Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme
Abstract
1. Introduction
2. Results and Discussion
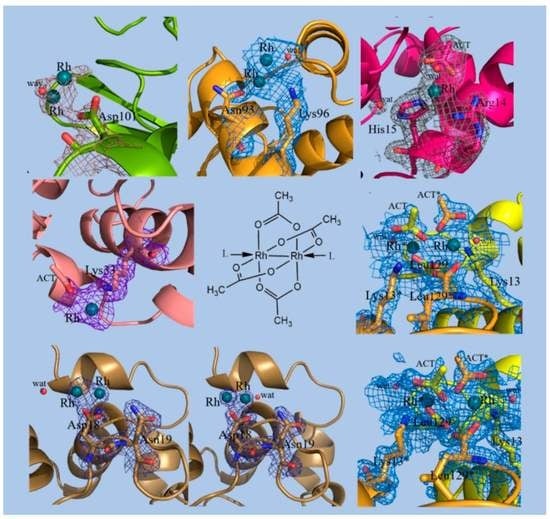
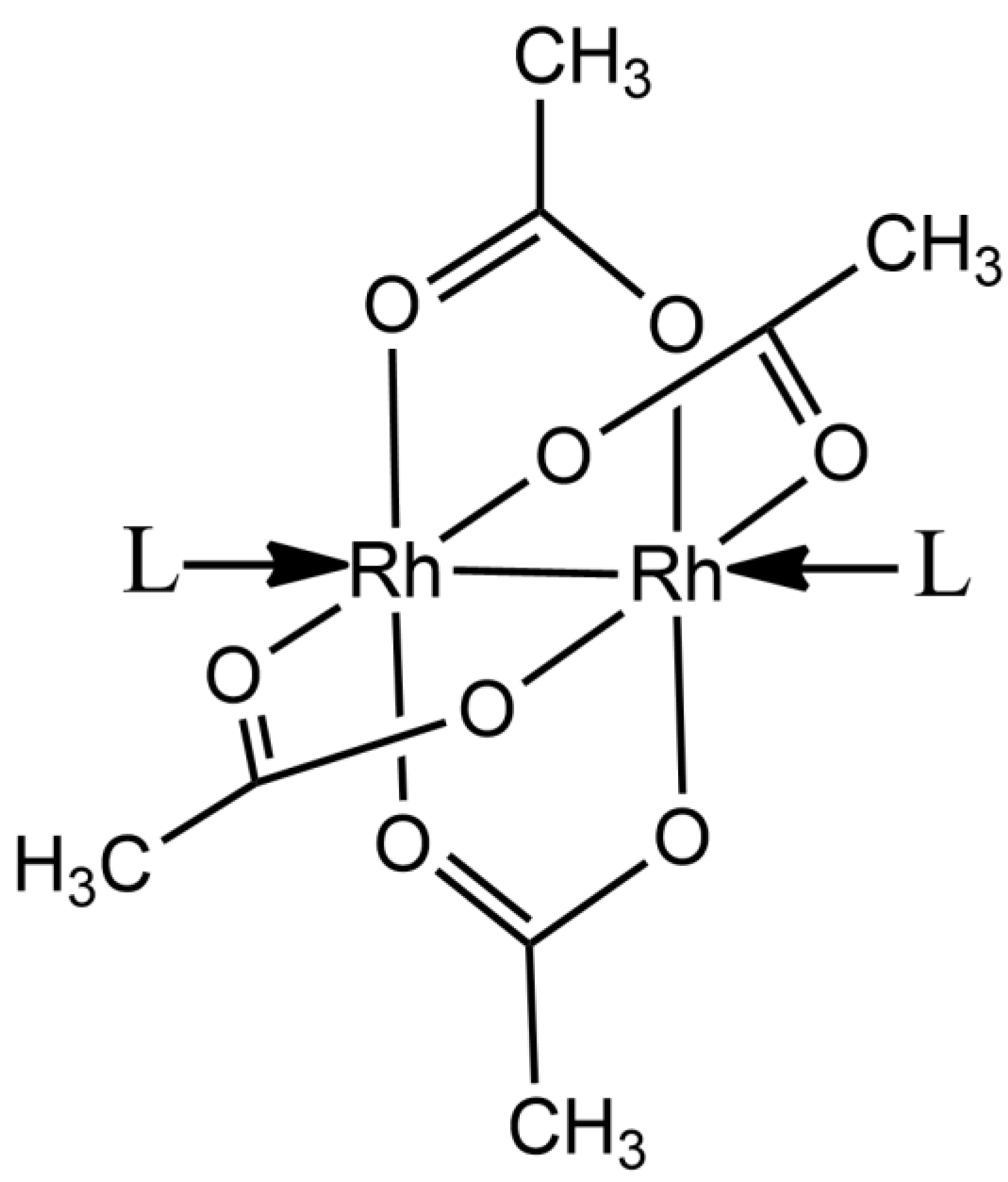
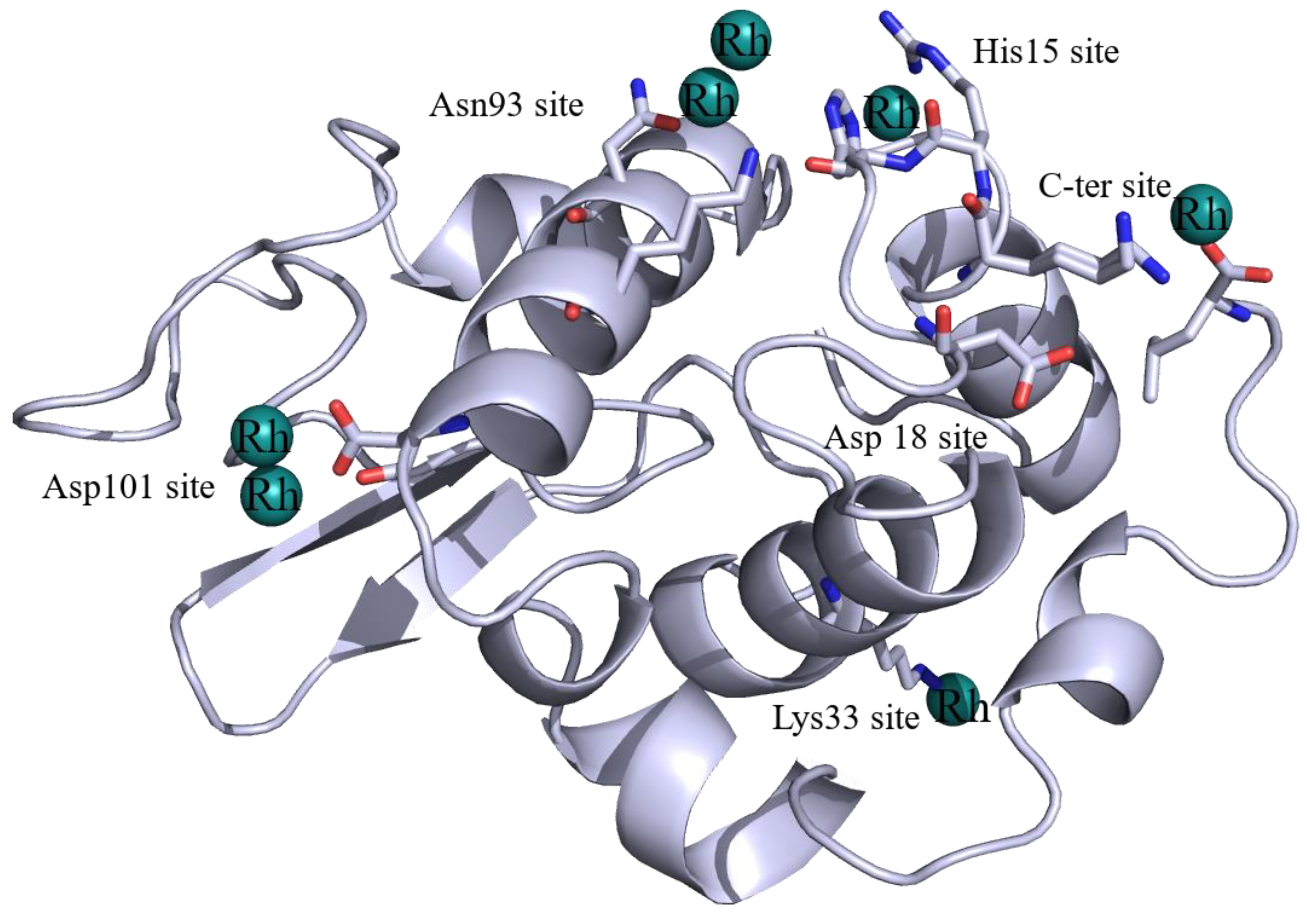
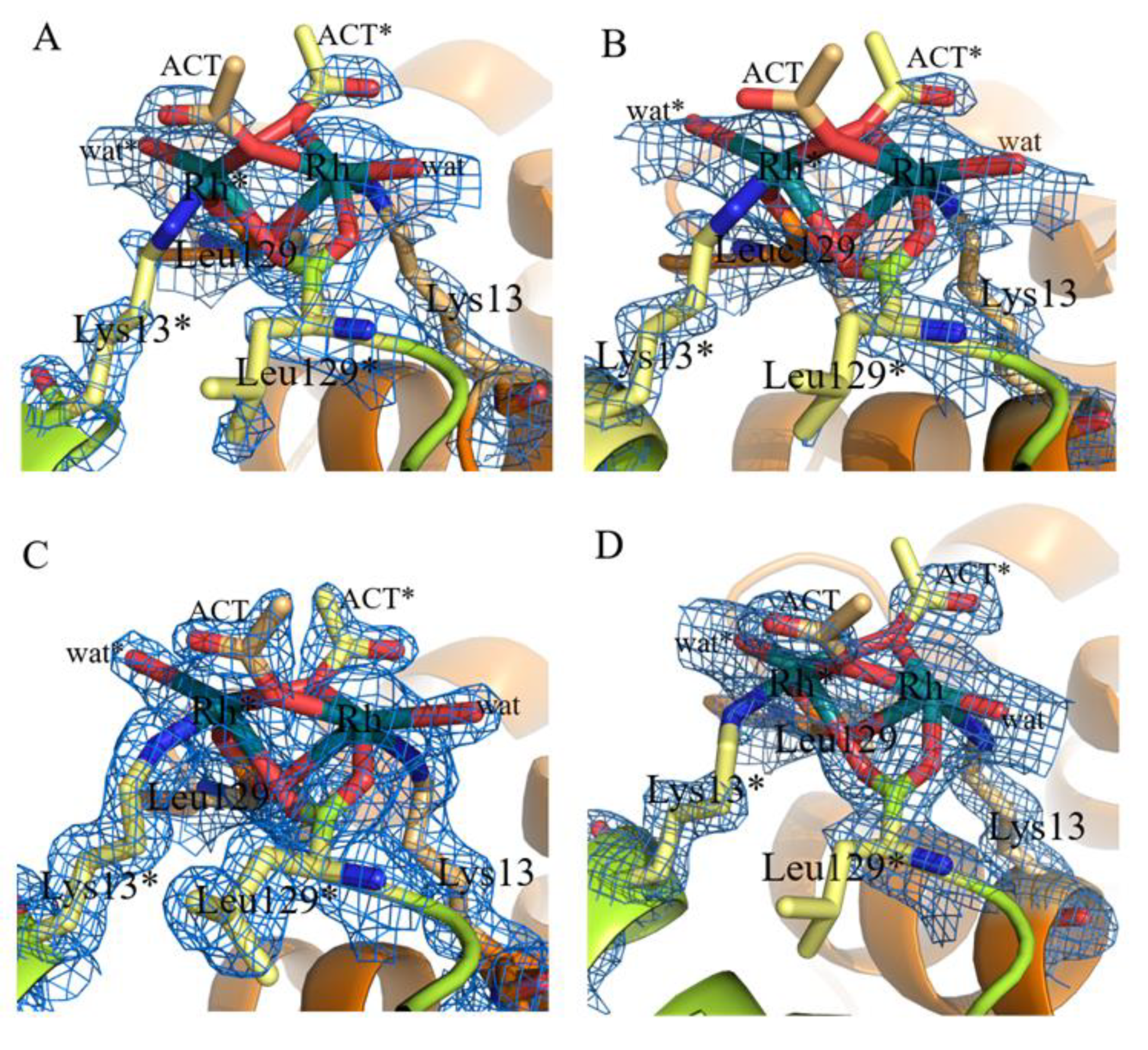
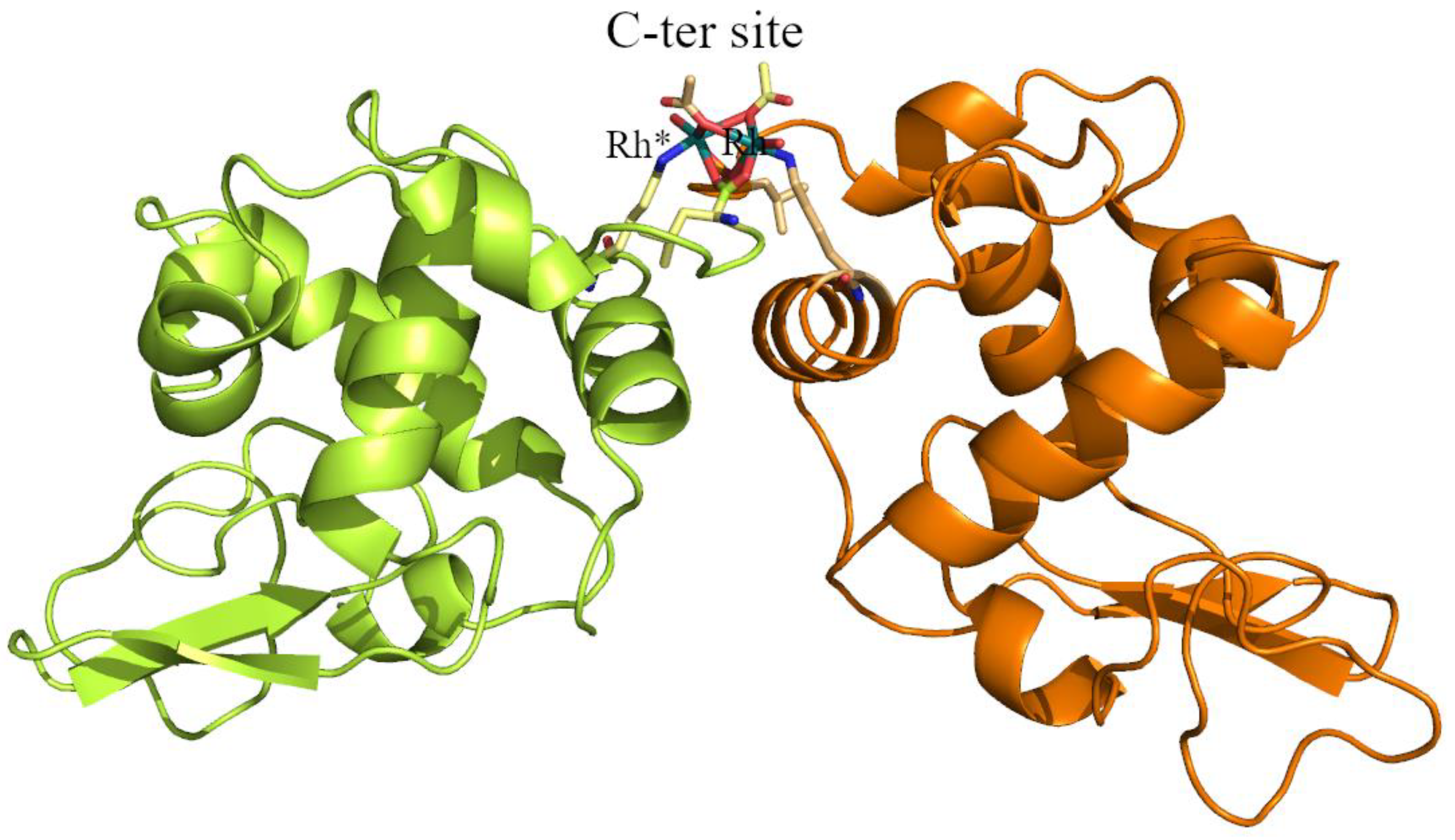
2.1. Structures of the Rh/HEWL Adducts Obtained by Cocrystallization
2.2. Structures of the Rh/HEWL Adduct Obtained by Soaking Procedure
2.3. Comparison with Literature Data
3. Materials and Methods
3.1. Preparation, Characterization in Solution and Crystallization of the Adduct Formed Upon Reaction of [Rh2(μ-O2CCH3)4] with HEWL
3.2. Data Collection and Refinement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | Circular dichroism |
| HEWL | Hen egg white lysozyme |
| HSA | Human serum albumin |
| PDB | Protein Data Bank |
| Rmsd | Root mean square deviation |
| RNase A | Bovine pancreatic ribonuclease |
| SDS | Sodium dodecyl sulfate |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| UV | Ultraviolet |
| UV–Vis | Ultraviolet–visible |
References
- Paulissenen, R.; Reimlinger, H.; Hayez, E.; Hubert, A.J.; Teyssie, P. Transition metal catalysed reactions of diazocompounds—II insertion in the hydroxylic bond. Tetrahedron. Lett. 1973, 14, 2233–2236. [Google Scholar] [CrossRef]
- Breslow, R.; Gellman, S.H. Intramolecular nitrene carbon-hydrogen insertions mediated by transition-metal complexes as nitrogen analogs of cytochrome P-450 reactions. J. Am. Chem. Soc. 1983, 105, 6728–6729. [Google Scholar] [CrossRef]
- Davies, H.M.L.; Beckwith, R.E. Catalytic Enantioselective C−H Activation by Means of Metal−Carbenoid-Induced C−H Insertion. Chem. Rev. 2003, 103, 2861–2904. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.M.L.; Manning, J.R. Catalytic C–H functionalization by metal carbenoid and nitrenoid insertion. Nature 2008, 451, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kuang, Y.; Wang, Z.; Zhu, J.; Wang, Y. Dirhodium(II)-catalyzed [3+2] cycloaddition of N-arylaminocyclopropane with alkyne derivatives. Beilstein. J. Org. Chem. 2019, 15, 542–550. [Google Scholar] [CrossRef]
- Doyle, M.P.; High, K.G.; Nesloney, C.L.; Clayton, T.W., Jr.; Lin, J. Rhodium(II) perfluorobutyrate catalyzed hydrosilylation of 1-alkynes. Trans addition and rearrangement to allylsilanes. Organometallics 1991, 10, 1225–1226. [Google Scholar] [CrossRef]
- Doyle, M.P.; Devora, G.A.; Nefedov, A.O.; High, K.G. Addition/elimination in the rhodium (II) perfluorobutyrate catalyzed hydrosilylation of 1-alkenes. Rhodium hydride promoted isomerization and hydrogenation. Organometallics 1992, 11, 549–555. [Google Scholar]
- Doyle, M.P.; Terpstra, J.W.; Winter, C.H.; Griffin, J.H. Rhodium (II) acetate catalyzed hydrocarbon oxidations by molecular oxygen. J. Mol. Catal. 1984, 26, 259–266. [Google Scholar] [CrossRef]
- Kataoka, Y.; Yano, N.; Hand, M.; Kawamoto, T. Intrinsic hydrogen evolution capability and a theoretically supported reaction mechanism of a paddlewheel-type dirhodium complex. Dalton. Trans. 2019, 48, 7302–7312. [Google Scholar] [CrossRef]
- Zyngier, S.; Kimura, E.; Najjar, R. Antitumor effects of rhodium(II) citrate in mice bearing Ehrlich tumors. Braz. J. Med. Biol. Res. 1989, 22, 397–401. [Google Scholar]
- Reibscheid, E.M.; Zyngier, S.; Maria, D.A.; Mistrone, R.J.; Sinisterra, R.D.; Couto, L.G.; Najjar, R. Antitumor effects of rhodium(II) complexes on mice bearing Ehrlich tumors. Braz. J. Med. Biol. Res. 1994, 27, 91–94. [Google Scholar] [PubMed]
- Bear, J.L.; Gray, H.B., Jr.; Rainen, L.; Chang, I.M.; Howard, R.; Serio, G.; Kimball, A.P. Interaction of rhodium(II) carboxylates with molecules of biologic importance. Cancer Chemother. Rep. 1975, 59, 611–620. [Google Scholar] [PubMed]
- Chang, I.; Woo, W.S. Effects of Rh2(O2CC2H5)4L2 on the replication of Ehrlich tumor cells in Vivo. Korean Biochem. J. 1976, 9, 175–180. [Google Scholar]
- Nothenberg, M.S.; Zyngier, S.B.; Giesbrecht, A.M.; Gambardella, M.T.P.; Santos, R.H.A.; Kimura, E.; Najjar, R. Biological activity and crystallographic study of a rhodium propionate-metronidazole adduct. J. Braz. Chem. Soc. 1994, 5, 23–29. [Google Scholar] [CrossRef]
- Bear, J.L. Rhodium compounds for antitumor use, in Precious Met. Proc. Jnt. Precious Met. Inst. Conf. 1986, 337–344. [Google Scholar]
- Hughes, R.G.; Bear, J.L.; Kimball, A.P. Synergistic effect of rhodium acetate and arabinosylcytosine on L1210. Proc. Am. Assoc. Cancer Res. 1972, 13, 120. [Google Scholar]
- Howard, R.A.; Kimball, A.P.; Bear, J.L. Mechanism of action of tetra-carboxylatodirhodium(II) in L1210 tumor suspension culture. Cancer Res. 1979, 39, 2568–2573. [Google Scholar]
- Erck, A.; Rainen, L.; Whileyman, J.; Chang, I.M.; Kimball, A.P.; Bear, J.L. Studies of rhodium(II) carboxylates as potential antitumor agents. Proc. Soc. Exp. Biol. Med. 1974, 145, 1278–1283. [Google Scholar] [CrossRef]
- Yang, H.; Swartz, A.M.; Park, H.J.; Srivastava, P.; Ellis-Guardiola, K.; Upp, D.M.; Lee, G.; Belsare, K.; Gu, Y.; Zhang, C.; et al. Evolving artificial metalloenzymes via random mutagenesis. Nat. Chem. 2018, 10, 318–324. [Google Scholar]
- Srivastava, P.; Yang, H.; Ellis-Guardiola, K.; Lewis, J.C. Engineering a dirhodium artificial metalloenzyme for selective olefin cyclopropanation. Nat. Commun. 2015, 6, 7789. [Google Scholar] [CrossRef]
- Szilvagyi, G.; Hollosi, M.; Tolgyesi, L.; Frelek, J.; Majer, Z. Dirhodium complexes of amino acid derivatives:separation and characterization by circular dichroism spectroscopy. Tetrahedron Asymmetry 2008, 19, 2594–2599. [Google Scholar] [CrossRef]
- Garcia, A.E.; Jalilehvand, F.; Niksirat, P.; Gelfand, B.S. Methionine binding to dirhodium(II) tetraacetate. Inorg. Chem. 2018, 57, 12787–12799. [Google Scholar] [CrossRef] [PubMed]
- Howard, R.A.; Spring, T.G.; Bear, J.L. The interaction of rhodium(II) carboxylates with enzymes. Cancer Res. 1976, 36, 4402–4405. [Google Scholar]
- Sorasaenee, K.; Fu, P.K.-L.; Angeles-Boza, A.M.; Dunbar, K.R.; Turro, C. Inhibition of transcription in vitro by anticancer active dirhodium(II) complexes. Inorg. Chem. 2003, 42, 1267–1271. [Google Scholar] [CrossRef] [PubMed]
- Chifotides, H.T.; Fu, P.K.-L.; Dunbar, K.R.; Turro, C. Effect of equatorial ligands of dirhodium(II,II) complexes on the efficiency and mechanism of transcription inhibition in Vitro. Inorg. Chem. 2004, 43, 1175–1183. [Google Scholar] [CrossRef]
- Trynda, L.; Pruchnik, F. Interactions of tetra-Ì-acetatodirhodium(II) with human serum albumin. J. Inorg. Biochem. 1995, 58, 69–77. [Google Scholar] [CrossRef]
- Esposito, B.P.; de Oliveira, E.; Zyngier, S.B.; Najjar, R. Effects of serum albumin in some biological properties of rhodium(II) complexes. J. Braz. Chem. Soc. 2000, 11, 447–452. [Google Scholar] [CrossRef]
- Chen, J.; Kostic, M. Binuclear transition-metal complexes as new reagents for selective cross-linking of proteins. Coordination of cytochrome c to dirhodium(II)-Ì-tetraacetate. Inorg. Chem. 1988, 27, 2682–2687. [Google Scholar] [CrossRef]
- Wongand, D.L.; Stillman, M.J. Metallothionin: An aggressive scavenger- The metabolism of rhodium(II) tetraacetate (Rh2(CH3CO2)4). ACS Omega 2018, 3, 16314–16327. [Google Scholar]
- Popp, B.V.; Chen, Z.; Ball, Z.T. Sequence-specific inhibition of a designed metallopeptide catalyst. Chem. Commun. 2012, 48, 7492–7494. [Google Scholar] [CrossRef]
- Zaykov, A.N.; Ball, Z.T. A general synthesis of dirhodium metallopeptides as MDM2 ligands. Chem. Commun. 2011, 47, 10927–10929. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, G.; Pratesi, A.; Messori, L.; Merlino, A. Protein interactions of dirhodium tetraaacetate: A structural study. Dalton. Trans. 2020, 49, 2412–2416. [Google Scholar] [CrossRef] [PubMed]
- Messori, L.; Marzo, T.; Fernandes Sanches, R.N.; Rehman, H.-U.; de Oliveira Silva, D.; Merlino, A. Unusual Structural Features in the Lysozyme Derivative of Tetrakis(acetato)chlorido Diruthenium(II,III) Complex. Angew. Chem. Int. Ed. 2014, 53, 6172–6175. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Abe, S.; Koshiyama, T.; Ohki, T.; Hikage, T.; Watanabe, Y. Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing RhIII Ions as the Model Surfaces. Chem. Eur. J. 2010, 16, 2730–2740. [Google Scholar] [CrossRef] [PubMed]
- Tanley, S.W.; Schreurs, A.M.; Kroon-Batenburg, L.M.; Meredith, J.; Prendergast, R.; Walsh, D.; Bryant, P.; Levy, C.; Helliwell, J.R. Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein. Acta Crystallogr. Sect. D 2012, 68 Pt 5, 601–612. [Google Scholar] [CrossRef]
- Messori, L.; Marzo, T.; Gabbiani, C.; Alvarez-Valdes, A.; Quiroga, A.; Merlino, A. Peculiar features in the crystal structure of the adduct formed between cis-PtI2(NH3)2 and hen egg white lysozyme. Inorg. Chem. 2013, 52, 13827–13829. [Google Scholar] [CrossRef]
- Vergara, A.; D’Errico, G.; Montesarchio, D.; Mangiapia, G.; Paduano, L.; Merlino, A. Interaction of anticancer ruthenium compounds with proteins: High-resolution X-ray structures and Raman microscopy studies of the adduct between hen egg white lysozyme and AziRu. Inorg. Chem. 2013, 52, 4157–4159. [Google Scholar] [CrossRef]
- Tabe, H.; Fujita, K.; Abe, S.; Tsujimoto, M.; Kuchimaru, T.; Kizaka-Kondoh, S.; Takano, M.; Kitagawa, S.; Ueno, T. Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold. Inorg. Chem. 2015, 54, 215–220. [Google Scholar] [CrossRef]
- Seixas, J.D.; Santos, M.F.A.; Mukhopadhyay, A.; Coelho, A.C.; Reis, P.M.; Veiros, L.F.; Marques, A.R.; NPenacho Gonçalves AM, L.; Romão, M.J.; Bernardes GJ, L.; et al. A contribution to the rational design of Ru(CO)3Cl2L complexes for in vivo delivery of CO. Dalton. Trans. 2015, 44, 5058–5075. [Google Scholar] [CrossRef]
- Santos-Silva, T.; Mukhopadhyay, A.; Seixas, J.D.; Bernardes, G.J.L.; Romão, C.C.; Romão, M.J. CORM-3 Reactivity toward Proteins: The Crystal Structure of a Ru(II) Dicarbonyl−Lysozyme Complex. J. Am. Chem. Soc. 2011, 133, 1192–1195. [Google Scholar] [CrossRef]
- Santos, M.F.A.; Seixas, J.D.; Coelho, A.C.; Mukhopadhyay, A.; Reis, P.M.; Romão, M.J.; Romão, C.C.; Santos-Silva, T. New insights into the chemistry of fac-[Ru(CO)3]2+ fragments in biologically relevant conditions: The CO releasing activity of [Ru(CO)3Cl2(1,3-thiazole)], and the X-ray crystal structure of its adduct with lysozyme. J. Inorg. Biochem 2012, 117, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Messori, L.; Scaletti, F.; Massai, L.; Cinellu, M.A.; Gabbiani, C.; Vergara, A.; Merlino, A. The Mode of Action of Anticancer Gold-Based Drugs: A Structural Perspective. Chem. Commun. 2013, 49, 10100–10102. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Messori, L.; Cinellu, M.A.; Marasco, D.; Sirignano, R.; Merlino, A. Interactions of Gold-based Drugs with Proteins: The structure and stability of the Adduct Formed in the reaction between Lysozyme and the Cytotoxic Gold(III) Compound Auoxo3. Dalton. Trans. 2014, 43, 17483–17488. [Google Scholar] [CrossRef]
- Panzner, M.J.; Bilinovich, S.M.; Youngs, W.J.; Leeper, T.C. Silver metallation of hen egg white lysozyme: X-ray crystal structure and NMR studies. Chem. Commun. 2011, 47, 12479–12481. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Ferreira, M.; Albuquerque, I.S.; Matak-Vinkovic, D.; Coelho, A.C.; Carvalho, S.M.; Saraiva, L.M.; Romão, C.C.; Bernardes, G.J.L. Spontaneous CO release from Ru(II)(CO)2-protein complexes in aqueous solution, cells, and mice. Angew. Chem. Int. Ed. 2015, 54, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T. Porous protein crystals as reaction vessels. Chemistry 2013, 19, 9096–9102. [Google Scholar] [CrossRef]
- Zobi, F.; Spingler, B. Post-protein-binding reactivity and modifications of the fac-[Re(CO)3]+ core. Inorg. Chem. 2012, 51, 1210–1212. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rychlewska, U.; Djuran, M.I.; Vasojevic, M.M.; Radanovic, D.D.; Ristanovic, V.M.; Radanovic, D.J. Hexadentate rhodium(III) complexes of 1,3-propanediamine-N,N′-diacetic-N,N′-di-3-propionic acid. Crystal structures of trans-(O5)-Na[Rh(1,3-pddadp)]·H2O and (+)589-trans-(O5O6)-Na[Rh(1,3-pddadp)]·3H2O and CD spectra correlation. Octahedral distortion of [Rh(edta-type)]− complexes in relation to the structure of the ligand and geometry of the complex. Inorg. Chim. Acta 2002, 328, 218. [Google Scholar]
- Garcia, A.E.; Jalilehvand, F.; Niksirat, P. Reactions of with thiols and thiolates: A structural study. J. Synchr. Rad. 2019, 26, 450–461. [Google Scholar] [CrossRef]
- Brink, A.; Helliwell, J. Formation of a highly dense tetra-rhenium cluster in a protein crystal and its implications in medical imaging. IUCr J. 2019, 6, 695–702. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Ferraro, G.; Pica, A.; Marquez, J.A.; Helliwell, J.H.; Merlino, A. Principles and methods used to grow and optimize crystals of protein–metallodrug adducts, to determine metal binding sites and to assign metal ligands. Metallomics 2017, 9, 1534–1547. [Google Scholar] [CrossRef] [PubMed]
- Ugone, V.; Sanna, D.; Ruggiu, S.; Sciortino, G.; Garribba, E. Covalent and non-covalent binding in vanadium–protein adducts. Inorg. Chem. Front. 2021. [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-rays diffraction data collected in oscillation mode. Method Enzymol. 1997, 276, 307−326. [Google Scholar]
- Battye, T.G.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G.W. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. Sect. D 2011, 67 Pt 4, 271–281. [Google Scholar] [CrossRef]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D 2011, 67, 293–302. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Vaney, J.M.C.; Maignan, S.; Ries-Kautt, M.; Ducriux, A. High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission. Acta Crystallogr. Sect. D 1996, 52, 505–517. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. Sect. D 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D 2004, 60, 2126−2132. [Google Scholar] [CrossRef]

| Crystal 1 | Crystal 2 | Crystal 3 | Crystal 4 | Crystal 5 | Crystal 6 | |
|---|---|---|---|---|---|---|
| Procedure | Cocrystallization | Cocrystallization | Cocrystallization | Cocrystallization | Soaking | Soaking |
| Crystallization conditions | 20% ethylene glycol, 0.100 M sodium acetate at pH 4.5, and 0.6 M sodium nitrate | |||||
| Data collection | ||||||
| Space group | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 |
| a = b (Å) | 77.964 | 77.948 | 78.360 | 78.604 | 78.007 | 78.930 |
| c (Å) | 37.291 | 37.380 | 37.610 | 37.307 | 37.245 | 37.120 |
| Resolution range (Å) | 25.48–1.94 | 26.99–1.62 | 78.36–1.40 | 55.58–1.32 | 35.19–1.74 | 55.81–1.65 |
| (1.97–1.94) | (1.65–1.62) | (1.47–1.40) | (1.37–1.32) | (1.83–1.74) | (1.80–1.65) | |
| Unique reflections | 8895 (426) | 15,018 (623) | 22,922 (3274) | 26,513 (1327) | 11,348 (567) | 11,800 (591) |
| Completeness (%) | 99.0 (97.1) | 98.7 (86.6) | 100 (100) | 95.4 (51.1) | 91.1 (36.0) | 94.0 (52.6) |
| Redundancy | 5.3 (3.7) | 9.0 (1.9) | 19.6 (16.6) | 25.8 (25.9) | 25.0 (27.6) | 24.5 (22.4) |
| † Rmerge (%) | 0.095 (0.440) | 0.068 (0.190) | 0.099 (2.00) | 0.047 (2.55) | 0.070 (3.12) | 0.120 (3.23) |
| Rpim | 0.045 (0.249) | 0.021 (0.152) | 0.023 (0.519) | 0.013 (0.720) | 0.020 (0.848) | 0.025 (0.680) |
| Average I/σ(I) | 14.9 (2.2) | 56.7 (3.4) | 10.2 (1.0) | 28.8 (1.1) | 24.7 (1.1) | 22.5 (1.3) |
| CC1/2 | 0.997 (0.808) | 0.999 (0.942) | 0.999 (0.701) | 1.00 (0.612) | 1.00 (0.474) | 1.00 (0.51) |
| Anomalous completeness (%) | 99.1 (94.1) | 96.8 (67.3) | 100 (100) | 95.3 (52.1) | 90.8 (36.6) | 93.8 (51.4) |
| Anom. Redundancy | 2.9 (2.0) | 5.0 (1.3) | 10.3 (8.5) | 13.8 (13.4) | 13.7 (14.5) | 13.4 (12.0) |
| Refinement | ||||||
| Resolution range (Å) | 25.48–1.94 | 26.99–1.62 | 55.41–1.40 | 55.58–1.32 | 35.19–1.74 | 55.81–1.65 |
| Anom. Redundancy | 2.9 (2.0) | 5.0 (1.3) | 10.3 (8.5) | 13.8 (13.4) | 13.7 (14.5) | 13.4 (12.0) |
| Refinement | ||||||
| Resolution range (Å) | 25.48–1.94 | 26.99–1.62 | 55.41–1.40 | 55.58–1.32 | 35.19–1.74 | 55.81–1.65 |
| Number of reflections (working set) | 8445 | 14,241 | 22,453 | 25,227 | 10,794 | 11,205 |
| Number of reflections (test set) | 8445 | 14,241 | 22,453 | 25,227 | 10,794 | 11,205 |
| R-factor/R-free (%) | 15.6/21.7 | 16.4/20.2 | 19.0/20.9 | 17.7/22.3 | 18.5/22.8 | 18.1/23.4 |
| N. of atoms | 1221 | 1231 | 1283 | 1275 | 1129 | 1139 |
| Average B-factors (Å2) | ||||||
| All atoms | 21.1 | 21.4 | 16.8 | 27.3 | 42.9 | 30.6 |
| Rh occupancy | 0.70/0.35/0.40/0.40 | 0.65/0.50/0.20/0.20 | 0.65/0.30 | 0.70/0.30 | 0.25/0.40/0.20 | 0.40/0.45/0.35/0.20/0.30/0.30/0.40 |
| Rh atoms | 31.7/33.7/67.9/75.3 | 30.8/46.9/39.3/42.5 | 23.1/26.7 | 35.5/41.7 | 81.1/73.9/65.2 | 59.6/69.0/66.9/49.3/68.7/57.4/79.8 |
| Root mean square deviations | ||||||
| Bond lengths (Å) | 0.009 | 0.013 | 0.013 | 0.013 | 0.008 | 0.008 |
| Bond angles (°) | 2.38 | 1.96 | 1.87 | 2.39 | 1.55 | 1.55 |
| Ramachandran statistics (Coot analysis) | ||||||
| N. of residues in | ||||||
| Allowed/disallowed regions | 4/0 | 4/0 | 3/0 | 3/0 | 4/0 | 3/0 |
| Crystal 1 | Crystal 2 | Rh/HEWL Adduct Ligands Crystal 3 | Crystal 4 | Crystal 5 | Crystal 6 | |
|---|---|---|---|---|---|---|
| His15 site | Rh (0.35) His15 (1) Arg14 (1) Act (1) H2O (0.35) H2O (0.65) | Rh (0.5) His15 (1) Arg14 (1) Act (1) H2O (0.5) | Rh (0.3) His15 (1) Arg14 (0.5) Act (1) | Rh (0.3) His15 (1) Arg14 (0.5) Act (1) H2O (0.3) H2O (0.3) | Rh (0.25) His15 (1) Arg14 (0.5) Act (1) H2O (1) | Rh (0.35) His15 (1) Arg14 (1) H2O (0.35) H2O (0.35) |
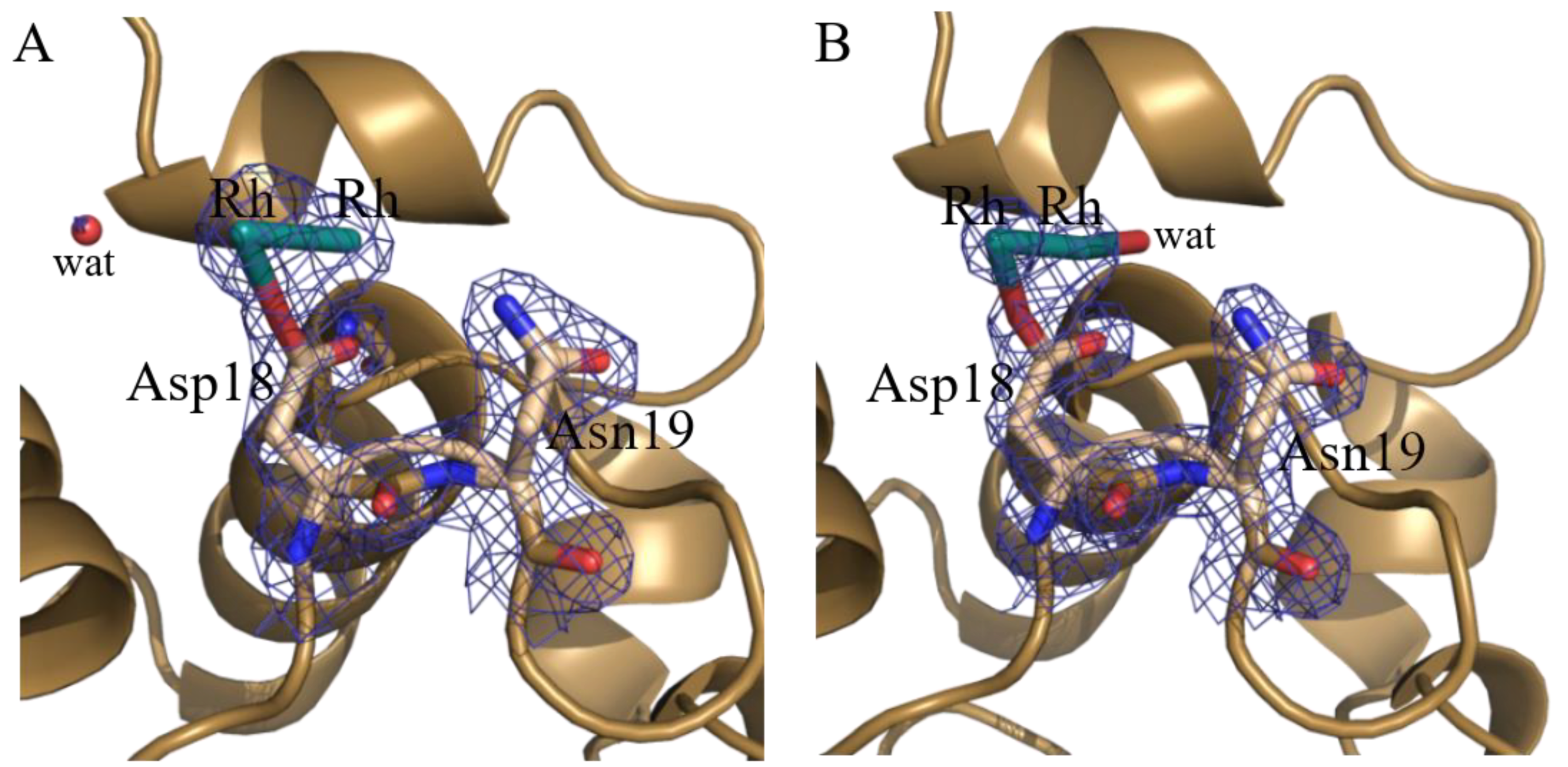
| Asp18 site | Rh (0.40) Rh (0.40) Asp18 (1) | Rh (0.20) Rh (0.20) Asp18 (1) H2O (0.20) | ||||
| C-ter site | Rh (0.65) | Rh (0.65) | Rh (0.65) | Rh (0.7) | Rh (0.4) Rh * (0.4) Leu129 (1) Leu129 * (1) Lys13 (1) Lys13 * (1) | Rh (0.45) Rh * (0.45) Leu129 (1) Leu129 * (1) Lys13 (1) Lys13 * (1) H2O (0.5) H2O * (0.5) |
| Rh * (0.65) | Rh * (0.65) | Rh * (0.65) | Rh* (0.7) | |||
| Leu129 (1) | Leu129 (1) | Leu129 (1) | Leu129 (1) | |||
| Leu129 * (1) | Leu129 * (1) | Leu129 * (1) | Leu129 * (1) | |||
| Lys13 (1) | Lys13 (1) | Lys13 (1) | Lys13 (1) | |||
| Lys13 * (1) | Lys13 * (1) | Lys13 * (1) | Lys13 * (1) | |||
| Act (0.60) | Act (0.65) | Act (0.65) | Act (0.7) | |||
| Act * (0.60) | Act * (0.65) | Act * (0.65) | Act * (0.7) | |||
| H2O (0.65) | H2O (0.65) | H2O (0.65) | H2O (0.7) | |||
| H2O * (0.65) | H2O * (0.65) | H2O * (0.65) | H2O * (0.7) | |||
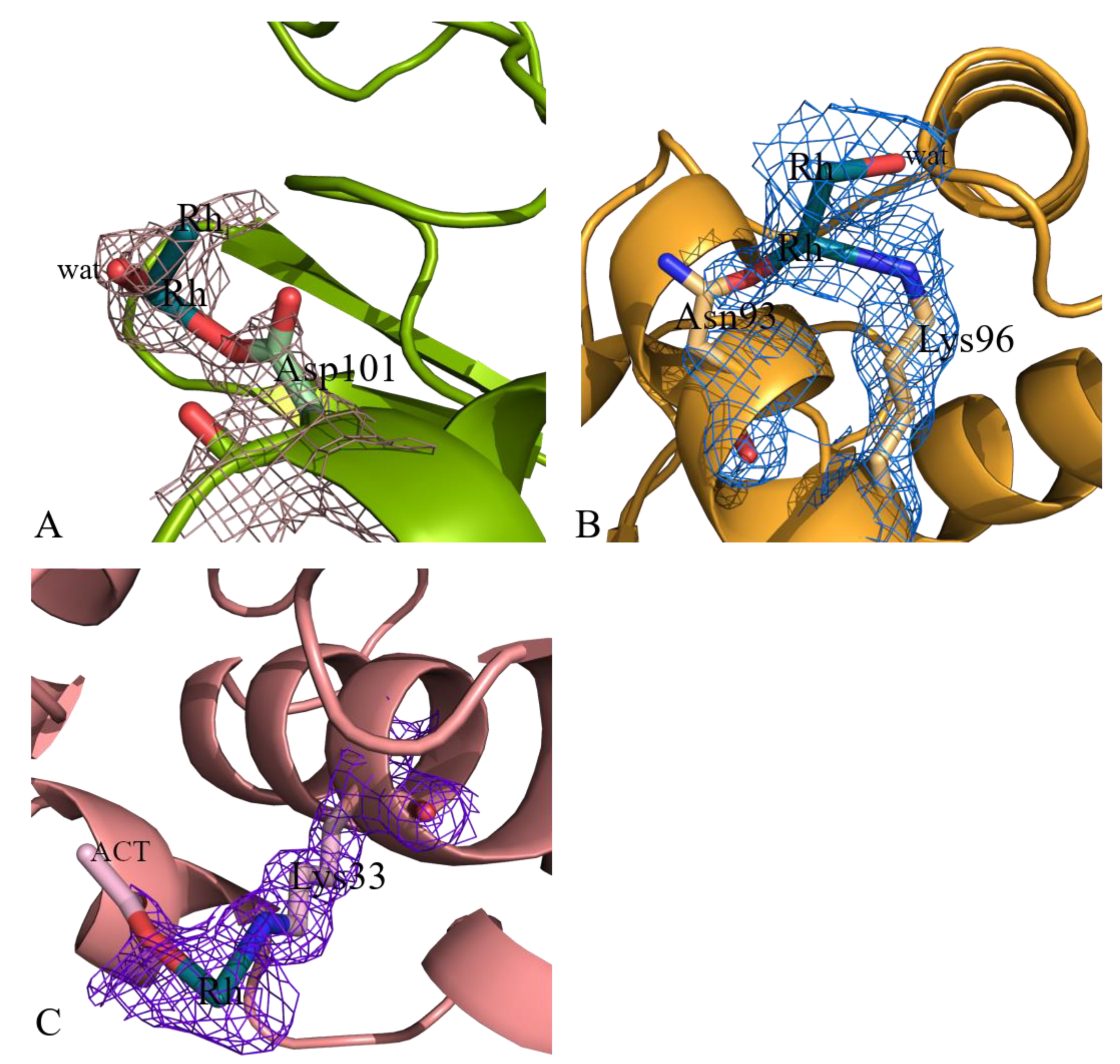
| Asp101 site | Rh (0.40) | |||||
| Rh (0.40) | ||||||
| Asp101 (1) | ||||||
| H2O (0.40) | ||||||
| Lys33 site | Rh (0.20) | |||||
| Lys33 (1) | ||||||
| Act (0.33) | ||||||
| Asn93 site | Rh (0.30) | |||||
| Rh (0.30) | ||||||
| Asn93 (1) | ||||||
| Lys96 (1) | ||||||
| H2O (1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loreto, D.; Ferraro, G.; Merlino, A. Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme. Int. J. Mol. Sci. 2021, 22, 1496. https://doi.org/10.3390/ijms22031496
Loreto D, Ferraro G, Merlino A. Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme. International Journal of Molecular Sciences. 2021; 22(3):1496. https://doi.org/10.3390/ijms22031496
Chicago/Turabian StyleLoreto, Domenico, Giarita Ferraro, and Antonello Merlino. 2021. "Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme" International Journal of Molecular Sciences 22, no. 3: 1496. https://doi.org/10.3390/ijms22031496
APA StyleLoreto, D., Ferraro, G., & Merlino, A. (2021). Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme. International Journal of Molecular Sciences, 22(3), 1496. https://doi.org/10.3390/ijms22031496