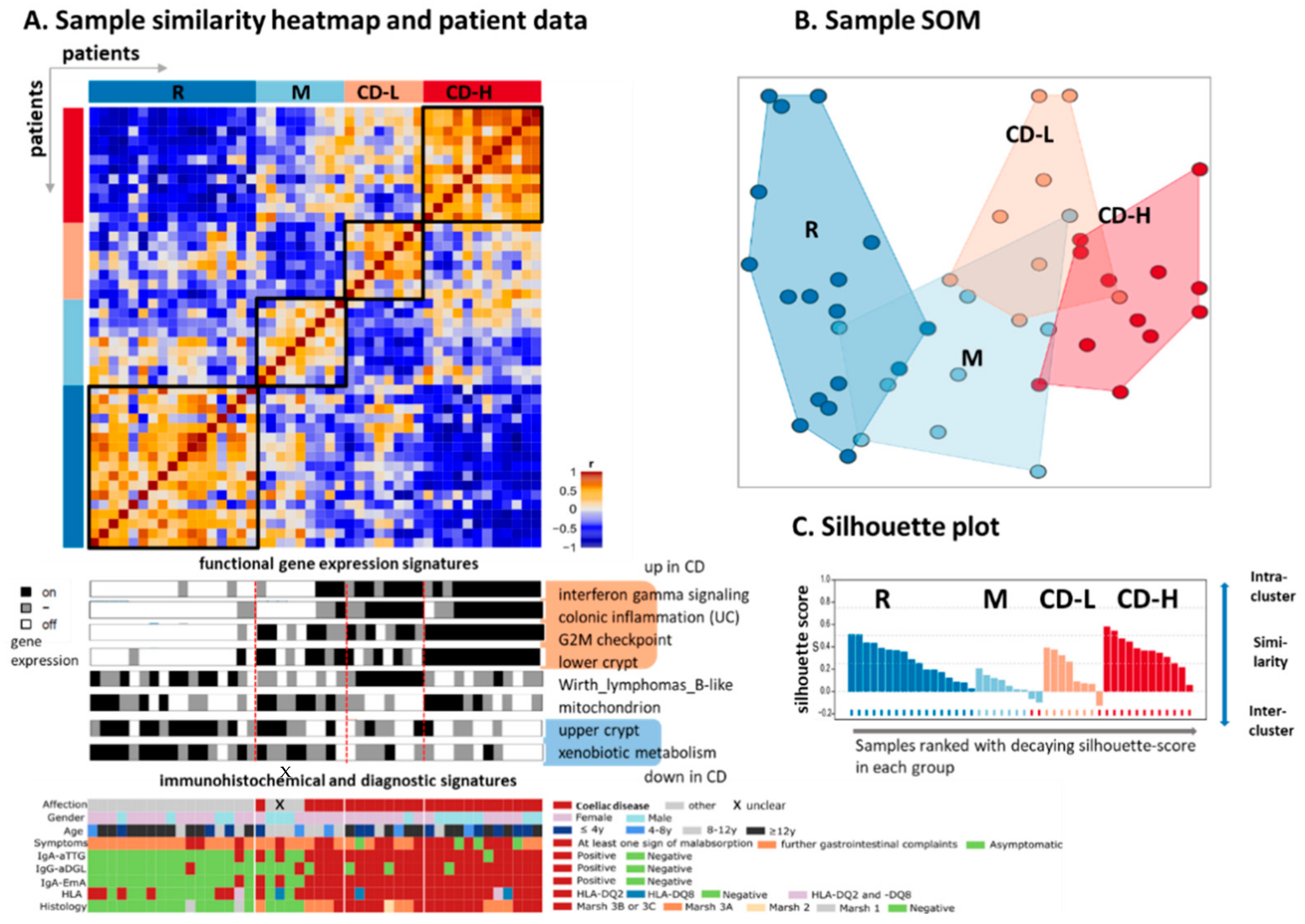
2.1. Clustering Identifies Three Transcriptional Subgroups of CD Patients
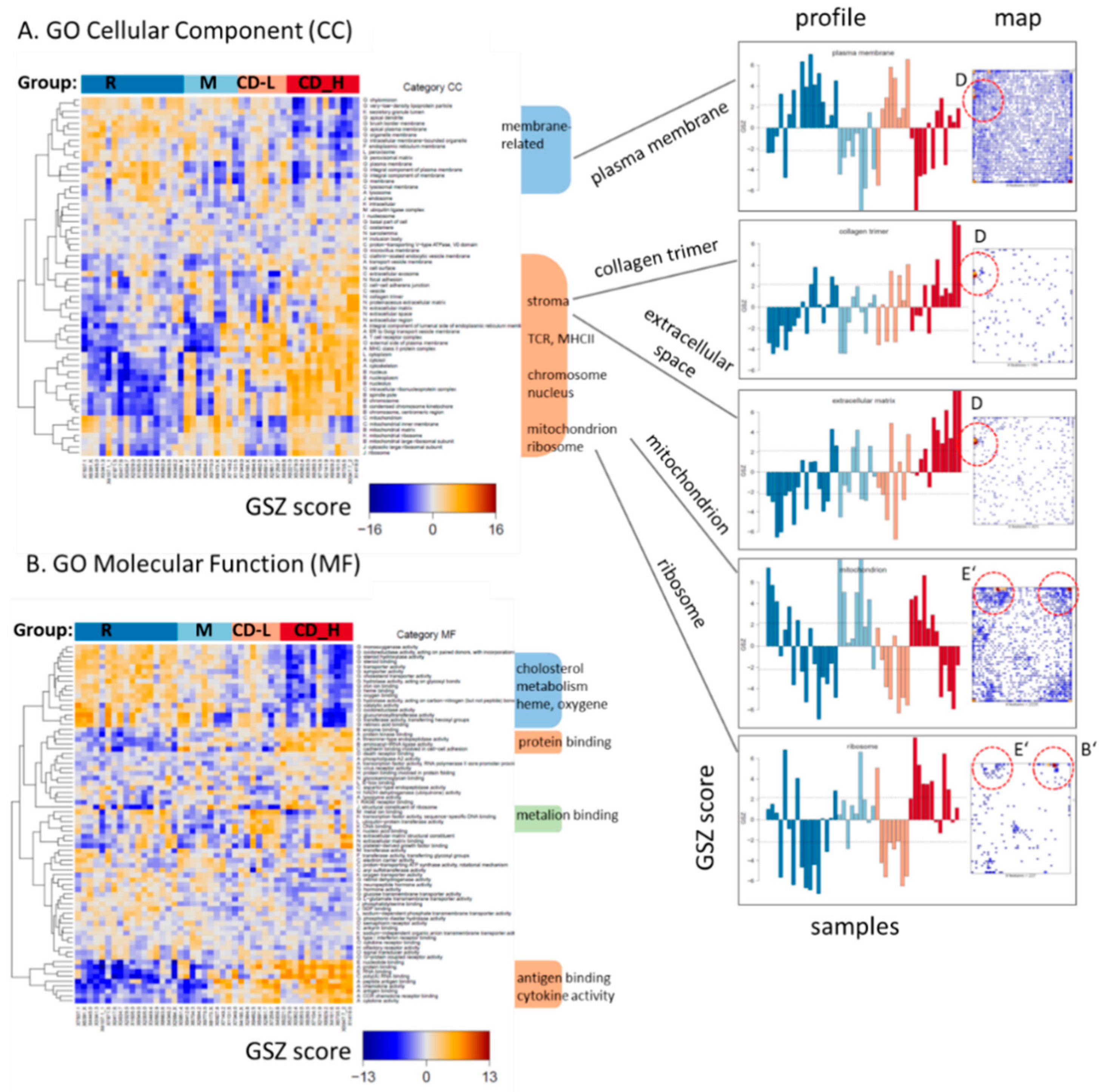
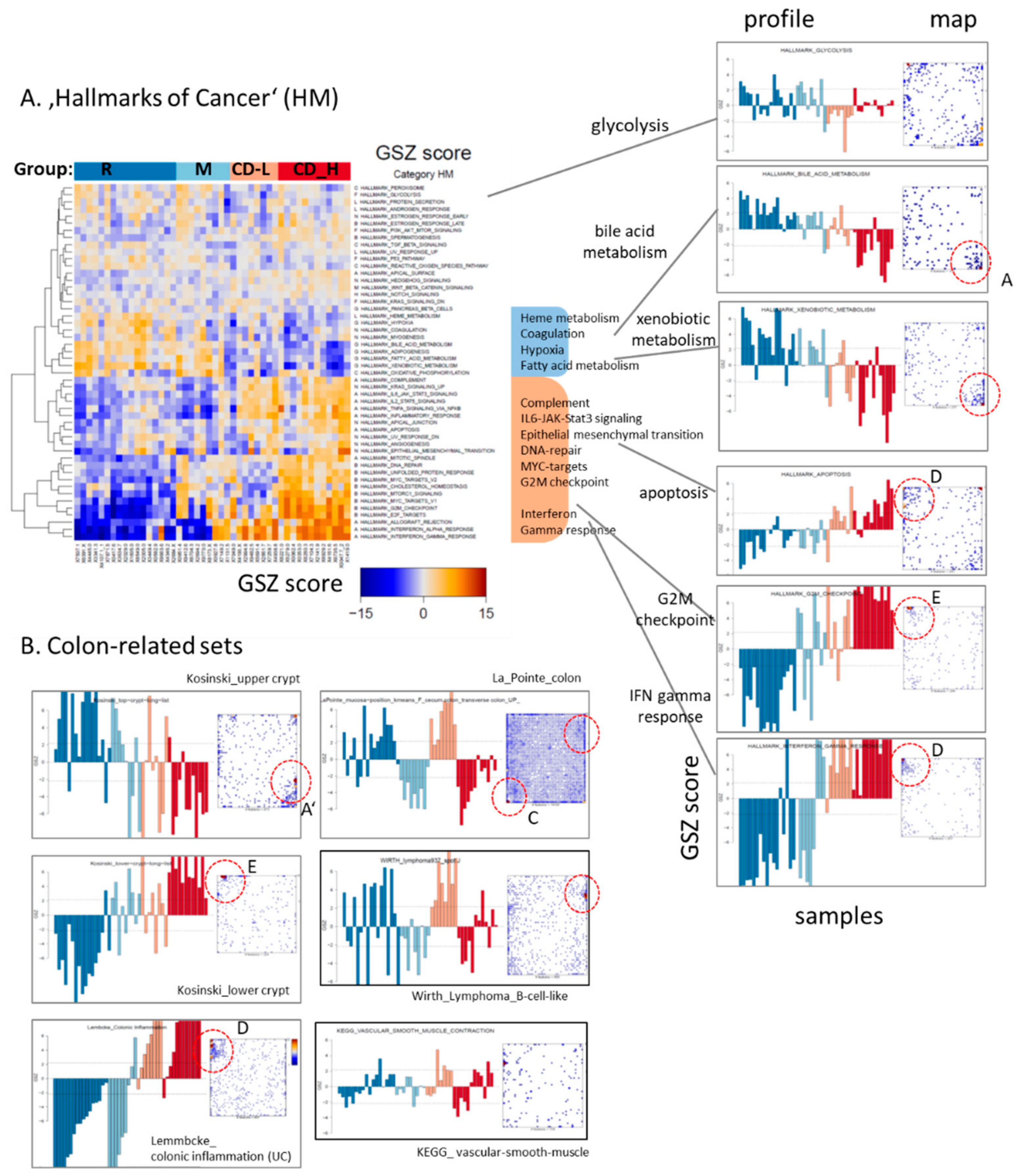
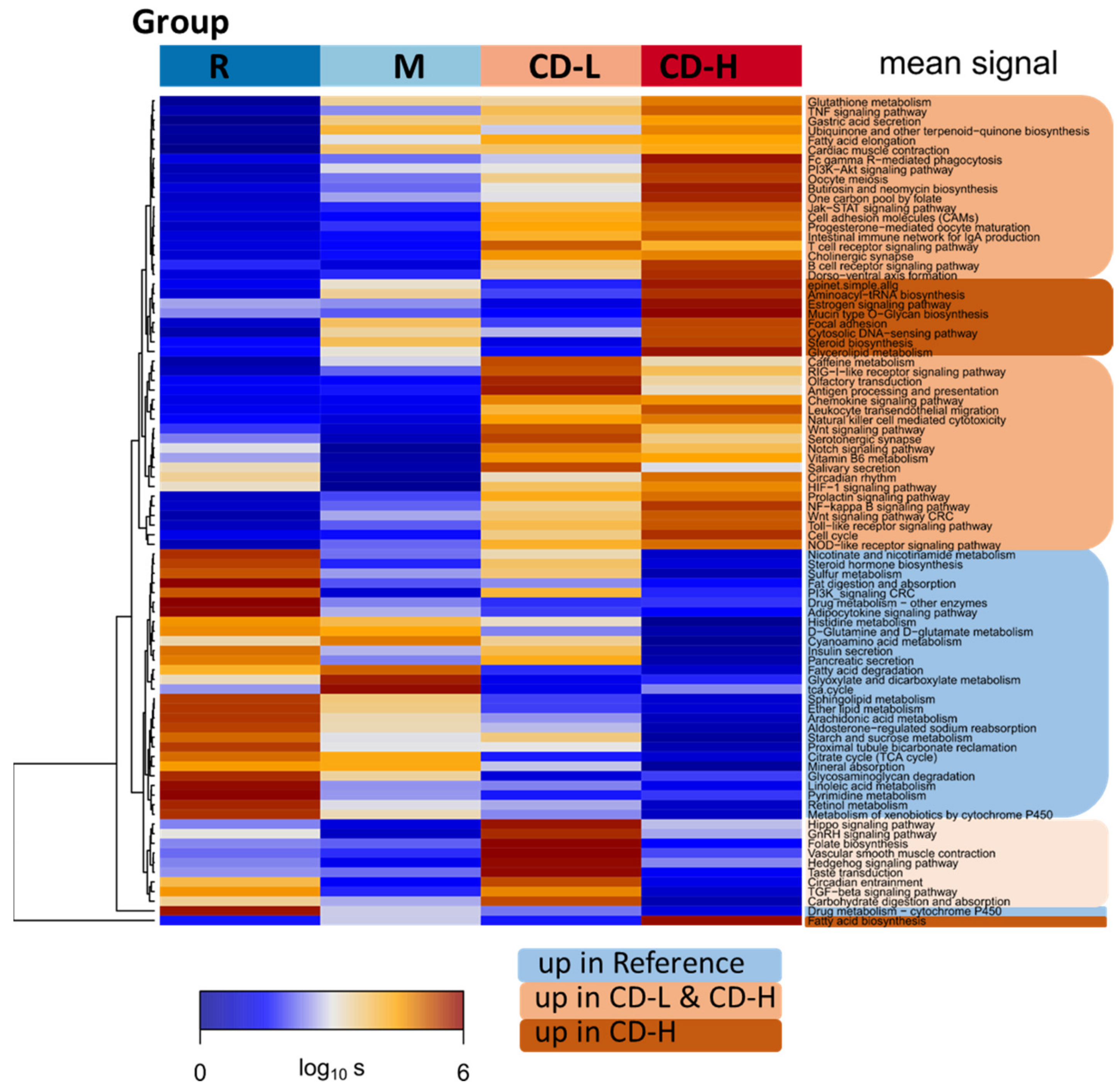
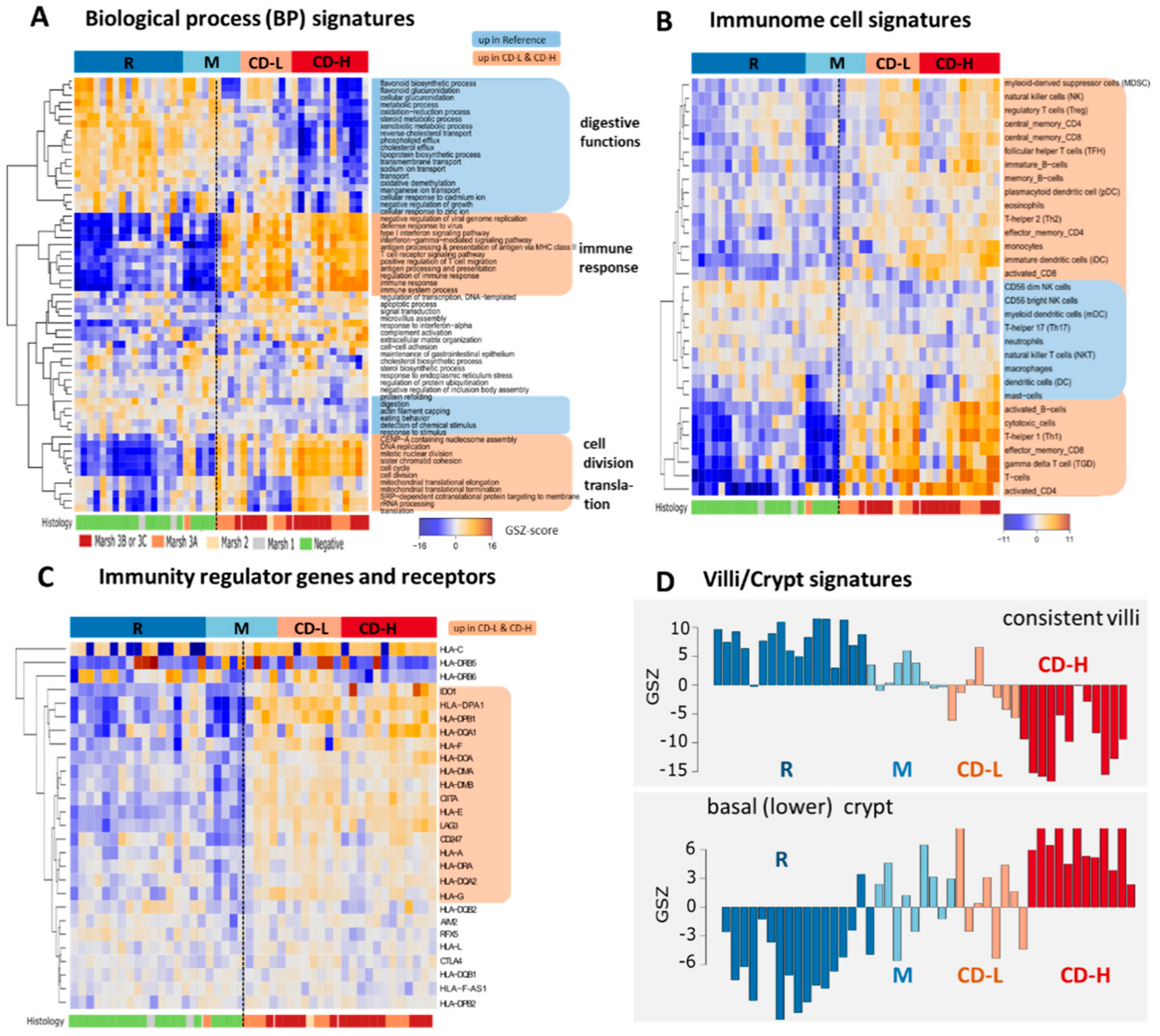
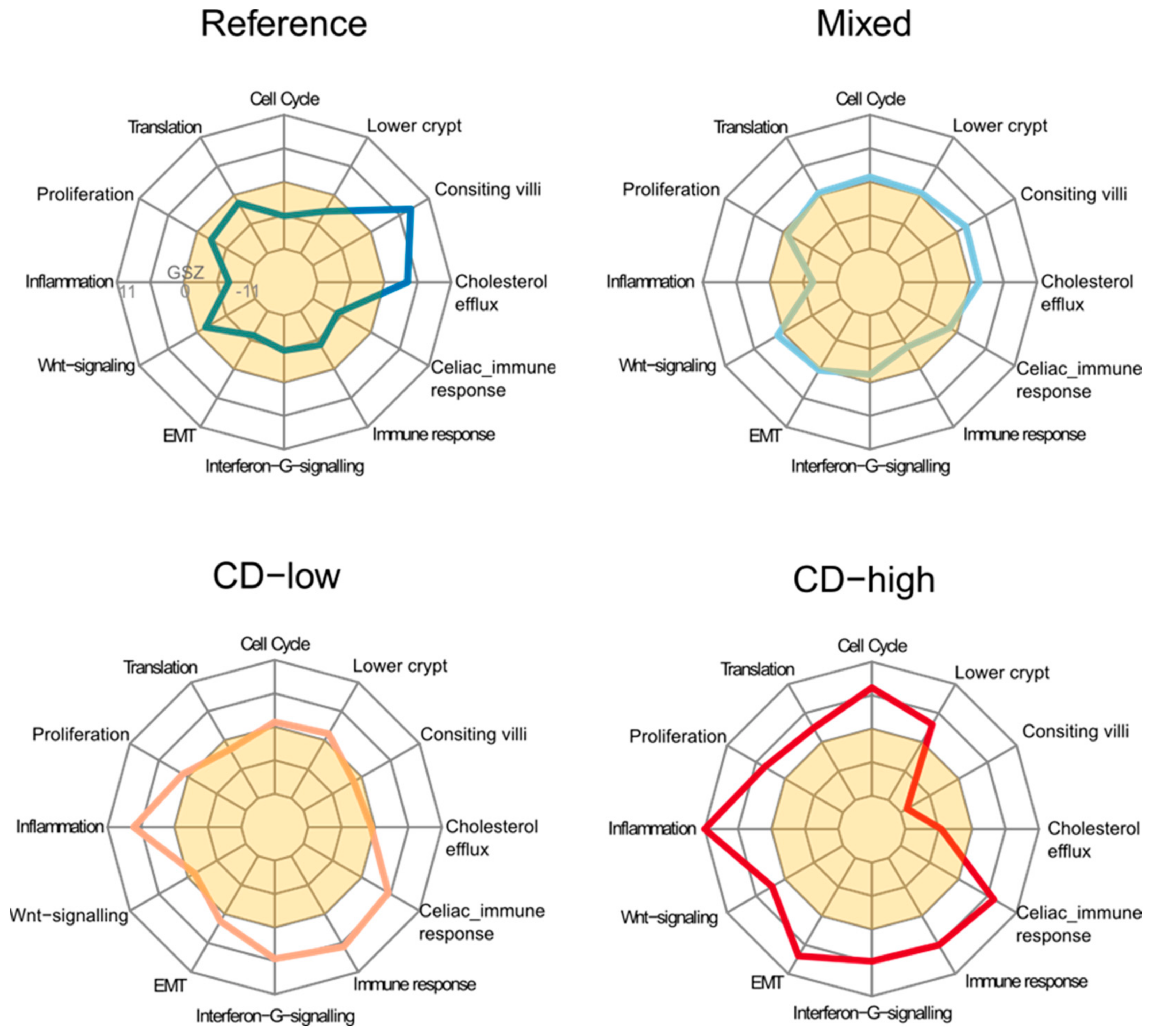
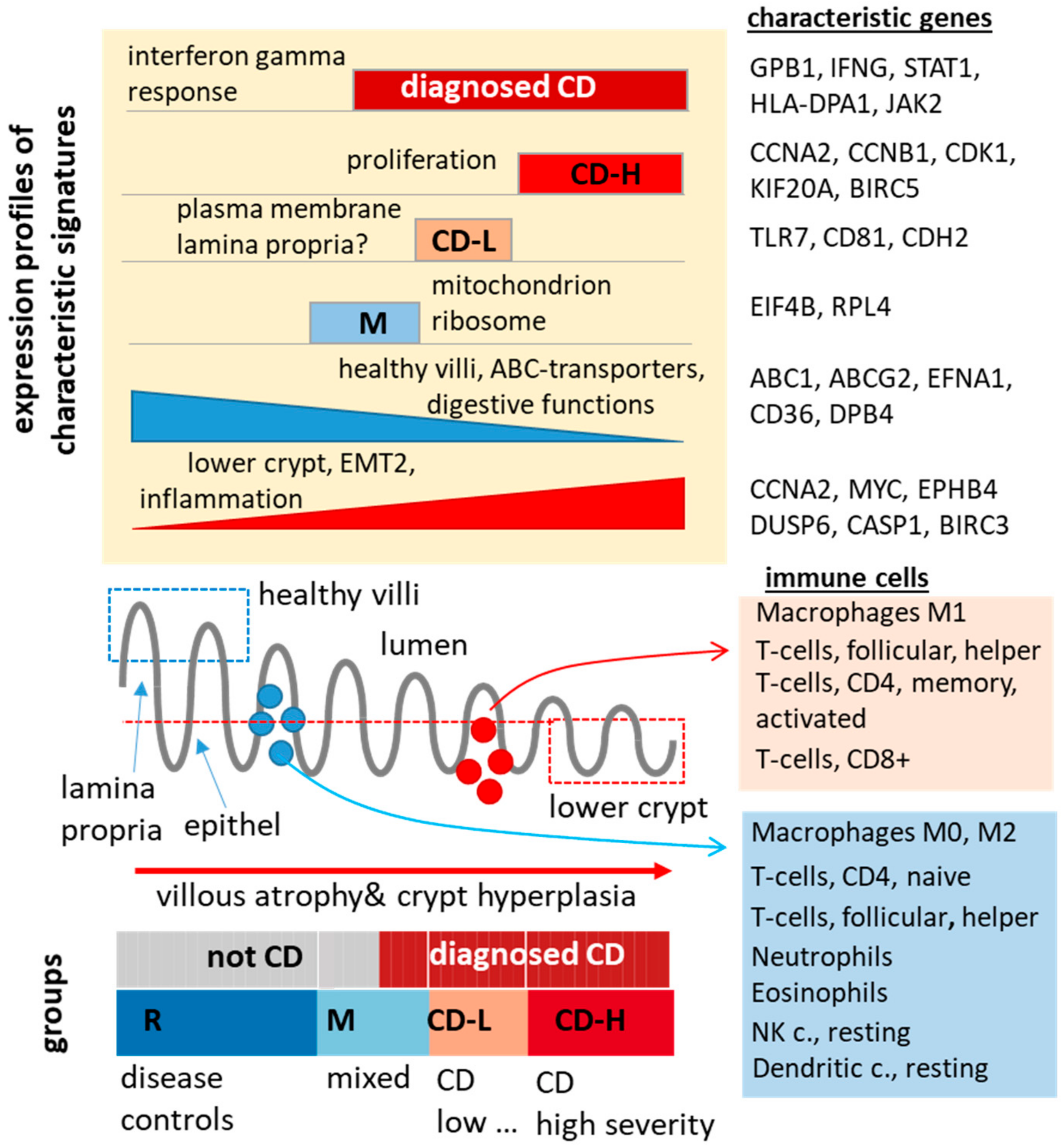
2.2. Expression Signatures Characterize the Functional State of the Small Intestine Epithelium
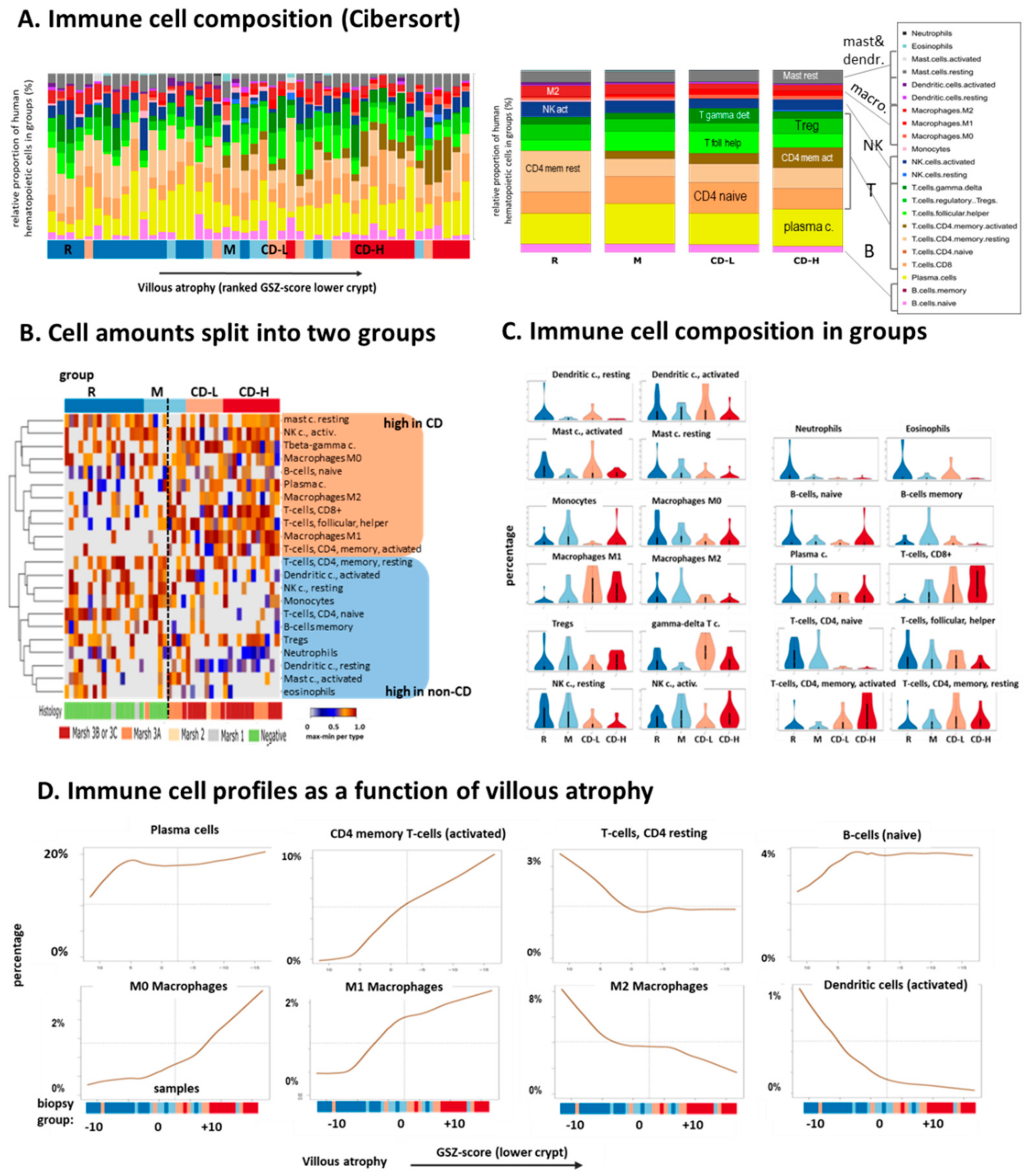
2.3. Immune Cell Infiltration
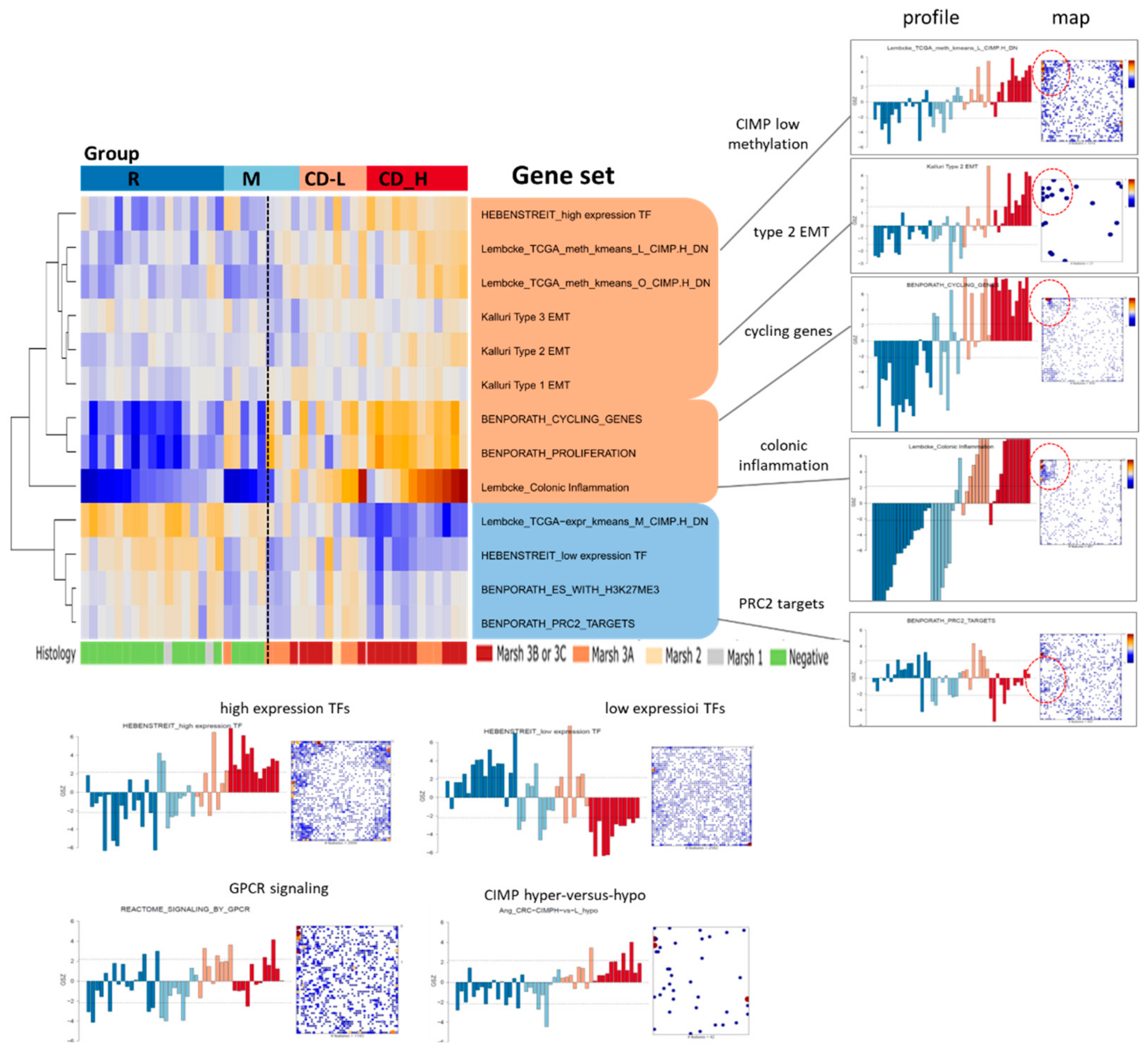
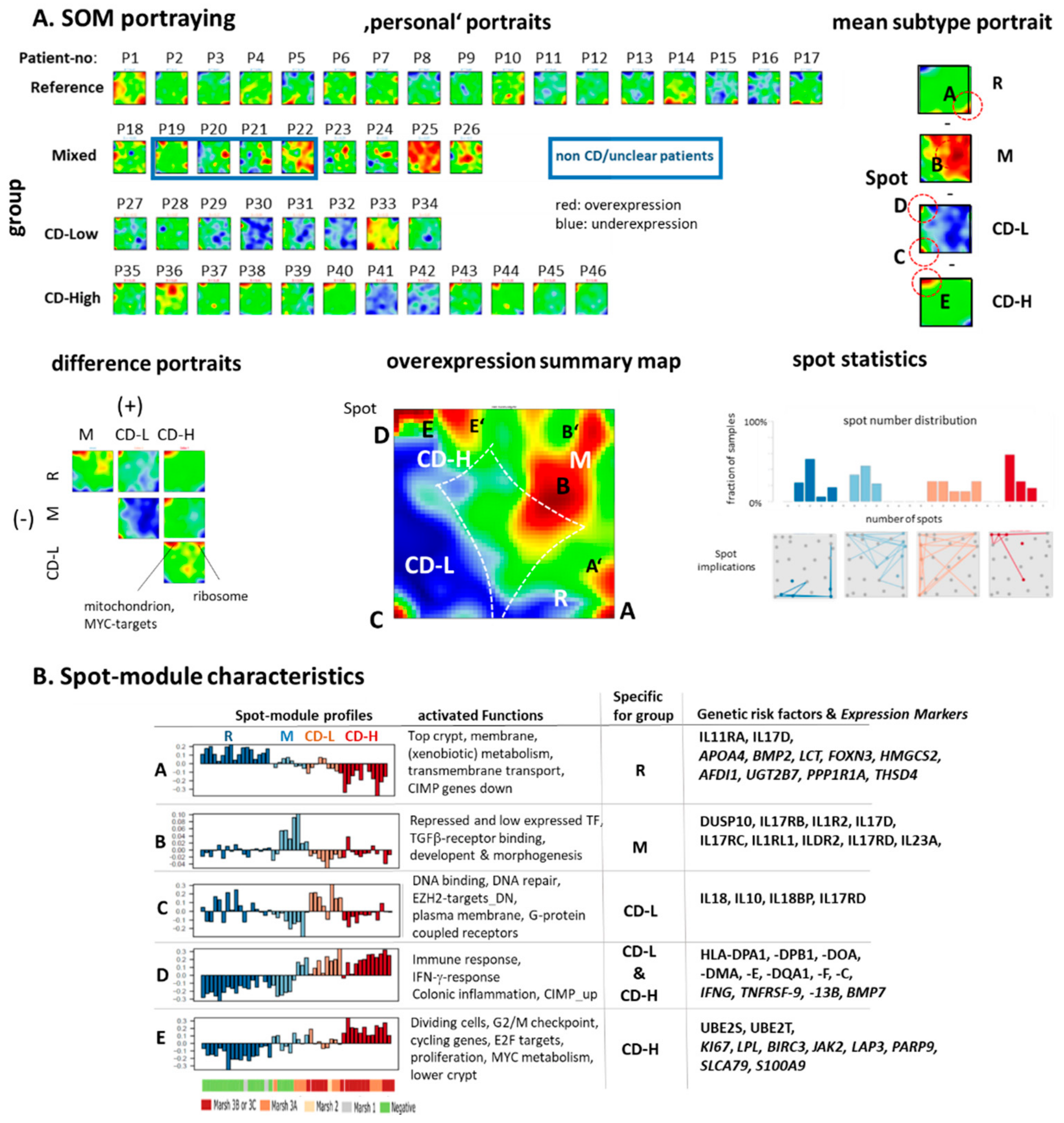
2.4. Transcriptome Portraits Dissect the Expression Landscape into Modules of Co-Regulated Genes Characterizing CD
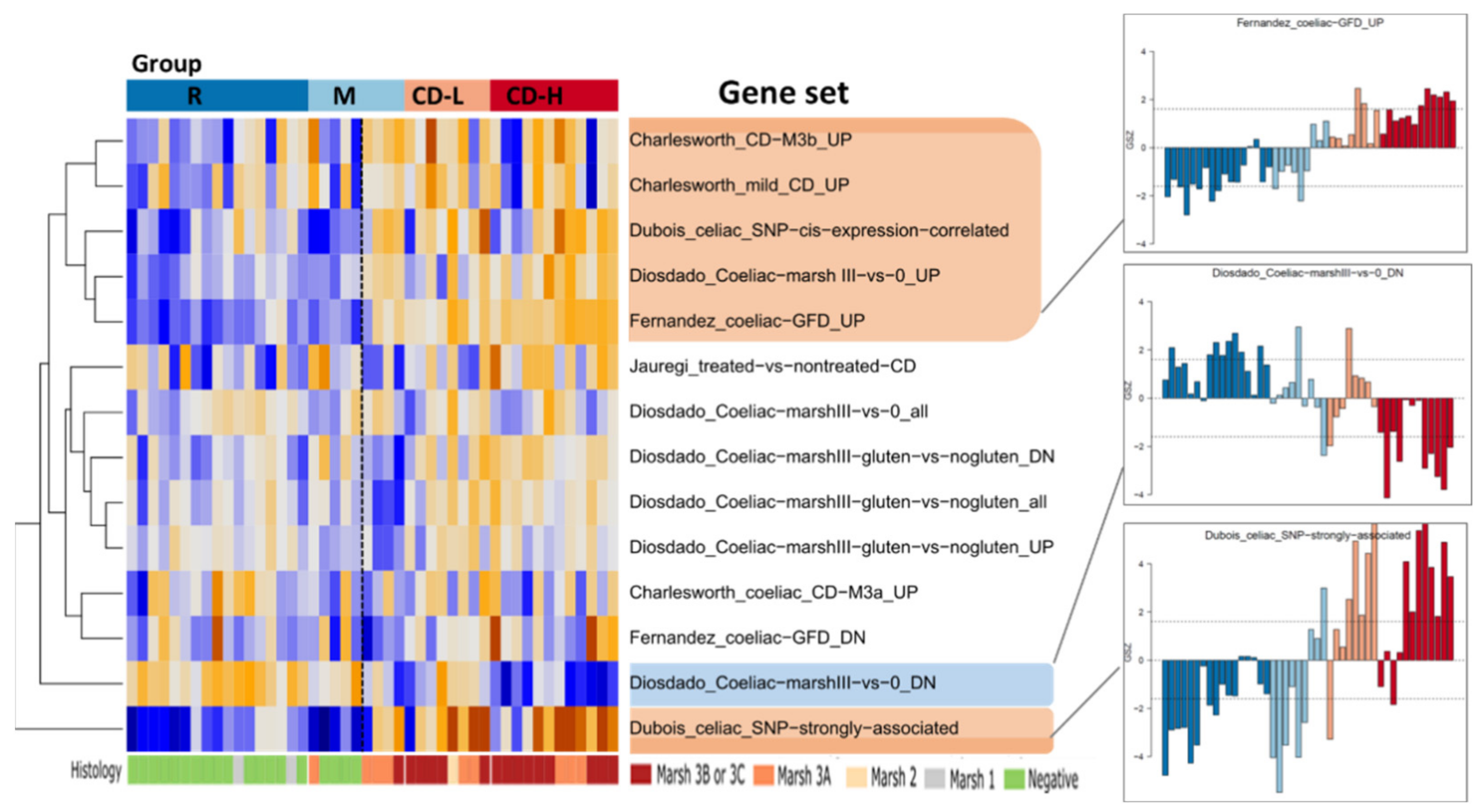
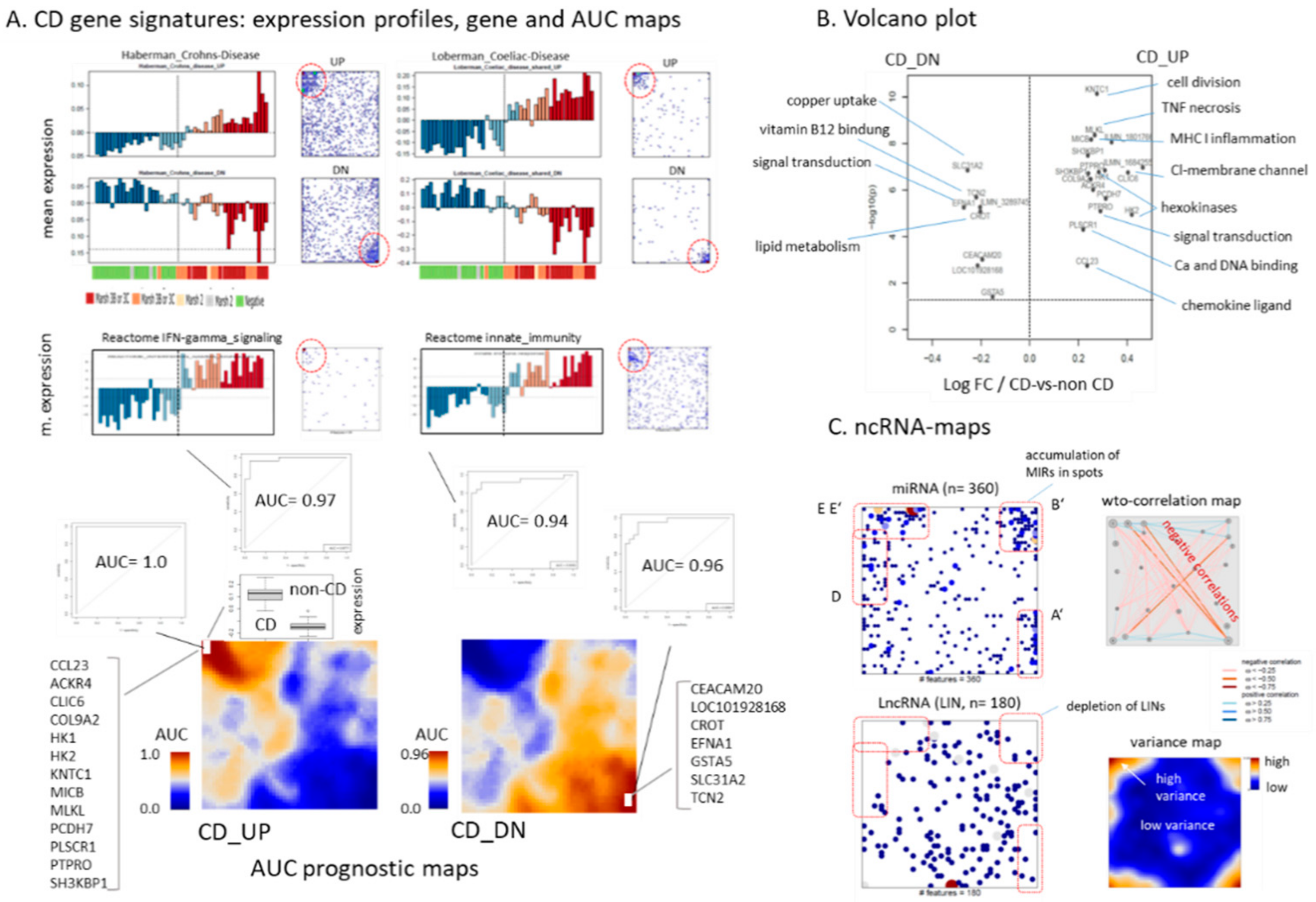
2.5. Expression Signatures and Markers of Coeliac Disease
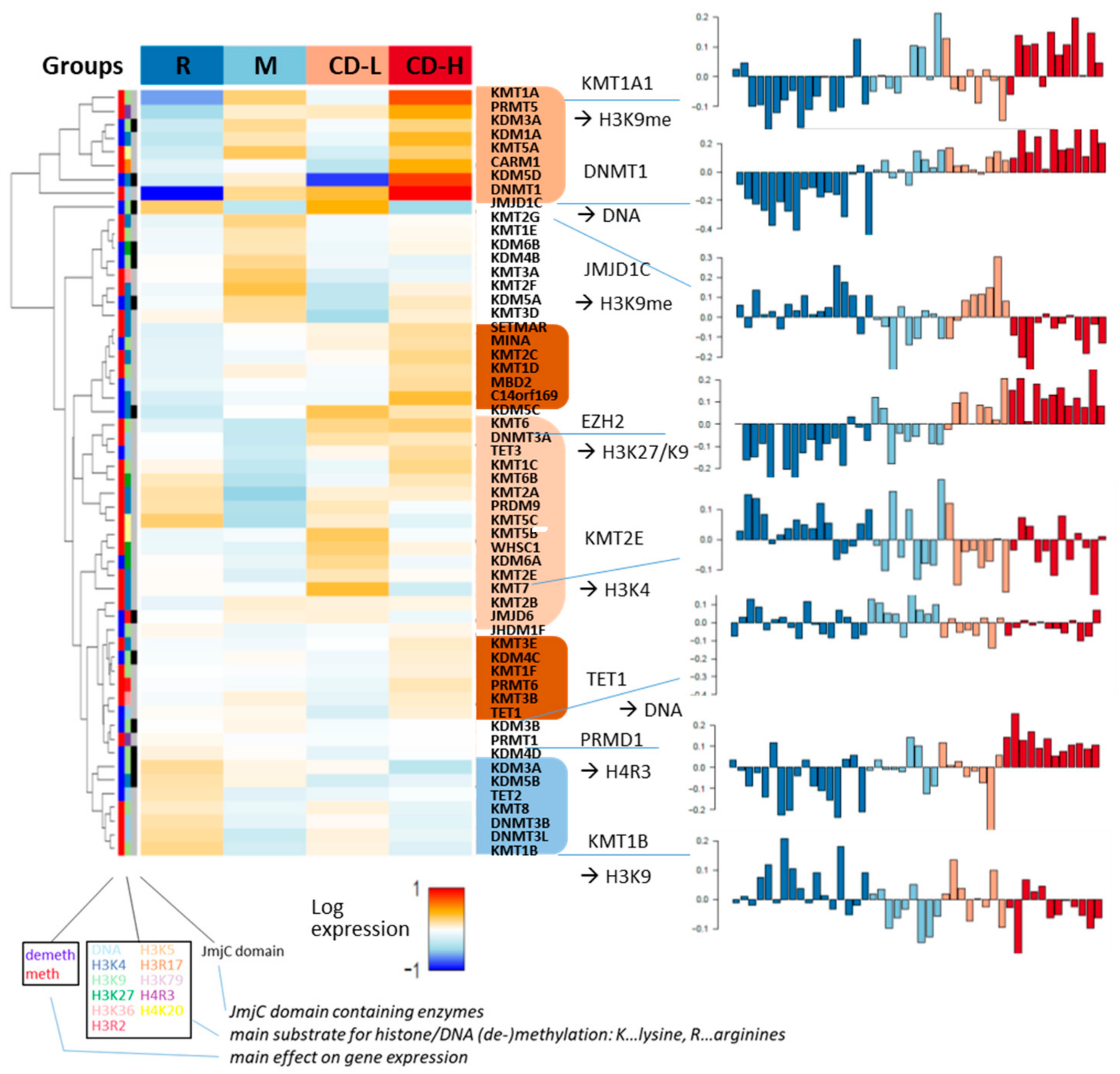
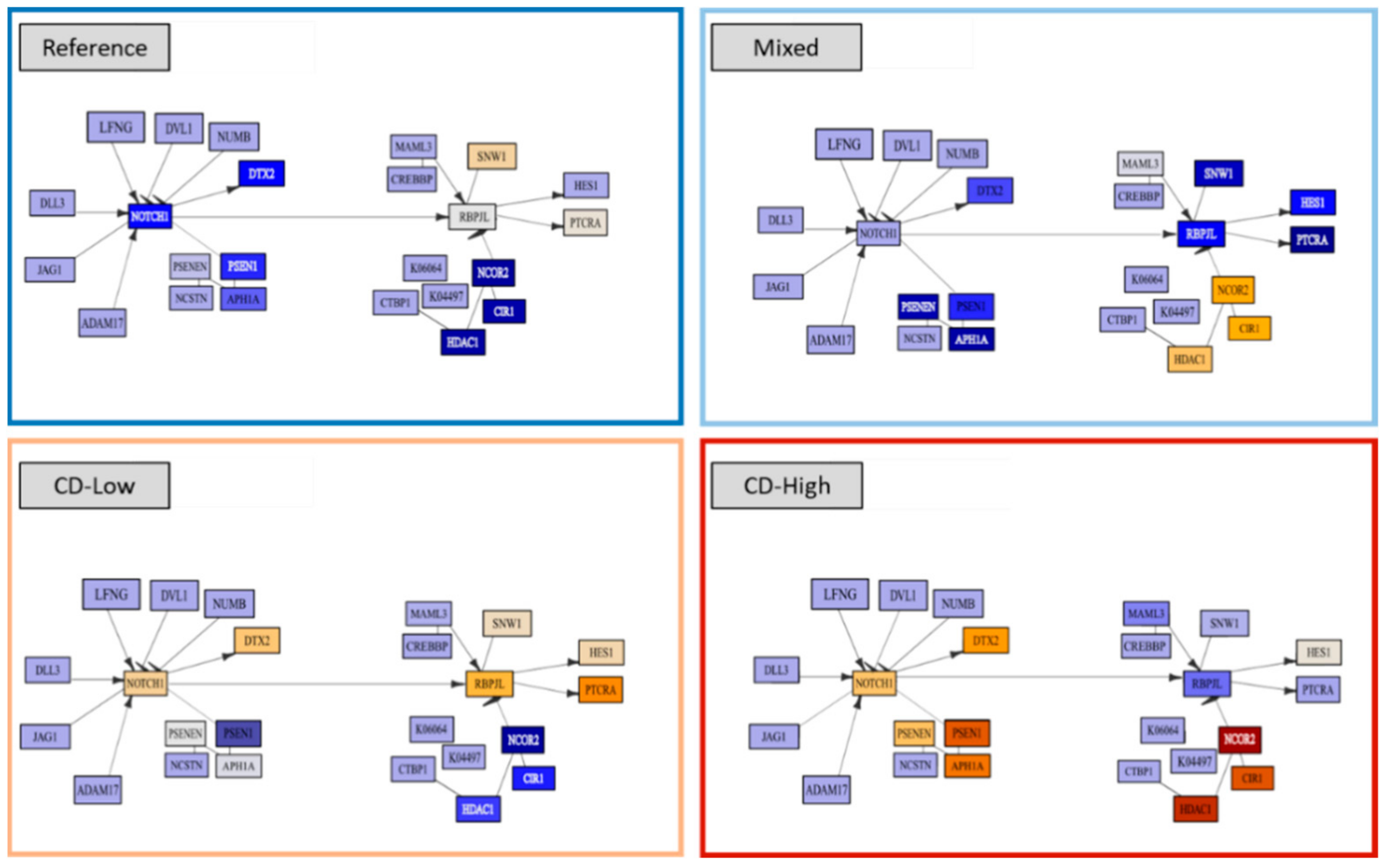
2.6. Noncoding RNAs and Epigenetics
2.7. Browsing the Transcriptome of Coeliac Biopsies
3. Discussion
4. Materials and Methods
4.1. Study Design and Patients
4.2. RNA Extraction and Microarray Measurements
4.3. Antibody Assays and HLA-Typing
4.4. Expression Analysis and SOM Portrayal
4.5. Identification of Transcriptional Subclasses
4.6. Functional Analysis
4.7. OposSOM Browser
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Spot | miRNA | LIN | |
|---|---|---|---|
| A | miR-34AHG, -555, -4260, -7114, -6778, -4469, -22, -6084, -37146125, -1248 | LINC00324 LINC00479 | down in CD |
| A’ | miR-614, -6887, -6778, -6887, -6872, -6791, -6778, -1199, -7846, -4651, -6729, -4640, -6787, -6836, -4329, -6734, -6721, -4687, -6833 | LINC-PINT LINC01547 LINC00319 LINC00909 LINC00852 | down in CD |
| D | miR-4435-2HG, -3606, -198, -5195, -1287, -4649, -5047, -1182, -568, -4720, -7704 | Up in CD | |
| E | miR-6732, -7108, -3917, -147B, -25, -664B, -4722, -147B, -636, -7107 | Up in CD |
References
- Dieli-Crimi, R.; Cénit, M.C.; Núñez, C. The genetics of celiac disease: A comprehensive review of clinical implications. J. Autoimmun. 2015, 64, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Alhassan, E.; Yadav, A.; Kelly, C.P.; Mukherjee, R. Novel Nondietary Therapies for Celiac Disease. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Kamboj, A.K.; Oxentenko, A.S. Clinical and Histologic Mimickers of Celiac Disease. Clin. Transl. Gastroenterol. 2017, 8, e114. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Arora, A.; Strand, T.A.; Leffler, D.A.; Catassi, C.; Green, P.H.; Makharia, G.K. Global Prevalence of Celiac Disease: Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2018, 16, 823–836.e2. [Google Scholar] [CrossRef] [PubMed]
- Händel, N.; Mothes, T.; Petroff, D.; Baber, R.; Jurkutat, A.; Flemming, G.; Wolf, J. Will the Real Coeliac Disease Please Stand Up? Coeliac Disease Prevalence in the German LIFE Child Study. J. Pediatric Gastroenterol. Nutr. 2018, 67, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Mubarak, A.; Spierings, E.; Wolters, V.; van Hoogstraten, I.; Kneepkens, C.M.F.; Houwen, R. Human Leukocyte Antigen DQ2.2 and Celiac Disease. J. Pediatric Gastroenterol. Nutr. 2013, 56, 428–430. [Google Scholar] [CrossRef]
- Romanos, J.; Rosén, A.; Kumar, V.; Trynka, G.; Franke, L.; Szperl, A.; Wijmenga, C. Improving coeliac disease risk prediction by testing non-HLA variants additional to HLA variants. Gut 2014, 63, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.; Kurppa, K.; Mearin, M.L.; Ribes-Koninckx, C.; Wessels, M. European Society Paediatric Gastroenterology, Hepatology and Nutrition Guidelines for Diagnosing Coeliac Disease 2020. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Montén, C.; Bjelkenkrantz, K.; Gudjonsdottir, A.H.; Browaldh, L.; Arnell, H.; Naluai, Å.T.; Agardh, D. Validity of histology for the diagnosis of paediatric coeliac disease: A Swedish multicentre study. Scand. J. Gastroenterol. 2016, 51, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.; Halvarsson, B.; Norström, F.; Myléus, A.; Carlsson, A.; Danielsson, L.; Sandström, O. Accuracy in Celiac Disease Diagnostics by Controlling the Small-bowel Biopsy Process. J. Pediatr. Gastroenterol. Nutr. 2011, 52, 549–553. [Google Scholar] [CrossRef]
- Villanacci, V.; Lorenzi, L.; Donato, F.; Auricchio, R.; Dziechciarz, P.; Gyimesi, J.; Mearin, M.L. Histopathological evaluation of duodenal biopsy in the PreventCD project. An observational interobserver agreement study. APMIS 2018, 126, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Kuitunen, P.; Kosnai, I.; Savilahti, E. Morphometric Study of the Jejunal Mucosa in Various Childhood Enteropathies with Special Reference to Intraepithelial Lymphocytes. J. Pediatr. Gastroenterol. Nutr. 1982, 1, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, R.P.G.; Agnew, L.L.; Scott, D.R.; Andronicos, N.M. Celiac disease gene expression data can be used to classify biopsies along the Marsh score severity scale. J. Gastroenterol. Hepatol. 2019, 34, 169–177. [Google Scholar] [CrossRef]
- Diosdado, B.; Wapenaar, M.C.; Franke, L.; Duran, K.J.; Goerres, M.J.; Hadithi, M.; Wijmenga, C. A microarray screen for novel candidate genes in coeliac disease pathogenesis. Gut 2004, 53, 944–951. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, B.M.; Feighery, C.; Coates, C.; O’Shea, U.; Delaney, D.; O’Briain, S.; Abuzakouk, M. The Absence of a Mucosal Lesion on Standard Histological Examination Does Not Exclude Diagnosis of Celiac Disease. Dig. Dis. Sci. 2008, 53, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Bragde, H.; Jansson, U.; Fredrikson, M.; Grodzinsky, E.; Söderman, J. Celiac disease biomarkers identified by transcriptome analysis of small intestinal biopsies. Cell. Mol. Life Sci. 2018, 75, 4385–4401. [Google Scholar] [CrossRef] [PubMed]
- Jauregi-Miguel, A.; Fernandez-Jimenez, N.; Irastorza, I.; Plaza-Izurieta, L.; Vitoria, J.C.; Bilbao, J.R. Alteration of Tight Junction Gene Expression in Celiac Disease. J. Pediatr. Gastroenterol. Nutr. 2014, 58, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Jimenez, N.; Castellanos-Rubio, A.; Plaza-Izurieta, L.; Irastorza, I.; Elcoroaristizabal, X.; Jauregi-Miguel, A.; Bilbao, J.R. Coregulation and modulation of NFκB-related genes in celiac disease: Uncovered aspects of gut mucosal inflammation. Hum. Mol. Genet. 2014, 23, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Juuti-Uusitalo, K.; Mäki, M.; Kaukinen, K.; Collin, P.; Visakorpi, T.; Vihinen, M.; Kainulainen, H. cDNA microarray analysis of gene expression in coeliac disease jejunal biopsy samples. J. Autoimmun. 2004, 22, 249–265. [Google Scholar] [CrossRef]
- Pascual, V.; Medrano, L.M.; López-Palacios, N.; Bodas, A.; Dema, B.; Fernández-Arquero, M.; Núñez, C. Different Gene Expression Signatures in Children and Adults with Celiac Disease. PLoS ONE 2016, 11, e0146276. [Google Scholar] [CrossRef] [PubMed]
- Dubois, P.C.A.; Trynka, G.; Franke, L.; Hunt, K.A.; Romanos, J.; Curtotti, A.; van Heel, D.A. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010, 42, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Wapenaar, M.C.; van Belzen, M.J.; Fransen, J.H.; Fariña Sarasqueta, A.; Houwen, R.H.J.; Meijer, J.W.R.; Wijmenga, C. The interferon gamma gene in celiac disease: Augmented expression correlates with tissue damage but no evidence for genetic susceptibility. J. Autoimmun. 2004, 23, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Sánchez, A.D.; Tan, I.L.; Gonera-de Jong, B.C.; Visschedijk, M.C.; Jonkers, I.; Withoff, S. Molecular Biomarkers for Celiac Disease: Past, Present and Future. Int. J. Mol. Sci. 2020, 21, 8528. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Hopp, L.; Arakelyan, A.; Kirsten, H.; Engel, C.; Wirkner, K.; Binder, H. The Human Blood Transcriptome in a Large Population Cohort and Its Relation to Aging and Health. Front. Big Data 2020, 3, 36. [Google Scholar] [CrossRef]
- Binder, H.; Willscher, E.; Loeffler-Wirth, H.; Hopp, L.; Jones, D.T.W.; Pfister, S.M.; Loeffler, M. DNA methylation, transcriptome and genetic copy number signatures of diffuse cerebral WHO grade II/III gliomas resolve cancer heterogeneity and development. Acta Neuropathol. Commun. 2019, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Hopp, L.; Loeffler-Wirth, H.; Nersisyan, L.; Arakelyan, A.; Binder, H. Footprints of Sepsis Framed Within Community Acquired Pneumonia in the Blood Transcriptome. Front. Immunol. 2018, 9, 1620. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Hopp, L.; Lembcke, K.; Wirth, H. Personalized Disease Phenotypes from Massive OMICs Data. In Big Data Analytics in Bioinformatics and Healthcare; Baoying, W., Ruowang, L., William, P., Eds.; IGI Global: Hershey, PA, USA, 2015; pp. 359–378. [Google Scholar]
- Olsen, J.; Gerds, T.A.; Seidelin, J.B.; Csillag, C.; Bjerrum, J.T.; Troelsen, J.T.; Nielsen, O.H. Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm. Bowel Dis. 2009, 15, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Mesirov, J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Angelova, M.; Charoentong, P.; Hackl, H.; Fischer, M.L.; Snajder, R.; Krogsdam, A.M.; Trajanoski, Z. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ciccocioppo, R.; D’Alo, S.; Di Sabatino, A.; Parroni, R.; Rossi, M.; Doglioni, C.; Corazza, G.R. Mechanisms of villous atrophy in autoimmune enteropathy and coeliac disease. Clin. Exp. Immunol. 2002, 128, 88–93. [Google Scholar] [CrossRef]
- Kosinski, C.; Li, V.S.W.; Chan, A.S.Y.; Zhang, J.; Ho, C.; Tsui, W.Y.; Chen, X. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl. Acad. Sci. USA 2007, 104, 15418–15423. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, N.S. Non–Gluten Sensitivity–Related Small Bowel Villous Flattening With Increased Intraepithelial Lymphocytes: Not All That Flattens Is Celiac Sprue. Am. J. Clin. Pathol. 2004, 121, 546–550. [Google Scholar] [CrossRef][Green Version]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- LaPointe, L.C.; Dunne, R.; Brown, G.S.; Worthley, D.L.; Molloy, P.L.; Wattchow, D.; Young, G.P. Map of differential transcript expression in the normal human large intestine. Physiol. Genom. 2008, 33, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453. [Google Scholar] [CrossRef] [PubMed]
- López Casado, M.Á.; Lorite, P.; Ponce de León, C.; Palomeque, T.; Torres, M.I. Celiac Disease Autoimmunity. Arch. Immunol. Et Ther. Exp. 2018, 66, 423–430. [Google Scholar] [CrossRef]
- Valle, J.; Morgado, J.M.T.; Ruiz-Martín, J.; Guardiola, A.; Lopes-Nogueras, M.; García-Vela, A.; Sánchez-Muñoz, L. Flow cytometry of duodenal intraepithelial lymphocytes improves diagnosis of celiac disease in difficult cases. United Eur. Gastroenterol. J. 2017, 5, 819–826. [Google Scholar] [CrossRef]
- Nijeboer, P.; van Gils, T.; Reijm, M.; Ooijevaar, R.; Lissenberg-Witte, B.I.; Bontkes, H.J.; Bouma, G. Gamma-Delta T Lymphocytes in the Diagnostic Approach of Coeliac Disease. J. Clin. Gastroenterol. 2019, 53, e208–e213. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Newell, E.W.; Glanville, J.; Fernandez-Becker, N.; Khosla, C.; Chien, Y.; Davis, M.M. Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proc. Natl. Acad. Sci. USA 2013, 110, 13073–13078. [Google Scholar] [CrossRef]
- Marafini, I.; Imeneo, M.; Monteleone, G. The Role of Natural Killer Receptors in Celiac Disease. Immunome Res. 2017, 13, 1–2. [Google Scholar] [CrossRef]
- Jaguin, M.; Houlbert, N.; Fardel, O.; Lecureur, V. Polarization profiles of human M-CSF-generated macrophages and comparison of M1-markers in classically activated macrophages from GM-CSF and M-CSF origin. Cell. Immunol. 2013, 281, 51–61. [Google Scholar] [CrossRef]
- Serena, G.; Huynh, D.; Lima, R.S.; Vise, L.M.; Freire, R.; Ingano, L.; Fasano, A. Intestinal Epithelium Modulates Macrophage Response to Gliadin in Celiac Disease. Front. Nutr. 2019, 6, 167. [Google Scholar] [CrossRef] [PubMed]
- Hebenstreit, D.; Fang, M.; Gu, M.; Charoensawan, V.; van Oudenaarden, A.; Teichmann, S.A. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol. Syst. Biol. 2011, 7, 497. [Google Scholar] [CrossRef] [PubMed]
- Dotsenko, V.; Oittinen, M.; Taavela, J.; Popp, A.; Peräaho, M.; Staff, S.; Viiri, K. Genome-Wide Transcriptomic Analysis of Intestinal Mucosa in Celiac Disease Patients on a Gluten-Free Diet and Postgluten Challenge. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 13–32. [Google Scholar] [CrossRef] [PubMed]
- Taavela, J.; Viiri, K.; Popp, A.; Oittinen, M.; Dotsenko, V.; Peräaho, M.; Isola, J. Histological, immunohistochemical and mRNA gene expression responses in coeliac disease patients challenged with gluten using PAXgene fixed paraffin-embedded duodenal biopsies. BMC Gastroenterol. 2019, 19, 189. [Google Scholar] [CrossRef]
- Tosi, R.; Vismara, D.; Tanigaki, N.; Ferrara, G.B.; Cicimarra, F.; Buffolano, W.; Auricchio, S. Evidence that celiac disease is primarily associated with a DC locus allelic specificity. Clin. Immunol. Immunopathol. 1983, 28, 395–404. [Google Scholar] [CrossRef]
- Sollid, L.M.; Markussen, G.; Ek, J.; Gjerde, H.; Vartdal, F.; Thorsby, E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J. Exp. Med. 1989, 169, 345–350. [Google Scholar] [CrossRef]
- Östensson, M.; Montén, C.; Bacelis, J.; Gudjonsdottir, A.H.; Adamovic, S.; Ek, J.; Torinsson-Naluai, Å. A Possible Mechanism behind Autoimmune Disorders Discovered By Genome-Wide Linkage and Association Analysis in Celiac Disease. PLoS ONE 2013, 8, e70174. [Google Scholar] [CrossRef]
- Garner, C.P.; Murray, J.A.; Ding, Y.C.; Tien, Z.; van Heel, D.A.; Neuhausen, S.L. Replication of celiac disease UK genome-wide association study results in a US population. Hum. Mol. Genet. 2009, 18, 4219–4225. [Google Scholar] [CrossRef]
- Loberman-Nachum, N.; Sosnovski, K.; Di Segni, A.; Efroni, G.; Braun, T.; BenShoshan, M.; Haberman, Y. Defining the Celiac Disease Transcriptome using Clinical Pathology Specimens Reveals Biologic Pathways and Supports Diagnosis. Sci. Rep. 2019, 9, 16163. [Google Scholar] [CrossRef] [PubMed]
- Haberman, Y.; BenShoshan, M.; Di Segni, A.; Dexheimer, P.J.; Braun, T.; Weiss, B.; Denson, L.A. Long ncRNA Landscape in the Ileum of Treatment-Naive Early-Onset Crohn Disease. Inflamm. Bowel Dis. 2018, 24, 346–360. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Wirth, H.; Çakir, M.; Hopp, L.; Binder, H. Analysis of MicroRNA Expression Using Machine Learning. Methods Mol. Biol. 2014, 1107, 257–278. [Google Scholar] [PubMed]
- Felli, C.; Baldassarre, A.; Masotti, A. Intestinal and Circulating MicroRNAs in Coeliac Disease. Int. J. Mol. Sci. 2017, 18, 1907. [Google Scholar] [CrossRef]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Thalheim, T.; Hopp, L.; Binder, H.; Aust, G.; Galle, J. On the Cooperation between Epigenetics and Transcription Factor Networks in the Specification of Tissue Stem Cells. Epigenomes 2018, 2, 20. [Google Scholar] [CrossRef]
- Hopp, L.; Nersisyan, L.; Löffler-Wirth, H.; Arakelyan, A.; Binder, H. Epigenetic Heterogeneity of B-Cell Lymphoma: Chromatin Modifiers. Genes 2015, 6, 1076. [Google Scholar] [CrossRef]
- Ang, P.; Loh, M.; Liem, N.; Lim, P.; Grieu, F.; Vaithilingam, A.; Soong, R. Comprehensive profiling of DNA methylation in colorectal cancer reveals subgroups with distinct clinicopathological and molecular features. BMC Cancer 2010, 10, 227. [Google Scholar] [CrossRef]
- Fernandez-Jimenez, N.; Garcia-Etxebarria, K.; Plaza-Izurieta, L.; Romero-Garmendia, I.; Jauregi-Miguel, A.; Legarda, M.; Bilbao, J.R. The methylome of the celiac intestinal epithelium harbours genotype-independent alterations in the HLA region. Sci. Rep. 2019, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- Loeffler-Wirth, H.; Reikowski, J.; Hakobyan, S.; Wagner, J.; Binder, H. oposSOM-Browser: An interactive tool to explore omics data landscapes in health science. BMC Bioinform. 2020, 21, 465. [Google Scholar] [CrossRef]
- Nersisyan, L.; Loeffler-Wirth, H.; Arakelyan, A.; Binder, H. Gene set- and pathway- centered knowledge discovery assigns transcriptional activation patterns in brain, blood and colon cancer—A bioinformatics perspective. J. Bioinform. Knowl. Min. 2016, 4, 46–70. [Google Scholar]
- Atlasy, N.; Bujko, A.; Brazda, P.B.; Janssen-Megens, E.; Bækkevold, E.S.; Jahnsen, J.; Stunnenberg, H.G. Single cell transcriptome atlas of immune cells in human small intestine and in celiac disease. bioRxiv 2019, 721258. [Google Scholar] [CrossRef]
- Wolf, J.; Petroff, D.; Richter, T.; Auth, M.K.H.; Uhlig, H.H.; Laass, M.W.; Mothes, T. Validation of Antibody-Based Strategies for Diagnosis of Pediatric Celiac Disease without Biopsy. Gastroenterology 2017, 153, 410–419.e17. [Google Scholar] [CrossRef] [PubMed]
- Löffler-Wirth, H.; Kalcher, M.; Binder, H. oposSOM: R-package for high-dimensional portraying of genome-wide expression landscapes on bioconductor. Bioinformatics 2015, 31, 3225–3227. [Google Scholar] [CrossRef]
- Wirth, H.; Löffler, M.; von Bergen, M.; Binder, H. Expression cartography of human tissues using self organizing maps. BMC Bioinform. 2011, 12, 306. [Google Scholar] [CrossRef]
- Hopp, L.; Wirth, H.; Fasold, M.; Binder, H. Portraying the expression landscapes of cancer subtypes: A glioblastoma multiforme and prostate cancer case study. Syst. Biomed. 2013, 1, 99–121. [Google Scholar] [CrossRef]
- Kunz, M.; Löffler-Wirth, H.; Dannemann, M.; Willscher, E.; Doose, G.; Kelso, J.; Schartl, M. RNA-seq analysis identifies different transcriptomic types and developmental trajectories of primary melanomas. Oncogene 2018, 37, 6136–6151. [Google Scholar] [CrossRef]
- Toronen, P.; Ojala, P.; Marttinen, P.; Holm, L. Robust extraction of functional signals from gene set analysis using a generalized threshold free scoring function. BMC Bioinform. 2009, 10, 307. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef]

| Patient Group | Reference | Mixed | CD-Low | CD-High |
|---|---|---|---|---|
| N (cases) | 17 | 9 | 8 | 12 |
| Diagnosis | ||||
| Coeliac disease | 0 (0.0%) | 5 (55.5%) | 8 (100%) | 12 (100%) |
| Disease controls | 17 (100%) | 3 (33.3%) | 0 (0.0%) | 0 (0.0%) |
| Unclear | 0 (0.0%) | 1 (14.2%) | 0 (0.0%) | 0 (0.0%) |
| Age | ||||
| Median (range) | 14 y (1–17) | 10 y (3–12) | 8 y (3–17) | 13 y (4–15) |
| Age interval | ||||
| 1y–4y | 3 (17.6%) | 1 (11.1%) | 2 (25.0%) | 5 (41.7%) |
| 5y–8y | 1 (5.9%) | 2 (22.2%) | 1 (12.5%) | 1 (8.3%) |
| 9y–12y | 3 (17.6%) | 6 (66.7%) | 1 (12.5%) | 1 (8.3%) |
| 13y–17y | 10 (58.5%) | 0 (0.0%) | 4 (50.0%) | 5 (41.7%) |
| Sex | ||||
| Female | 13 (76.5%) | 6 (66.7%) | 7 (87.5%) | 5 (41.7%) |
| Gastrointestinal complaints | ||||
| at least one sign of clear malabsorption (1) asymptomatic (2) | 17 (100%) | 8 (88.9%) | 6 (75.0%) | 9 (75%) |
| 0 (0.0%) | 3 (33.3%) | 3 (37.5%) | 7 (58.3%) | |
| 0 (0.0%) | 0 (0.0%) | 1 (12.5%) | 3 (25.0%) | |
| Symptoms since(3) | ||||
| <3 months | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (8.3%) |
| 3–24 months | 11 (64.7%) | 4 (4.44%) | 4 (50.0%) | 5 (41.7%) |
| >24 months | 5 (29.4%) | 4 (44.4%) | 2 (25.0%) | 3 (25%) |
| IgA-aTTG | ||||
| positive | 1 (5.9%) | 6 (66.7%) | 8 (100%) | 11 (91.7%) (4) |
| IgG-EmA | ||||
| Positive | 1 (5.9%) | 6 (66.7%) | 8 (100%) | 12 (100%) |
| IgG-aDGL | ||||
| positive | 1 (5.9%) | 4 (44.4%) | 6 (75.0%) | 11 (91.7%) |
| HLA-Type | ||||
| negative | 10 (58.8%) | 2 (22.2%) | 0 (0.0%) | 0 (0.0%) |
| DQ2 (5) | 7 (41.2%) | 6 (66.7%) | 7 (87.5%) | 11 (91.7%) |
| DQ8 | 0 (0.0%) | 1 (11.1%) | 1 (12.5%) | 1 (8.3%) |
| DQ2 and DQ8 | 1 (5.9%) | 0 (0.0%) | 0 (0.0%) | 1 (8.3%) |
| Marsh stage | ||||
| Normal | 15 (88.2%) | 4 (44.4%) | 0 (0.0%) | 0 (0.0%) |
| Marsh 1 | 2 (11.8%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Marsh 2 | 0 (0.0%) | 0 (0.0%) | 1 (12.5%) | 0 (0.0%) |
| Marsh 3A | 0 (0.0%) | 4 (44.4%) | 2 (25.0%) | 3 (25.0%) |
| Marsh 3B/C | 0 (0.0%) | 1 (11.1%) | 5 (62.5%) | 9 (75.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolf, J.; Willscher, E.; Loeffler-Wirth, H.; Schmidt, M.; Flemming, G.; Zurek, M.; Uhlig, H.H.; Händel, N.; Binder, H. Deciphering the Transcriptomic Heterogeneity of Duodenal Coeliac Disease Biopsies. Int. J. Mol. Sci. 2021, 22, 2551. https://doi.org/10.3390/ijms22052551
Wolf J, Willscher E, Loeffler-Wirth H, Schmidt M, Flemming G, Zurek M, Uhlig HH, Händel N, Binder H. Deciphering the Transcriptomic Heterogeneity of Duodenal Coeliac Disease Biopsies. International Journal of Molecular Sciences. 2021; 22(5):2551. https://doi.org/10.3390/ijms22052551
Chicago/Turabian StyleWolf, Johannes, Edith Willscher, Henry Loeffler-Wirth, Maria Schmidt, Gunter Flemming, Marlen Zurek, Holm H. Uhlig, Norman Händel, and Hans Binder. 2021. "Deciphering the Transcriptomic Heterogeneity of Duodenal Coeliac Disease Biopsies" International Journal of Molecular Sciences 22, no. 5: 2551. https://doi.org/10.3390/ijms22052551
APA StyleWolf, J., Willscher, E., Loeffler-Wirth, H., Schmidt, M., Flemming, G., Zurek, M., Uhlig, H. H., Händel, N., & Binder, H. (2021). Deciphering the Transcriptomic Heterogeneity of Duodenal Coeliac Disease Biopsies. International Journal of Molecular Sciences, 22(5), 2551. https://doi.org/10.3390/ijms22052551








