Abstract
In this study, we found that the loss of OmpR, the response regulator of the two-component EnvZ/OmpR system, increases the cellular level of Fur, the master regulator of iron homeostasis in Y. enterocolitica. Furthermore, we demonstrated that transcription of the fur gene from the YePfur promoter is subject to negative OmpR-dependent regulation. Four putative OmpR-binding sites (OBSs) were indicated by in silico analysis of the fur promoter region, and their removal affected OmpR-dependent fur expression. Moreover, OmpR binds specifically to the predicted OBSs which exhibit a distinct hierarchy of binding affinity. Finally, the data demonstrate that OmpR, by direct binding to the promoters of the fecA, fepA and feoA genes, involved in the iron transport and being under Fur repressor activity, modulates their expression. It seems that the negative effect of OmpR on fecA and fepA transcription is sufficient to counteract the indirect, positive effect of OmpR resulting from decreasing the Fur repressor level. The expression of feoA was positively regulated by OmpR and this mode of action seems to be direct and indirect. Together, the expression of fecA, fepA and feoA in Y. enterocolitica has been proposed to be under a complex mode of regulation involving OmpR and Fur regulators.
Keywords:
Yersinia enterocolitica; OmpR regulator; Fur repressor; iron homeostasis; fecA; fepA; feoA 1. Introduction
Yersinia enterocolitica is a Gram-negative bacterium that exhibits a dual lifestyle, existing as both a pathogen of a broad range of animals and a saprophyte widespread in nature [1,2]. In humans, it causes yersiniosis, an important zoonotic gastrointestinal disease that occurs worldwide [3]. Y. enterocolitica is a heterogeneous species comprising six biotypes (1A, 1B, 2, 3, 4 and 5) that vary in pathogenicity due to the synthesis of different virulence factors [4,5]. The survival and growth of pathogenic bacteria outside and inside the host organism depends on iron availability [6]. However, at physiological pH in an aerobic environment, ferric ions (Fe3+) are essentially insoluble and unavailable for use by bacterial cells. Conversely, at low pH and in anaerobic environments, iron can switch from the insoluble ferric form to the more soluble ferrous (Fe2+) form. Y. enterocolitica and other pathogens encounter an additional obstacle in their quest for inorganic iron in the form of iron-binding compounds in the host organism, which control the availability of this essential trace nutrient [7]. To obtain the quantities of iron necessary for growth, pathogenic Y. enterocolitica have developed diverse and complex acquisition systems to acquire iron from different sources [8,9,10,11,12,13,14,15]. To obtain ferric iron, highly pathogenic strains of Y. enterocolitica (biotype 1B) synthesize the siderophore yersiniabactin, a high-affinity Fe3+-specific chelator [16]. Less pathogenic strains are unable to synthesize yersiniabactin and therefore import foreign siderophores complexed with Fe3+ via TonB-dependent OM transporters/receptors such as FecA, FepA, FoxA and FcuA [10,17]. Another mechanism operating in Y. enterocolitica is a system for acquiring iron from heme or hemoproteins, which employs the OM receptor HemR [8,15]. The uptake of ferrous iron can be achieved independently of siderophores and Gram-negative bacteria are equipped with an Fe2+ active transport system located in the cytoplasmic membrane (Feo system) [18]. Ferrous iron is thought to diffuse freely through the OM porins into the periplasm from where it is transported into the cytoplasm via the FeoABC system [19]. Systems homologous to FeoABC have been identified in pathogenic Yersinia species [20].
The concentration of iron in the environment controls the growth and cellular metabolism of bacteria [21]. However, while an iron shortage restricts bacterial growth, excessive intracellular accumulation of iron can be deleterious, damaging bacteria through the generation of reactive oxygen species by the Fenton reaction [22]. The precise control of iron acquisition, storage and utilization that ensures bacterial survival in a changing environment is coordinated by the Ferric-uptake regulator (Fur). Fur is an Fe2+-responsive transcription factor that controls the expression of genes involved in diverse cellular mechanisms associated with iron homeostasis. The Fur-dependent regulation of gene expression has been best studied in Escherichia coli K-12 [23,24]. Under iron-replete conditions, Fur binds ferrous iron (Fe2+), leading to its activation and binding of a 19-bp consensus DNA sequence (Fur box), located in the promoters of iron-regulated genes/operons required for iron acquisition or transport, which represses their transcription [25,26]. Under iron starvation conditions, the repression of Fur-regulated genes is relieved by dissociation of the ferrous ion from the Fur protein [27].
Since the original description of Fur in E. coli K-12 [28,29], this regulator has been recognized in many other bacterial species including human pathogens. The functions of Fur extend far beyond the regulation of iron homeostasis and include important roles in the defense against oxidative stress [30,31], carbon metabolism [9,32] and the acid tolerance response [33]. Moreover, Fur seems to be involved in other functions that are unrelated to the regulation of transcription and occur irrespective of the iron status [34]. Genome-wide computational analyses have revealed a number of potentially Fur-controlled genes, which indicates that Fur has an extremely broad range of influence [24]. Thus, the full picture of Fur activity is much more complex than was previously thought [35,36,37,38].
Based on experimental and bioinformatic analyses, Fur homologs have been identified in pathogenic Yersinia species, i.e., the plague bacillus Y. pestis and enteropathogen Y. pseudotuberculosis [14,39,40]. Almost all of the iron scavenging mechanisms in Y. pestis were shown to be controlled by Fur [41]. In addition, the identification of a Fur regulon in Y. pestis affirmed its role as a global regulator of gene expression [41,42]. In Y. enterocolitica, all genes required for the synthesis and transport of yersiniabactin, encoded within a high-pathogenicity island (HPI) found in Y. enterocolitica biotype 1B and responsible for its increased pathogenicity, are repressed by Fur under iron-replete conditions [14,43]. Other Y. enterocolitica genes subject to Fur-dependent regulation in response to iron concentrations encode outer membrane receptors and transport systems for the foreign siderophores ferrichrome, ferrioxamine B and E and enterobactin [10,17].
In Gram-negative bacteria, active Fe2+-Fur protein levels are regulated by fur gene expression and the intracellular labile Fe2+ pool [44]. The expression of fur is subject to complex regulation in response to environmental conditions such as the iron concentration, the available sources of carbon, oxidative stress and pH [45,46,47,48]. It has been shown that fur transcription in E. coli is autoregulated, and iron starvation slightly increases fur expression. Moderate autoregulation by the Fur protein occurs in most Enterobacteriaceae [46,49,50]. In E. coli, OxyR and SoxRS, two oxidative stress response regulators, activate fur expression [47]. Another important layer of fur regulation in E. coli is post-transcriptional downregulation involving the non-coding sRNA RyhB [51].
Two-component signal transduction systems (TCSs) are widespread in prokaryotes and play vital roles in adaptation to environmental changes by modulating bacterial gene expression [52,53]. The EnvZ/OmpR system of E. coli, an archetype of TCSs, comprises the EnvZ transmembrane sensor kinase and the transcriptional response regulator OmpR. As a classical response regulator, OmpR has an N-terminal receiver domain with a conserved Asp residue at position 55 and a C-terminal helix-turn-helix domain for DNA binding. It can regulate transcription in a positive or negative way by binding to consensus sequence elements that occur at different locations in promoter regions [54,55,56]. It has been shown that the EnvZ/OmpR system modulates the expression of numerous genes in response to environmental changes such as altered osmolarity, pH and nutrient content [57,58,59,60,61,62]. The transcriptional regulator OmpR has been best studied for its role in the inverse osmoregulation of the outer membrane porins OmpC and OmpF in E. coli. This is achieved by the binding of this factor, with different affinity, to multiple OmpR-binding sites within the regulatory regions of the ompC and ompF genes [54,56]. A large body of research has demonstrated that OmpR influences a wide variety of other cellular processes in E. coli [63,64,65,66], enteropathogenic Salmonella and Shigella [67,68], as well as pathogenic species of Yersinia [69,70,71]. The relationship between virulence and the activity of the OmpR protein has been described for Y. enterocolitica [72,73,74,75]. These studies showed that Y. enterocolitica OmpR can serve a variety of functions that are often specific to this organism, which highlights the pleiotropic role of this regulatory protein [76,77,78]. The results of studies in Y. enterocolitica emphasize the importance of OmpR in controlling the expression of several transcriptional regulators. The expression of FlhDC, the master regulator of motility genes, appears to be positively regulated by OmpR in Y. enterocolitica [79], which is in contrast to the negative role played by OmpR in E. coli [64]. The same positive regulation of flhDC by OmpR has also been demonstrated in Y. pseudotuberculosis [70]. Marked inhibition of AcrR, a regulator of multidrug transporter gene expression, was also noted in Y. enterocolitica [77]. Finally, KdgR, a negative regulator of oligogalacturonide-uptake genes in Y. enterocolitica, was identified as a direct target for positive OmpR-dependent regulation [78]. Thus, OmpR is involved in the control of diverse responses to environmental changes by altering the expression of downstream transcriptional regulators and functions as a global regulator in Y. enterocolitica.
Recently, differential proteomic analysis of Y. enterocolitica strain Ye9 (bio-serotype 2/O:9) revealed the role of OmpR in modulating (positively or negatively) the abundance of many proteins, including three iron transport system receptors: FepA for the uptake of ferrienterobactin, FecA for ferric dicitrate and HemR for heme compounds [80]. These results prompted us to investigate the link between OmpR and iron acquisition in Y. enterocolitica. It seemed reasonable to hypothesize that OmpR might influence the expression of iron-regulated genes by modulating expression of the Fur regulator. Thus, we investigated the role of OmpR in fur expression and examined the influence of diverse environmental factors on OmpR-dependent fur regulation. Subsequently, we determined whether OmpR can influence fur transcription by direct binding to the fur regulatory region. Lastly, we examined how OmpR-dependent regulation of fur affects the expression of Fur regulon genes.
By the application of reporter gene transcriptional fusions and RT-qPCR analysis, we show that OmpR inhibits the transcription of fur in Y. enterocolitica. Among the environmental factors tested, only pH and osmolarity influenced fur expression, but irrespectively of the activity of OmpR. Electrophoretic mobility shift assays showed that OmpR inhibits fur expression through binding to specific motifs present in the fur regulatory region. Quantification of a Fur-FLAG hybrid protein by Western blotting confirmed that the level of the Fur protein is negatively regulated by OmpR. The inhibition of fur transcription by OmpR was also found to occur in E. coli. Moreover, we examined whether OmpR could affect the expression of the fecA, fepA and feoA genes, members of the Y. enterocolitica Fur regulon, encoding proteins that participate in the transport of ferric and ferrous iron into cells. Our data demonstrate that the transcription of these genes is subject to a complex mode of regulation involving the OmpR and Fur regulators.
2. Results
2.1. Characterization of the Fur Promoter Region in Y. enterocolitica Strain Ye9N
Analysis of the shotgun genome sequence of Y. enterocolitica strain Ye9N (2/O:9) (NCBI/GenBank Acc. No. NZ_JAALCX000000000.1) localized the fur gene, encoding the Fur repressor (an ORF of 447 nt, 148 aa), downstream of fldA, encoding flavodoxin I (175 aa), and upstream of chb, encoding chitobiase (891 aa) (Figure 1A). In silico analysis revealed that the fldA-fur-chb arrangement is highly conserved among Y. enterocolitica strains of different bio-serotypes and other pathogenic Yersinia species, i.e., Y. pestis and Y. pseudotuberculosis (Figure S1A). The fldA and fur ORFs are in the same transcriptional orientation, whereas chb is oriented divergently. The relatively long fldA-fur intergenic region (368 bp) suggests the potential for regulatory interactions. A putative promoter located upstream of fur (Pfur) was identified using the software BPROM [81]. The −10 and −35 core promoter motifs are located 84 and 107 nt upstream of the start codon of fur and show partial resemblance to the respective consensus sequences for E. coli σ70 RNA polymerase (TATAAT and TTGACA) (Figure 1B). This analysis also revealed the presence of one putative Fur-binding site (Fur boxYe9, 5′-TATAATGAGACGCAATTGA-3′) in the fur promoter, overlapping the −10 sequence, with 58% identity to the E. coli Fur box consensus sequence, a 19-nt inverted repeat (5′-GATAATGATAATCATTATC-3′) [25]. Interestingly, four possible 20-nt OmpR-binding sites (OBSs) were also identified in the Y. enterocolitica fur promoter region (Figure 1B). These putative OBSs, named M1, M2, M3 and M4, are centered at −331, −227, −191 and −20 bp relative to the start codon of fur and, respectively, exhibit 45%, 60%, 55% and 50% identity to the E. coli consensus sequence proposed by Maeda et al. 1991 [82]. Moreover, two of four OBSs (M2 and M4) and a Fur box were revealed in the fur regulatory region of Y. pestis and Y. pseudotuberculosis (Figure S1B).
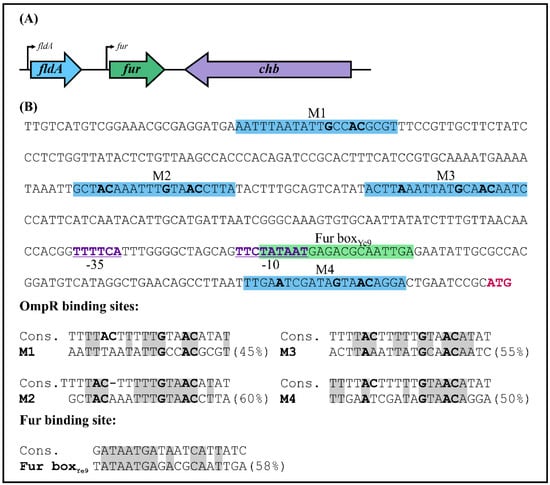
Figure 1.
Chromosomal organization of the Y. enterocolitica fur locus. (A) Genes comprising the Y. enterocolitica fur locus. The directions of transcription are indicated by arrows. The encoded products are: fur—Fur (ferric uptake regulator); fldA—flavodoxin I; chb—chitobiase. (B) The fur promoter region of Y. enterocolitica Ye9N. The nucleotide sequence from −368 to +3 nt relative to the first nucleotide of the fur ATG start codon (colored pink) is shown. The −10 and −35 motifs of the fur promoter are marked purple, bold and underlined. The putative OmpR-binding sites (M1, M2, M3, M4) are highlighted by a blue background, while the potential Fur-binding site (Fur boxYe9) is highlighted by a green background. The putative OmpR- and Fur-binding sites are shown aligned to the E. coli consensus sequences (% identity values are shown). Identical nucleotides in the compared sequences are shaded gray. In both the promoter region sequence and alignments, nucleotides important for OmpR binding are in bold.
2.2. OmpR Inhibits the Activity of the Ye9 Fur Promoter
In order to establish the role of OmpR in fur regulation, Y. enterocolitica strains Ye9F (WT) and AR4F (ompR deletion mutant) harboring a single-copy chromosomal Pfur’::lacZYA transcriptional fusion were constructed using a mobilizable suicide plasmid. The Ye9 fur promoter region was cloned upstream of a promoterless lacZYA operon in vector pFUSE. The resulting plasmid pFUSEfur was transferred to strains Ye9N and AR4 by conjugation and exconjugants harboring plasmid co-integrates produced by a single crossover event between the Y. enterocolitica sequences present on the plasmid and the bacterial fur locus were selected. Correct integration was confirmed by PCR and sequencing (data not shown). β-galactosidase activity was measured in early stationary phase cultures of the resulting strains Ye9F and AR4F grown at 26 °C under iron-limiting (DPD, 150 µM) or iron-replete conditions (FeCl3, 10 µM). The results presented in Figure 2A demonstrate that β-galactosidase activity in the OmpR-deficient mutant AR4F was increased 1.7-fold compared to Ye9F, suggesting that fur transcription is repressed in an OmpR-dependent manner. The addition of iron chelator DPD to Ye9F cultures resulted in slightly higher reporter gene expression compared to those supplemented with FeCl3. However, a lack of response to iron availability was observed in the ompR mutant. To confirm the negative OmpR-dependent regulation of fur, plasmid pBOmpR carrying a gene coding for the His-tagged OmpR protein was used to complement the ompR mutation in strain AR4F. The β-galactosidase activity in AR4F/pOmpR was decreased 1.6-fold compared to AR4F (Figure 2A). The introduction of empty vector pBBR1 MCS-3 did not affect the lacZ expression level (data not shown). Thus, complementation of the ompR mutation resulted in the inhibition of fur expression, confirming that OmpR is required for the repression of fur.
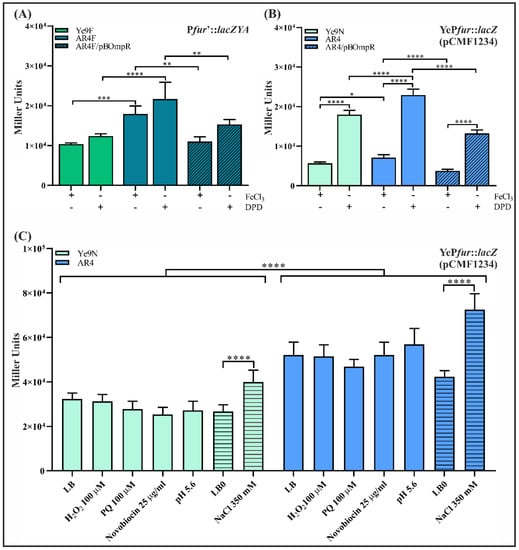
Figure 2.
OmpR-dependent regulation of fur transcription in Y. enterocolitica Ye9N. (A) Expression of a chromosomally encoded Pfur’::lacZYA transcriptional fusion in strains Ye9F (WT) and AR4F (ΔompR) and complemented strain AR4F/pBOmpR (expresses OmpR from Ye9). β-Galactosidase activity was measured in strains grown to early stationary phase in LB medium at 26 °C, under iron-limiting (150 μM DPD) or iron-replete (10 μM FeCl3) conditions. (B) Expression of plasmid-encoded YePfur::lacZ transcriptional fusion in strains Ye9N (WT) and AR4 (ΔompR) and complemented strain AR4/pBOmpR (expresses OmpR from Ye9). β-Galactosidase activity was measured in strains grown in LB medium supplemented with FeCl3 or DPD, as above. (C) The influence of environmental factors on the expression of a YePfur::lacZ transcriptional fusion encompassing the Pfur region with four potential OmpR-binding motifs (pCMF1234) in strains Ye9N (WT) and AR4 (ΔompR). The strains were grown to late exponential phase in LB (smooth bars) or LB0 (hatched bars) medium at 26 °C, then different stress factors were added before overnight incubation. The presented data (A–C) represent mean β-galactosidase activity values with SD from three independent experiments, each performed using at least triplicate cultures of each strain. Significance was calculated using one-way ANOVA (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
The effect of OmpR on the expression of fur was also examined in the wild-type strain Ye9N and in the strain lacking ompR (AR4) by RT-qPCR (Figure S1). To determine whether iron influences fur transcript levels, RT-qPCR was performed using RNA isolated from the same strains grown under iron-limiting (DPD) and iron-replete (FeCl3) conditions. A visible (~2-fold) increase in the level of fur mRNA was detected in the strain lacking ompR. This corroborated the results of the lacZ reporter gene study showing OmpR-dependent inhibition of fur. In addition, fur transcript levels were not significantly changed by iron content (Figure S2).
Finally, to confirm that OmpR affects fur expression through the regulation of fur promoter activity, we generated a transcriptional fusion of the Ye9 fur promoter region (YePfur), containing four predicted OmpR-binding sequences, with the lacZ gene in vector pCM132Gm. The resulting plasmid pCMF1234 was introduced into strains Ye9N and AR4. Quantification of β-galactosidase activity along the bacterial growth curve showed that fur promoter activity in both strains increased around 2-fold in the early stationary phase compared with exponentially growing cells (Figure S3). The strains carrying pCMF1234 were grown to the early stationary phase at 26 °C in LB supplemented with FeCl3 (10 µM) or an iron chelator (DPD, 150 µM), then β-galactosidase activity was measured (Figure 2B). According to these measurements, the activity of YePfur was slightly higher in the ΔompR mutant strain compared to Ye9N in the ron-replete medium, and this difference was clearer under iron-limiting conditions. Complementation of the ompR mutation in strain AR4 by introducing plasmid pBOmpR led to a significantly reduced level of YePfur activity, confirming the role of OmpR in the inhibition of Ye9 fur expression. Interestingly, in these experiments using the plasmid-encoded YePfur::lacZ transcriptional fusion, we observed ~3-fold higher reporter activity in iron-limiting (DPD) than in iron-replete (FeCl3) conditions, for all studied strains (Figure 2B).
2.3. Effect of Environmental Signals on OmpR-Dependent Ye9 Fur Expression
The data presented above show that the expression of fur in Y. enterocolitica wild-type strain Ye9 is negatively regulated by OmpR and iron availability. The presence of a Fur-binding box in the fur promoter region (Figure 1) suggests that autoregulation of fur can occur. We next examined the OmpR-dependent regulation of fur expression under various environmental stresses: low pH, high osmolarity, novobiocin, paraquat and H2O2. It has been shown previously that OmpR modulates the expression of numerous genes in response to altered osmolarity and pH in E. coli and Salmonella [57,58,59,60,61,83]. In Salmonella, it was hypothesized that DNA relaxation due to the presence of novobiocin might promote OmpR binding to DNA to modulate expression of regulators SsrA and HilC encoded by the pathogenicity islands SPI-2 and SPI-1, respectively [84]. On the other hand, E. coli fur expression was shown to be responsive to oxidative stress generated by treatment with hydrogen peroxide or paraquat [47].
To determine the effects of these stresses on Ye9 fur expression, the plasmid-encoded YePfur::lacZ transcriptional fusion (pCMF1234) was used. In order to attribute changes in fur expression to OmpR, these experiments were carried out using both the wild type (Ye9N/pCMF1234) and ΔompR mutant (AR4/pCMF1234) grown in LB medium and in iron-limiting (150 µM DPD) conditions.
The addition of hydrogen peroxide (100 µM H2O2) or the superoxide generator paraquat (PQ, 100 µM) to the LB medium had no effect on fur expression in the strains Ye9N/pCMF1234 and AR4/pCMF1234 (Figure 2C). To ascertain whether the expression of fur responds to high osmolarity, both strains were grown in LB0 medium (10 g/L Tryptone, 5 g/L Yeast Extract) with or without the addition of 350 mM NaCl. High osmolarity appeared to slightly increase (~1.5-fold for Ye9N and ~1.7 for AR4) fur expression irrespective of the presence of OmpR (Figure 2C). We did not observe any changes in YePfur activity in either strain grown in LB medium at low pH (pH 5.6). The presence of novobiocin, known to influence DNA topology in an OmpR-dependent manner [84], also had no effect on fur promoter activity in the studied strains (Figure 2C). No significant differences in fur expression were observed in response to the applied environmental stimuli in the absence of iron (data not shown). It is worth mentioning that during incubation of the tested strains under the studied conditions, i.e., in the presence of PQ, H2O2, novobiocin, low pH or altered osmolarity, we saw no significant differences in the growth of cultures.
2.4. Fur Abundance in Y. enterocolitica Is Modulated by the OmpR Regulator and Certain Environmental Conditions
To study the role of OmpR and iron content in modulating the abundance of the Fur protein, an in-frame translational fusion of Fur with a 3×FLAG epitope at the C-terminal end was constructed in the suicide vector pDS132. The obtained plasmid pDSFur-FLAG was introduced into the chromosome of Ye9N and its isogenic mutant AR4 (ΔompR) by homologous recombination to replace the respective wild-type fur alleles. The correctness of the obtained fusions was confirmed by PCR and DNA sequencing (data not shown). These strains were grown in LB medium or LB supplemented with FeCl3 or DPD and the Fur-3×FLAG protein was detected in whole cell lysates by Western blotting using an anti-FLAG antibody (Figure 3A). The epitope-tagged Fur protein showed strong and specific reactivity with the anti-FLAG antibody. In the wild-type strain, the amount of Fur-3×FLAG increased by ~95% under iron-limiting conditions (LB + DPD), compared with iron-replete conditions (LB + FeCl3) or LB alone, confirming the regulatory role of iron in modulating the abundance of Fur. Fur-3×FLAG was present in higher amounts in the ompR deletion mutant (AR4Fflag) than in the wild-type Ye9Fflag strain, indicating that OmpR has an inhibitory effect on the level of the Fur protein (Figure 3A). However, the level of de-repression caused by the absence of OmpR varied according to the growth medium, i.e., a ~120% increase occurred in LB with FeCl3, ~100% in LB with DPD and only ~20% in LB medium. Complementation of the ompR mutation in the strain AR4Fflag with the plasmid pBOmpR resulted in a decrease in the amount of Fur-3×FLAG independently of the iron content of the medium, which is consistent with an inhibitory effect of OmpR on fur expression (Figure 3A). The Western blotting results were validated by probing the same blots with an antiserum specific for the cytoplasmic molecular chaperone DnaK to confirm equal protein loading. The intensity of the DnaK bands did not differ significantly between the tested samples.
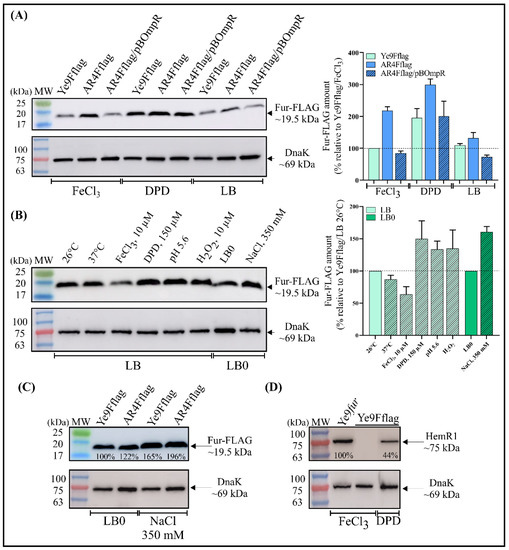
Figure 3.
Western blot analysis of Fur expression in Y. enterocolitica Ye9N. (A–C) Levels of Fur-3×FLAG were analyzed in Y. enterocolitica cells grown in LB at 26 °C (unless indicated) for 24 h until stationary phase. Western blotting of cell extracts was performed with a monoclonal mouse anti-FLAG epitope antibody. As a loading control, the blots were also probed with an antiserum specific for cytoplasmic molecular chaperone DnaK. The Fur-FLAG band is ~19.5 kDa in size and DnaK ~69.0 kDa. (A, left panel) Comparison of Fur levels in the following strains: Ye9Fflag (Ye9N wild type with chromosomal fur::3×flag translational fusion), AR4Fflag (ΔompR mutant AR4 with chromosomal fur::3×flag translational fusion), AR4Fflag/pBOmpR (AR4Fflag complemented with a plasmid carrying the wild-type ompR allele). The strains were grown in LB or in LB supplemented with FeCl3 (10 μM) or DPD (150 μM). (A, right panel) Quantification of the amount of Fur from three independent Western blots is shown as graphs with mean ± SEM. The amount of Fur measured in Ye9Fflag grown in LB + FeCl3 was set to 100%. (B, left panel) The strain Ye9Fflag was grown at a different temperature (26 vs. 37 °C), iron concentration (FeCl3 vs. DPD), reduced pH (5.6), increased oxygen stress (H2O2) or higher osmolarity (LB0 vs. LB0 plus 350 mM NaCl). (B, right panel) Quantification of the amount of Fur from three independent Western blots is shown as graphs with mean ± SEM. The amount of Fur measured in Ye9Fflag grown in LB at 26 °C was set to 100%. (C) Y. enterocolitica strains differing in OmpR content were grown at 26 °C in LB0 and in LB0 supplemented with 350 mM NaCl. The percentage values indicate the protein band intensities relative to Ye9Fflag grown in LB0. (D) Levels of HemR1 protein were analyzed in Y. enterocolitica cells grown in LB supplemented with FeCl3 or DPD at 26 °C for 24 h until stationary phase. Western blotting of cell extracts was performed with a polyclonal anti-HemR1 antibody (upper panel), and the blots were also probed with an anti-DnaK antibody as a loading control (lower panel). The HemR1 band is ~ 75.0 kDa in size. HemR1 levels were compared in strains Ye9Fflag and Ye9fur (fur deletion mutant of Ye9N, Δfur::Gm). The percentage values indicate the protein band intensities relative to Ye9fur. The presented results (A–D) are representative of at least three independent experiments. The immunoreactive bands were quantified by densitometric analysis using ImageQuant TL analysis software (GE Healthcare). MW—3-color prestained protein marker (DNA Gdańsk).
We next examined the influence of different environmental factors (temperature, iron content, low pH, oxidative stress or high osmolarity) on Fur-3×FLAG synthesis in Ye9Fflag. As shown in Figure 3B, iron clearly reduced the abundance of Fur-3×FLAG, while a higher temperature (37 vs. 26 °C) had a slight inhibitory effect on Fur-3×FLAG synthesis. In contrast, low pH (5.6), oxidative stress caused by hydrogen peroxide (100 µM H2O2) and high osmolarity (350 mM NaCl) led to increased Fur production (Figure 3B).
The abundance of Fur-3×FLAG was then checked in strains differing in OmpR content, i.e., Ye9Fflag (WT) and AR4Fflag (ΔompR), grown in LB0 and LB0 supplemented with NaCl (350 mM) (Figure 3C). Higher levels of Fur-3×FLAG were observed in the high-osmolarity medium (350 mM NaCl) regardless of the presence of OmpR, which suggested that osmoregulation occurs independently of this regulator.
Finally, to confirm the functionality of the Fur-3×FLAG fusion protein, expression of the outer membrane protein HemR1 was examined. It was shown previously that production of HemR1 in Y. enterocolitica is significantly influenced by iron and Fur content and is undetectable in wild-type strain Ye9N grown under iron-replete conditions and highly abundant in iron-depleted conditions [15]. Western blotting with an anti-HemR1 antibody showed an elevated level of HemR1 in the strain Ye9Fflag, synthesizing Fur-3×FLAG, grown under iron-limiting conditions, whereas no HemR1 could be observed in iron-replete conditions (Figure 3D). In strain Ye9N with the Δfur mutation, HemR1 production was notably increased, confirming that Fur is involved in iron-dependent inhibition of HemR1 protein production. These data confirm the regulatory function of Fur/iron and show that the presence of the 3×FLAG peptide did not alter the activity of the Fur regulator.
2.5. The Effect of OmpR and Successive Deletion of the Fur Regulatory Region on the Transcriptional Activity of the YePfur Promoter
The presence of four potential OmpR-binding sites within the fur regulatory region (M1, M2, M3, M4; Figure 4A) prompted us to investigate whether the activity of the fur promoter depends on these cis-acting sequences. The fur regulatory region encompassing the four putative OBSs (fragment F1234) was successively deleted upstream of the predicted core promoter motifs to produce DNA fragments of different lengths that harbor a decreasing number of OBSs, i.e., fragments F234 (M2, M3, M4), F34 (M3, M4) and F4 (M4). In addition, fragment F0, lacking all OBSs, was obtained by removing OBS M4 located downstream of the −10 core promoter motif. These fragments were used to create transcriptional fusions with the lacZ reporter gene in the vector pCM132Gm, producing constructs pCMF234, pCMF34, pCMF4 and pCMF0, respectively (Figure 4A). The four plasmids were introduced into the wild-type Y. enterocolitica Ye9N and these strains, together with Ye9N harboring plasmid pCMF1234, carrying the fragment encompassing all four OBSs, were used to study the transcriptional activity of fur in cells grown to the early stationary phase in iron-replete and iron-limiting conditions. Regardless of the number of OBSs, YePfur::lacZ expression was always higher under iron-limiting conditions, which indicated that all studied regulatory fragments contain cis-acting sequences responsive to iron. Since all transcriptional fusions retained the Fur-box sequence, this suggested that the regulator Fur was responsible for the observed iron-dependent repression. However, sequential deletion of three putative OBSs from the distal 5′-end of the fur regulatory region (M1, M2, M3) did not significantly affect the expression of YePfur::lacZ. Interestingly, shortening of the fur regulatory region by removal of OBS M4, located downstream of the −10 promoter motif (i.e., construct pCMF0), led to a ~50% increase in β-galactosidase activity in both iron-replete and iron-limiting conditions, which suggests some role for this cis-regulatory sequence in the inhibition of YePfur activity (Figure 4A).
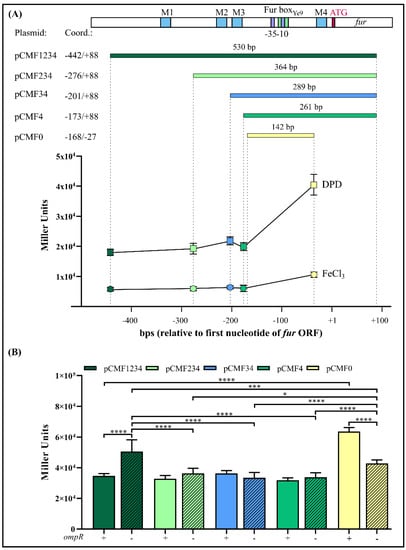
Figure 4.
Effect of successive deletion of the Y. enterocolitica Ye9N fur regulatory region on the expression of fur. (A) At the top, a schematic presentation of the Y. enterocolitica fur promoter region (YePfur) with the location of potential OmpR-binding motifs (M1, M2, M3 and M4) marked. Below this, the different fragments of YePfur used to construct transcriptional fusions with lacZ in pCM132Gm are shown. The sizes of the fragments are given and coordinates assigned, with +1 corresponding to the first nucleotide of the fur ATG start codon. At the bottom, the expression of the Pfur::lacZ transcriptional fusions from the following plasmids is plotted: pCMF1234, pCMF234, pCMF34, pCMF4 and pCMF0. β-Galactosidase activity was measured in Ye9N (WT) transformants grown to early stationary phase in LB medium at 26 °C, under iron-limiting (150 μM DPD) or iron-replete (10 μM FeCl3) conditions. Data represent mean activity values with SD from three independent experiments. The obtained differences are statistically significant with p < 0.0001. Significance was calculated using one-way ANOVA. (B) Expression of Pfur::lacZ fusions encompassing different fragments of YePfur in strains Ye9N (WT, +ompR, smooth bars) and AR4 (ΔompR mutant, −ompR, hatched bars). β-Galactosidase activity was measured in the strains grown to early stationary phase in LB medium at 26 °C. The presented data represent mean β-galactosidase activity values with SD from three independent experiments, each performed using at least triplicate cultures of each strain. Significance was calculated using one-way ANOVA (* p < 0.05, *** p < 0.001, **** p < 0.0001).
To elucidate the role of OmpR in the regulation of the fur promoter, the YePfur::lacZ transcriptional fusions described above were analyzed in the wild-type strain Ye9N and the isogenic ΔompR mutant AR4 (Figure 4B), grown to the early stationary phase in LB medium and LB with DPD. As was previously shown, expression of the YePfur::lacZ fusion with all four OmpR-binding sites present (i.e., fragment F1234) was significantly higher in the mutant AR4, lacking OmpR, compared with the wild-type strain Ye9N, both grown in LB medium. However, when the promoter region was deprived of the regulatory fragment containing OBS M1, no influence of OmpR on the expression of the YePfur::lacZ fusion was observed. Moreover, further successive deletion of the fur regulatory region harboring M2 and M3 in the constructed fusions did not lead to changes in β-galactosidase levels in the ompR+ compared to the ompR- background. Interestingly, when the fur regulatory region fused to lacZ-lacked sequences downstream of the −10 core promoter motif including the potential OBS M4, upregulation of expression was observed in strain Ye9N compared to the other fusions. However, less upregulation was noted in the ΔompR mutant strain carrying the same fusion. Considering the transcriptional activity of the different YePfur::lacZ fusions, OmpR-mediated repression of fur was most affected by deletion of putative OBS M4. Interestingly, the same pattern of OmpR-dependent expression of fur, determined using reporter gene fusions, was observed in cells grown under iron-limiting conditions (data not shown). Taken together, these results suggest that fur repression is mediated by iron/Fur and the OmpR regulator, which probably primarily interacts with site M4. The presence of iron does not appear to influence OmpR-dependent fur regulation.
2.6. Interaction of OmpR with the Full Length and Truncated Fur Regulatory Regions of Y. enterocolitica
The four potential OmpR-binding sites in the fur promoter region of Y. enterocolitica indicated by in silico analysis (M1, M2, M3, M4) display 45%, 60%, 55% and 50% identity to the E. coli consensus OmpR-binding site, respectively (Figure 1B). To determine whether these OBSs function as binding sites for OmpR in vitro, we performed electrophoretic mobility shift assays (EMSAs) using native polyacrylamide gels to detect binding of the purified OmpR-His6 protein to fur regulatory region DNA fragments containing different numbers of potential OmpR target sequences (Figure 5). First, EMSAs were performed using the 445-bp F1234 fragment encompassing the whole fur regulatory region (−442 to +3). This fragment, containing four potential OBSs (M1, M2, M3, M4) (Figure 6A), was PCR-amplified with the primer pair EFurYe1/EFurYe2 (Table S2) and incubated with increasing amounts of the His-tagged OmpR protein. Four shifted bands of decreasing mobility were observed as the OmpR concentration was increased from 0.65 to 3.25 µM, indicating the presence of multiple OmpR-binding sites. Next, a competitive bandshift assay was performed to study the ability of OmpR to alter the electrophoretic mobility of fragment F1234 when a control 304-bp 16S rDNA fragment was included in the binding reaction (Figure 6B). Again, slower migrating OmpR/DNA complexes were observed in the presence of OmpR at concentrations of 0.65 μM and above. OmpR had clearly bound to the fur promoter fragment encompassing four putative OBSs but failed to interact with the 16S rDNA fragment used as a negative control.
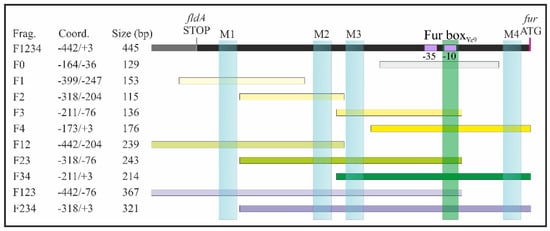
Figure 5.
DNA fragments of the Y. enterocolitica Ye9N fur regulatory region used in electrophoretic mobility shift assay (EMSA) experiments. The position of DNA fragments within the extended Y. enterocolitica fur regulatory region is shown. The fragments contain one (yellow bars), two (green bars), three (purple bars) or four (black bar) putative OmpR-binding sites, or no sites (gray bar). Light blue rectangles indicate the location of the four potential OmpR-binding motifs M1, M2, M3 and M4. The putative Fur boxYe9 is marked in green, and the −10 and −35 core promoter motifs are marked in violet. The DNA fragment names, sizes and coordinates relative to the first nucleotide of the fur ATG start codon (marked in pink) are given. All fragments were amplified with primers listed in Supplementary Table S2.
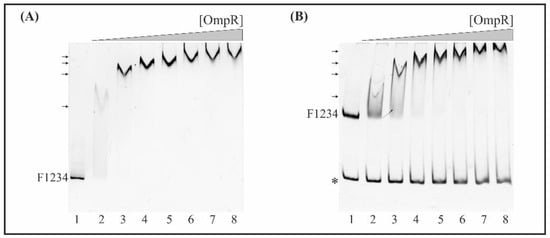
Figure 6.
EMSA of the interaction between OmpR-His6 with the F1234 DNA fragment encompassing the whole fur regulatory region including four putative OmpR-binding sites. (A) Quantitative EMSA. The 445-bp fur promoter fragment F1234 was incubated without protein (lane 1) or with increasing amounts of purified OmpR: 0.65 (lane 2), 1.3 (lane 3), 1.95 (lane 4), 2.6 (lane 5), 3.25 (lane 6), 3.9 (lane 7), 4.55 μM (lane 8). (B) Competitive EMSA. Mixtures of the 445-bp fur promoter fragment F1234 and a 304-bp fragment of 16S rDNA (*) were incubated with increasing amounts of purified OmpR as in (A). The OmpR/DNA complexes were resolved on a native polyacrylamide gel and detected by SYBR Green staining. Shifted bands containing the fur fragment F1234 are marked by arrows.
Subsequently, EMSAs were performed using fragments of the fur regulatory region lacking OBS M1 or M4, located at the 5′- and 3′-ends of the sequence, respectively. These assays revealed that fragment F234 (−318 to +3), lacking OBS M1 (Figure 7A), and fragment F123 (−442 to −76), lacking M4 (Figure 7B), possessed OmpR-binding ability. However, in both cases, a smaller number of step-shifted slower migrating OmpR/DNA complexes appeared in the presence of OmpR at a higher concentration (1.31 μM), compared to fragment F1234 (−442 to +3) (Figure 6). The 304-bp 16S rDNA, used as the negative control in both cases, was not shifted. These data suggest that both M1 and M4 are responsible for the gel mobility shift pattern observed with the fragment F1234.
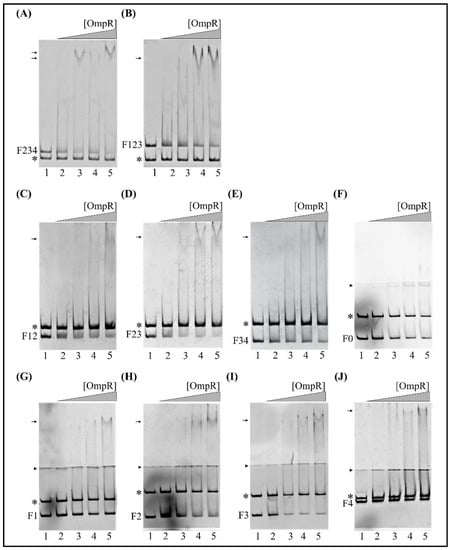
Figure 7.
EMSAs of the interaction between OmpR-His6 and sub-fragments of the fur regulatory region carrying different numbers of potential OmpR-binding sites. (A,B) EMSAs with fur regulatory region DNA fragments comprising three potential OmpR-binding sites (OBSs): (A) lacking motif M1 from the 5′-end (F234, 321 bp) and (B) lacking motif M4 from the 3′-end (F123, 367 bp). (C–E) EMSAs with DNA fragments comprising two potential OBSs: (C) F12 (239 bp), (D) F23 (243 bp) and (E) F34 (214 bp). (F) EMSA with a DNA fragment lacking all OBSs. (G–J) EMSAs with DNA fragments containing one OBS: (G) F1 (153 bp), (H) F2 (115 bp), (I) F3 (136 bp) and (J) F4 (176 bp). In all cases, the DNA fragments were incubated without protein (lane 1) or with 0.65 (lane 2), 1.3 (lane 3), 1.95 (lane 4) or 2.6 μM (lane 5) OmpR-His6. Free DNA and protein/DNA complexes (arrows) are indicated. Fragments of 16S rDNA (*) were included in the reaction mixtures with the fur regulatory region fragments and served as a negative control. The OmpR/DNA complexes were resolved on a native polyacrylamide gel and detected by SYBR Green staining. In the EMSAs with very short DNA fragments (F–J), electrophoresis was performed on native polyacrylamide gels with two layers, i.e., a large pore 4.2% gel on the top and small pore 8.4% gel on the bottom, for better visualization of shifted complexes (arrowheads indicate the interface between the two gel concentrations).
Next, we performed EMSAs to study the ability of OmpR to directly interact with the two potential OBSs within fragments F12 (−442 to −204, M1 and M2), F23 (−318 to −76, M2 and M3) and F34 (−211 to +3, M3 and M4), in the presence of the 304-bp 16S rDNA control fragment. Native polyacrylamide gel electrophoresis of binding reactions containing equimolar amounts of these DNA fragments demonstrated the formation of slower migrating OmpR/F12, OmpR/F23 and OmpR/F34 complexes (Figure 7C–E). The complex OmpR/F23 was already present at a protein concentration of 1.3 μM (Figure 7D, lane 3). All of these OmpR/DNA complexes were specific, and their appearance was associated with the simultaneous disappearance of fragments F12, F23 and F34. OmpR was unable to bind the 16S rDNA negative control fragment. Notably, the 129 bp fragment F0 (−164 to −36), lacking all predicted OBSs, was not shifted at any of the OmpR concentrations employed (Figure 7F).
Finally, to determine the comparative hierarchy of OmpR binding by the putative OBSs, we tested their individual contributions to the binding of OmpR to the fur regulatory region. For this purpose, DNA fragments of ~100 to 200 bp long carrying single OBSs were used in EMSAs with increasing amounts of OmpR. For improved visualization of slower migrating OmpR/DNA complexes, electrophoresis was performed on native polyacrylamide gels cast in two layers, with a large pore gel on the top and a small pore gel on the bottom.
The 153-bp DNA fragment F1 (−399 to −247), containing the most upstream fur regulatory region including OBS M1, exhibited a mobility shift at 2.6 µM OmpR (Figure 7G, lane 5). A lower concentration of OmpR (1.95 µM) started to shift the 115-bp fragment F2 (−318 to −204) encompassing OBS M2 (Figure 7H, lane 4), and the 136-bp fragment F3 (−211 to −76) containing M3 (Figure 7I, lane 4). In turn, the 176-bp fragment F4 (−173 to +3) containing OBS M4, located downstream of the fur core promoter, was shifted at 2.6 µM OmpR (Figure 7J, lane 5). The 211-bp 16S rDNA control fragment, present in each reaction together with the studied targets, was not shifted. Analysis of the EMSA results for these single OBS target fragments established the binding hierarchy as M2 and M3 > M1 and M4.
2.7. E. coli Fur Expression Is Negatively Regulated by OmpR
In light of the results obtained for Y. enterocolitica, we decided to examine the regulatory region of E. coli fur for the presence of potential OmpR-binding sites. In silico analysis revealed that the organization of fur in E. coli K-12 strain MG1655 (GenBank Acc. No. NZ_CP009273.1) is similar to that of Y. enterocolitica Ye9N (GenBank Acc. No. NZ_JAALCX000000000.1), but with slight differences (Figure 8A). Immediately upstream of E. coli fur, there is the ORF uof encoding a short leader polypeptide, which is absent in Y. enterocolitica. Upstream of uof, there is the fldA ORF, as is also the case in the Y. enterocolitca fur locus. Downstream of E. coli fur, the chiQ ORF encoding chitosugar-induced lipoprotein is present instead of chb, encoding chitobiase in Y. enterocolitica. The fur gene of E. coli may be co-transcribed with fldA or with uof from their respective promoters. In addition, two fur gene promoters are recognized (fura and furb), so potentially four transcriptional units carrying fur might be induced under certain growth conditions (Figure 8A) [47,85,86].
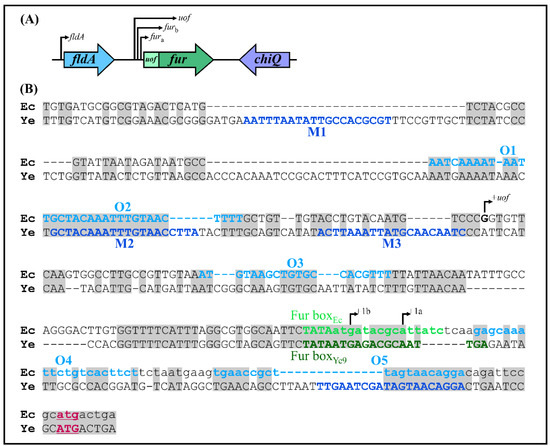
Figure 8.
Comparison of the fur loci of Y. enterocolitica and E. coli. (A) Chromosomal organization of the E. coli fur locus. The direction of transcription from the fldA, uof and the two fur promoters is indicated by arrows (based on 47 and 85). The following products are encoded by these genes: fldA—flavodoxin I, uof—Fur leader peptide, fur—Fur (ferric uptake regulator), chiQ—chitosugar-induced lipoprotein. (B) Alignment of the fur regulatory regions of E. coli and Y. enterocolitica. Chromosomal sequences of E. coli (Ec) and Y. enterocolitica (Ye) extending from the end of the fldA gene to the start codon of fur (plus five nucleotides from the end of the uof gene present in E. coli) were aligned using BlastN. Identical nucleotides are shaded gray. The putative transcription start sites of E. coli fur (a and b) and uof are indicated by +la, + lb and +1uof, respectively. Putative OmpR-binding sites (M1–M4) from Y. enterocolitica are in dark blue text, while putative E. coli OmpR-binding sites (O1–O5) are in light blue. The Fur-binding sites (Fur boxEc and Fur boxYe9) are in light and dark green text, respectively. The sequence of E. coli uof is written in lowercase text. The fur translation start codons are in bold, underlined and colored pink.
DNA sequence alignment revealed that the fur promoter regions of E. coli (289 bp) and Y. enterocolitica (376 bp) are 86% identical, with multiple nucleotide deletions in the former sequence (Figure 8B). Five putative OmpR-binding sites O1, O2, O3, O4 and O5, respectively, exhibiting 55, 55, 50, 50 and 55% identity to the E. coli consensus, are present in the E. coli fur regulatory region (Figure 9A), suggesting some influence of OmpR on fur expression in this bacterium. Two of the OBSs, O1 and O2, are positioned upstream of the uof transcription start site (+1uof), whereas O3 is located upstream of the fur transcription start sites (+1fura and +1furb) and O4 and O5 downstream of them (the OBSs O3, O4 and O5 are within the uof ORF). The Fur box overlaps the Pfura −10 core promoter motif.
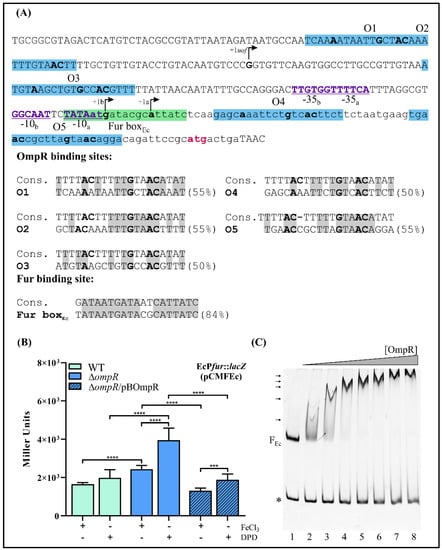
Figure 9.
OmpR-dependent fur regulation in E. coli. (A) The fur promoter region of E. coli (based on 47 and 85). The nucleotide sequence from nt −284 to +12 relative to the first nucleotide of the fur ATG start codon. The putative OmpR-binding sites are highlighted by a blue background, while the Fur-binding site (Fur boxEc) is highlighted by a green background. The translation start codon of fur is colored pink. The −10 and −35 elements of the fur promoter are in bold, underlined and colored purple. The transcription start sites of E. coli fur (a and b) and uof are indicated by +la, +lb and +1uof, respectively. The lowercase sequence represents the uof ORF. The putative OmpR- and Fur-binding sites are shown aligned to the E. coli consensus sequences (% identity values are shown). Identical nucleotides in the compared sequences are shaded gray. In both the promoter region sequence and alignments, the nucleotides important for OmpR binding are in bold. (B) Expression of a plasmid-encoded EcPfur::lacZ transcriptional fusion (pCMFEc) in E. coli strains BW25113 (WT) and JW3368-1 (ΔompR) and complemented strain JW3368-1/pBOmpR (expresses OmpR from Y. enterocolitica Ye9). β-Galactosidase activity was measured in strains grown at 37 °C to early stationary phase in LB medium, under iron-limiting (150 μM DPD) or iron-replete (10 μM FeCl3) conditions. The presented data represent mean activity values with SD from three independent experiments carried out in triplicate. Significance was calculated using one-way ANOVA (*** p < 0.001, **** p < 0.0001). (C) EMSA of the interaction between Y. enterocolitica OmpR-His6 and the E. coli fur promoter region (FEc) depicted in Figure 9A. The 304-bp fur promoter fragment FEc and the 211-bp fragment of 16S rDNA (*, from Y. enterocolitica Ye9) were incubated together in reaction mixtures without protein (lane 1) or with an increasing amount of purified OmpR: 0.65 (lane 2), 1.3 (lane 3), 1.95 (lane 4), 2.6 (lane 5), 3.25 (lane 6), 3.9 (lane 7), 4.55 μM (lane 8). The OmpR/DNA complexes were resolved on a native polyacrylamide gel and detected by SYBR Green staining. Shifted bands containing the fur fragment FEc are marked by arrows.
This in silico analysis and the results obtained for Y. enterocolitica prompted us to check the OmpR-dependent regulation of fur expression in E. coli. The EcPfur::lacZ transcriptional fusion in vector pCM132Gm was constructed similarly to the YePfur::lacZ fusion. Briefly, a 250-bp DNA fragment comprising 242 bp upstream of the fur start codon plus 8 bp of the fur ORF (encompassing all five putative OmpR-binding sites) was cloned in pCM132Gm. The obtained plasmid pCMFEc was introduced into the wild-type strain BW25113 and ompR deletion mutant JW3368-1. These strains were grown at 37 °C in LB supplemented with FeCl3 (10 µM) or DPD (150 µM). β-Galactosidase assays performed on cells in the early stationary phase showed that expression from the E. coli fur promoter was ~1.5-fold higher in the ΔompR mutant than in the WT strain in iron-replete conditions and ~2-fold higher in iron-limiting conditions (Figure 9B). Since the predicted amino acid sequences of the OmpR regulators of Y. enterocolitica and E. coli share 98% identity (data not shown), this led us to use OmpR of Y. enterocolitica to complement the ΔompR mutation in E. coli. When OmpR was expressed from plasmid pBOmpR in the E. coli ΔompR mutant background, fur promoter activity was decreased to the wild-type level. This confirmed the negative role of OmpR in fur regulation and showed that Y. enterocolitica OmpR can functionally substitute the OmpR of E. coli (Figure 9B). The presence of iron had no influence on EcPfur::lacZ expression in the wild-type strain, which contrasts with the clear effect in the ΔompR mutant strain, under the tested conditions. Finally, to reveal whether OmpR interacts with the fur regulatory region of E. coli, we performed EMSAs using a DNA fragment containing all predicted OBSs (O1, O2, O3, O4, O5). Increasing amounts of Y. enterocolitica OmpR-His6 were incubated with the 304-bp E. coli fur regulatory region fragment (FEc) and a 211-bp fragment of Y. enterocolitica Ye9N 16S rDNA was included in the binding reactions as a negative control. As shown in Figure 9C (lane 2), OmpR/DNA complexes of decreasing mobility were observed on a native polyacrylamide gel with an increasing concentration of OmpR, starting from 0.65 μM. Multiple shifted bands were observed as the OmpR concentration was increased up to 3.9 µM, suggesting that OmpR interacts with several sites within the fur regulatory region of E. coli.
2.8. Effect of OmpR on the Expression of Selected Fur Regulon Members in Y. enterocolitica
The next step in our studies was to examine whether the OmpR-dependent regulation of Fur might affect the expression of Fur regulon genes in Y. enterocolitica. Several genes belonging to the Fur regulon were chosen based on global analyses of iron assimilation and fur regulation performed previously in Y. pestis and E. coli [24,41,42] and a differential proteomic analysis of OmpR-regulated proteins in Y. enterocolitica [80]. Three genes controlled by Fur were selected: fecA and fepA encoding OM receptor proteins FecA and FepA necessary for transport of ferric dicitrate and ferrienterobactin, respectively, and feoA encoding an inner membrane protein required for Fe2+ transport (FeoABC system).
Initially, the levels of the fecA, fepA and feoA mRNAs were assessed by RT-qPCR in strains Ye9N (the wild-type strain), Ye9fur (Δfur), AR4 (ΔompR) and AR4fur (ΔompRΔfur). The Δfur::Gm mutants of Y. enterocolitica Ye9N (Ye9fur) and AR4 (AR4fur) were constructed by homologous recombination as described previously [15]. The obtained data show an increase in the mRNA transcript level of 2.4-fold for fecA and 34-fold for fepA in the Ye9fur strain (Δfur background) compared to the wild-type strain, indicating Fur-mediated repression of both genes (Figure 10). In strain AR4 (ΔompR background), the fecA and fepA transcript levels were increased 4.7-fold and 4.3-fold, respectively, relative to the wild-type strain, indicating that OmpR has a negative impact on fecA and fepA expression. Interestingly, the upregulation of fecA in the ΔompR background was 2-fold higher than that observed in the Δfur background and did not really change when these mutations were combined (ΔompRΔfur), indicating that the inhibitory effect of OmpR exceeds the repressive activity of Fur and can be observed regardless of Fur (Figure 10). In contrast, the strain devoid of Fur (Δfur background) exhibited 8-fold higher fepA expression than that lacking OmpR (ΔompR background). However, the combination of both mutations (ΔompRΔfur) led to a 2-fold decrease in the fepA transcript level, suggesting that OmpR may have a positive impact on fepA expression in the absence of Fur (Figure 10). Regarding the expression of the third tested gene, a 2.1-fold increase in feoA transcripts was observed in a Δfur background relative to the wild-type strain, indicating inhibition of feoA expression by Fur (Figure 10). In turn, the feoA mRNA level decreased slightly in the ΔompR genetic background in comparison to the wild type, indicating a weak positive influence of OmpR. Interestingly, in the double mutant (ΔompRΔfur), a ~4-fold decrease in the feoA mRNA transcript relative to the Δfur background was observed, resulting in levels equivalent to those in the ΔompR background. Thus, it seems that upregulation of feoA by OmpR may abolish Fur repression.
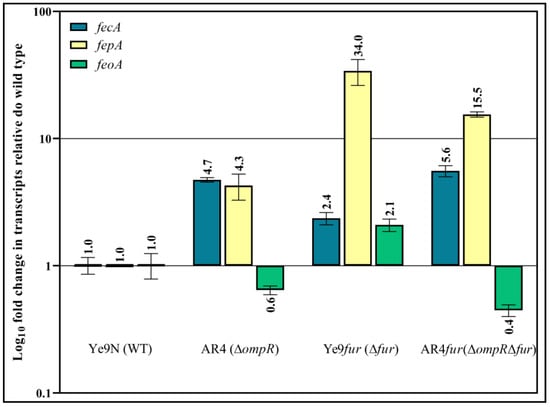
Figure 10.
OmpR- and Fur-dependent regulation of fecA, fepA and feoA transcription in Y. enterocolitica Ye9N. Effect of OmpR on Fur regulon members expression determined by RT-qPCR. This analysis was performed using RNA prepared from cells of the wild-type strain (Ye9N), the ΔompR mutant (AR4), the Δfur mutant (Ye9fur) and the double ΔfurΔompR mutant (Ye9furompR), grown to early stationary phase in LB medium. Relative fecA, fepA, feoA and ftnA transcript levels, normalized to the amount of 16S rRNA, are shown, taking the mRNA level in Ye9N as 1. The mean value and SD obtained from at least three independent experiments are indicated for each strain.
To clarify whether OmpR of Y. enterocolitica may participate in the direct regulation of fecA, fepA and feoA gene expression, we first analyzed their promoter regions in silico to identify putative OmpR-binding sites. One potential OBS was found in the promoter region of fecA, located at −40 to −21 nt relative to the start codon (overlapping the −10 core promoter motif) and two putative OBSs were identified in the promoter region of fepA (at −210 to −191 nt and −144 to −125 nt relative to the start codon) (Figure 11, appropriate left panels). Each OBS exhibits 50% identity to the consensus OmpR-binding site. We failed to detect any potential OBSs in the analyzed feoA promoter region. With regard to the Fur box, potential examples were identified in each of the three analyzed genes, overlapping the −10 or −35 core promoter motifs (Figure 11, left panels).
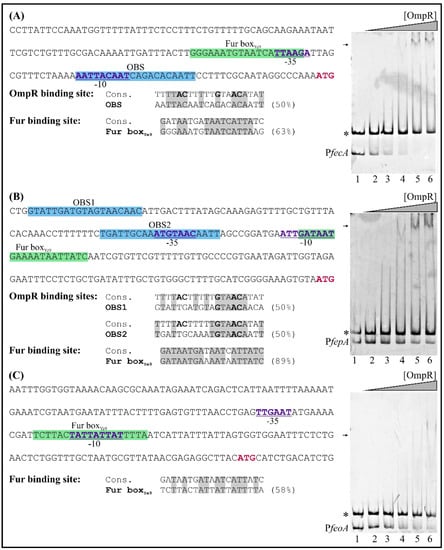
Figure 11.
Binding of OmpR-His6 to the fecA, fepA and feoA regulatory regions examined using EMSAs. Left panels show the promoter of fecA (A) from −159 to +3 nt relative to the ATG start codon, fepA (B) from −213 to +3 nt relative to the ATG start codon and feoA (C) from −200 to +16 nt relative to the ATG start codon, while right panels are the results of EMSA experiments. The sequences shaded blue correspond to the putative OmpR-binding sites. The potential Fur-binding site (Fur boxYe9) is highlighted by a green background. The fecA, fepA and feoA start codons (ATG) are shown in bold and marked pink. Beneath the sequence, the putative binding sites are compared with the consensus OmpR and Fur-binding motifs of E. coli. The percentage identity to these sequences is shown. Identical nucleotides in the compared sequences are shaded gray. Nucleotides important for OmpR binding are in bold. The PfecA (189 bp), PfepA (247 bp) and PfeoA (227 bp) promoter fragments and the 304-bp fragment of 16S rDNA (*, from Y. enterocolitica Ye9) were incubated together in reaction mixtures without protein (lane 1) or with an increasing amount of purified OmpR: 0.6 (lane 2), 1.2 (lane 3), 2.4 (lane 4), 4.8 (lane 5), 6 μM (lane 6). The OmpR/DNA complexes were resolved on a native polyacrylamide gel and detected by SYBR Green staining. Shifted bands containing the fecA/fepA/feoA promoter region are marked by arrows.
Regardless of the results of the in silico analysis, the promoter regions of the fecA, fepA and feoA genes were examined for OmpR binding using EMSAs. DNA fragments were incubated with increasing concentrations of OmpR (Figure 11, right panels). Shifted OmpR/DNA complexes were clearly produced when 189-bp fecA (−162 to +27 relative to the start codon) and 247-bp fepA (−305 to −59 relative to the start codon) fragments interacted with OmpR at a concentration of 2.4 µM (Figure 11, appropriate right panels). No shifting of an unrelated control DNA fragment (304-bp, 16S rDNA) was observed, confirming that these OmpR/DNA complexes were specific. In addition, their appearance was associated with the simultaneous disappearance of the fecA and fepA DNA fragments. The 227-bp feoA promoter region fragment (−195 to +32 relative to the start codon) interacted weakly with OmpR, and faint shifted OmpR/DNA complexes appeared at a concentration of 4.8 µM. Thus, this regulator appears to bind with low affinity to the studied feoA promoter region. Together, these EMSA results indicate that OmpR can interact directly with the fecA and fepA promoter regions in vitro, but the interaction with the feoA promoter seems to be much weaker.
3. Discussion
OmpR has been identified as a transcriptional regulator of various genes involved in the control of diverse cellular processes and functions in bacteria including Y. enterocolitica [58,68,75,87,88]. Based on the results of proteomic analyses, the production of a number of membrane proteins involved in the uptake and transport of compounds into Y. enterocolitica cells, including several that participate in iron or heme acquisition, is subject to control by the regulator OmpR [80]. The acquisition and storage of iron are modulated by the Fur regulator to maintain iron homeostasis necessary for bacterial survival [89]. In light of these findings, we initiated studies aimed at identifying any correlation between the function of the OmpR regulator and the expression of Fur in Y. enterocolitica. Despite the importance of Fur in the regulation of iron/heme acquisition in pathogenic Yersinia [8,9,15,41,90], the control of Fur biosynthesis is poorly understood. Analysis of the fur promoter region of Y. enterocolitica strain Ye9N identified one putative Fur-binding sequence and four predicted OmpR-binding sequences, suggesting a potential role for these regulators in the modulation of fur expression. In the course of this study, we identified OmpR as an inhibitor of fur expression, acting independently of Fur. Moreover, the activity of the YePfur promoter was found to be weakly inhibited by iron, implying a negative autoregulatory role for Fur. The fur genes of other Gram-negative bacteria are often, but not always, negatively autoregulated in the presence of iron, due to the binding of Fur to the Fur-box motif [25,49,91,92]. However, the activity of the fur promoters of Campylobacter jejuni and Burkholderia cepacia was not influenced by iron availability [45,93].
To ascertain whether the Y. enterocoltica Pfur is responsive to environmental cues, several factors known to influence OmpR and/or Fur level/activity in E. coli and Salmonella were tested [58,60,61]. OmpR is a component of the EnvZ/OmpR TCS, known to be one of the major regulatory systems in E. coli controlling the expression of outer membrane porins in response to osmolarity [55,94]. In an environment of high osmolarity, EnvZ phosphorylates the regulatory protein OmpR, leading to its activation and inverse regulation of the E. coli ompC and ompF genes [95]. In Y. enterocolitica, OmpR appears to be required for the inhibition of fur expression, so it might be expected that a decrease in the activity of YePfur should be observed under high-osmolarity conditions. However, we found that Y. enterocolitica fur expression increases in conditions of high osmolarity and independently of the presence of OmpR. We presume that other mechanisms may be involved in the observed regulation, i.e., changes in DNA supercoiling and the activity of nucleoid structuring proteins such as H-NS, known to be involved in the osmoregulation of some genes in E. coli [96,97]. In addition, it has been shown that some genes of the OmpR regulon are not responsive to changes in osmolarity [98,99,100]. Interestingly, recent studies have provided evidence for the involvement of Fur as part of the complex circuit that controls the response to osmotic stress in halophilic bacteria [101]. Low pH was recognized as an environmental signal that influences the expression of several genes in an OmpR-dependent manner in many bacterial species [102,103,104]. However, low pH did not have a significant impact on YePfur activity. We also showed that YePfur activity is not influenced by novobiocin, a compound known to induce DNA relaxation. Changes in DNA topology may influence the activity of some promoters in an OmpR-dependent manner [84], but this does not seem to apply to YePfur. An analysis of OmpR-dependent genes within Proteobacteria suggested that the OmpR regulon is not highly conserved across bacterial species and different regulatory networks exist to respond to the various environmental stress conditions encountered [105]. We observed no effect of hydrogen peroxide or the superoxide generator paraquat on YePfur promoter activity. This is in contrast to the situation in E. coli where fur gene expression is activated by both stressors. Previously, OxyR was identified as a regulator that senses elevated levels of hydrogen peroxide to induce fur expression in E. coli, while SoxRS was found to increase fur expression in response to paraquat [47]. The absence of any observable effect of oxidative stress on the fur transcription of Y. enterocolitica parallels the results of studies investigating the fur genes of B. cepacia, B. pseudomallei and P. aeruginosa [93,106,107]. Since bacteria experience the continual fluctuation of numerous environmental parameters, which alters the expression of many genes including regulators, this would explain the difficulty in identifying a single factor mediating changes in the expression level of fur. When Y. enterocolitica passes from the external environment into the host body, conditions such as temperature, pH, iron and oxygen availability and osmolarity change comprehensively. Therefore, a combination of these factors might be responsible for modulating fur expression.
To clarify whether OmpR participates in the production of the Fur regulator, Western blot analysis was performed. The results show that OmpR reduces Fur protein levels, in agreement with the fur promoter activity test data. In addition, iron had a greater effect on the level of the Fur protein than on fur transcription, suggesting the presence of another regulatory mechanism affecting iron-dependent fur expression. The increase in Fur protein abundance produced by high osmolarity correlated with the upregulation of fur transcription under these conditions. Interestingly, low pH and H2O2 led to increased Fur synthesis that was not reflected at the transcriptional level. It has previously been shown that the effect of low pH on gene expression is complex and interconnected with other regulatory factors. Changes in mRNA abundance, and hence in the level of protein synthesis, may be the consequence of mRNA stabilization or destabilization [108]. Enhanced RNA stability at acidic pH, not only the RNA phosphodiester bond but also the aminoacyl-(t)RNA and peptide bonds, has been demonstrated [109]. Notably, the translation process and the regulation of mRNA stability are preferential targets of H2O2 in eukaryotic organisms [110].
Four potential OmpR-binding sites (M1–M4) were identified in the fur promoter region of Y. enterocolitica. These share considerable similarity with the OmpR consensus binding sequence defined based on sites identified within the E. coli ompF and ompC promoter regions [82]. The putative OmpR-binding sites in YePfur share several features, in particular the central GXXAC motif shown to be critical for OmpR binding [56,111]. In the case of OBS M2, a conserved AC base pair located 9 nt away from the AC element of the central motif GXXAC was also present. This sequence configuration was previously demonstrated to promote the stability of OmpR binding [56,111]. Recently, by applying ChIP-chip analysis, SELEX-chip screening and ChIPMunk methods, other binding site motifs bound by OmpR with different affinities were identified in E. coli [58,105,112]. Thus, the available data indicate that OmpR has moderate binding sequence specificity.
The study of the relative contribution of four putative OmpR-binding sites on Pfur activity revealed that deletion of the fur regulatory region from the 5′-end encompassing OBS M1, M2 and M3 did not affect the promoter activity. However, significant upregulation of fur occurred when the deletion was extended to include OBS M4, located downstream of the −10 core promoter motif, suggesting that M4 is most important for OmpR-dependent inhibition of fur expression. Unexpectedly, the extent of this upregulation of fur observed in the wild-type strain was reduced in the ΔompR mutant background, concomitantly with the lack of OmpR, which suggests a role for other transcriptional regulators whose expression could be under OmpR control or some interplay with OmpR at the DNA sequence level. Upon analyzing other YePfur::lacZ fusions in the absence of OmpR, we found that the removal of OBS M1, but not M2 and/or M3, led to the inhibition of fur expression, suggesting the presence of a cis-regulatory sequence for a putative transcriptional activator. In E. coli, the OxyR and SoxRS regulators co-activate fur expression in response to oxidative stress [47,105]. Our in silico analysis identified potential binding sites for these activators within the Y. enterocolitica fur promoter region. Between OBS M1 and M2, putative SoxR- and SoxS-binding motifs with 58% and 70% identity to the E. coli consensus sequences were revealed (data not shown) [105,113]. Moreover, an OxyR-binding site (OxBS) that overlaps OBS M2 was recognized (data not shown). The nucleotide sequence of Y. enterocolitica OxBS exhibits 100% identity to the E. coli OxBS sequence found in the fur promoter [47]. These potential regulatory sequences and OxyR and/or SoxRS might be responsible for activation of fur expression in Y. enterocolitica. However, confirmation of this hypothesis will require further detailed study, especially since the activation of fur expression by oxidative stress was not detected under the conditions tested. We cannot exclude the possibility that the observed effect of DNA sequence deletions on the expression from YePfur is the result of changing the distance between regulatory sites, which alters the number of turns of the DNA helix, thus affecting the interaction of bound factors and influencing promoter activity, as has been demonstrated for the promoters of ilvGp2 and the araBAD operon [114,115].
EMSAs were performed to demonstrate the ability of OmpR to bind to the fur promoter region in vitro and establish the OmpR-binding hierarchy of the OBSs. The analysis revealed that OBSs M2 and M3, with a higher degree of identity to the consensus binding site, were most important for OmpR binding in vitro. In addition, only M2 among the four OBSs has a second AC nucleotide doublet at the correct distance from the TGTAAC motif, recognized as the most highly conserved and critical sequence for OmpR binding [111]. The EMSA results allow us to establish the hierarchy of OmpR binding within the fur regulatory sequences as M2, M3 > M1, M4. Together, these data suggest that fur repression may depend on the presence of all four OBSs, which could bind OmpR molecules that interact with each other in a complex way. The regulation of fur expression by OmpR might be based on hierarchical and cooperative binding of this protein to OBSs in the fur regulatory sequence, as was observed previously for ompC and ompF of E. coli [116,117,118,119,120]. Progressive occupancy of the OBSs in the fur promoter region could provide differential modulation of the OmpR-mediated repression. The occurrence of multiple repressor binding sites has been described for OmpR in the csgD promoter [121] and for regulators CpxR [122] and CytR [123]. To summarize, our results show that OmpR inhibits fur expression by direct binding to YePfur and that the other putative regulatory proteins may influence OmpR-mediated regulation to ensure the appropriate abundance of Fur in Yersinia cells.
In the light of our findings regarding OmpR and fur expression in Y. enterocolitica, we were curious whether OmpR participates in the regulation of fur in E. coli. Despite sequence divergence in the fur regulatory regions of Y. enterocolitica and E. coli, in silico analysis indicated five potential OmpR-binding sites in the latter. Experiments conducted with an EcPfur::lacZ transcriptional fusion confirmed that OmpR inhibits fur promoter activity in E. coli. EMSAs demonstrated in vitro binding of OmpR to an E. coli fur promoter region fragment encompassing all putative OBSs. The slower migrating stepshift OmpR/DNA complexes observed at higher OmpR concentrations confirmed the presence of more than one OBS. Interestingly, Genomic SELEX screening in E. coli identified a potential OmpR-binding site upstream of the uof sequence present in the fur regulatory region [112]. It is also noteworthy that microarray analysis has demonstrated the involvement of the EnvZ/OmpR system in the expression of a number of Fur-regulated genes in E. coli, particularly those concerned with enterobactin synthesis and transport [87].
Given the negative regulation of Fur by OmpR in Y. enterocolitica, we next examined whether OmpR could affect the expression of genes belonging to the Y. enterocolitica Fur regulon, involved in the transport of ferric and ferrous iron into cells. The genes selected for this analysis were fecA and fepA, encoding OM receptors of ferric siderophore-based iron transport systems, and feoA of the FeoABC system specific for ferrous iron transport, which have been characterized in both E. coli and Yersinia [89,124,125,126]. RT-qPCR data show that all three studied genes are repressed by Fur in Y. enterocolitica cells grown aerobically, which is in agreement with the results of studies in E. coli [18,127,128,129]. Interestingly, a recent study on the transcriptional activity of feoABC in Y. pestis revealed that Fur-mediated regulation of this locus occurs during microaerobic but not aerobic growth [20]. The most obvious transcriptional change (repression by Fur) was observed for the fepA gene encoding the FepA receptor for ferric enterobactin, the most potent siderophore of E. coli [130]. This may be due to the presence of a Fur box bearing most similarity to the 19-bp consensus sequence (a mismatch of only two base pairs compared to eight mismatches in the case of the Fur boxes of fecA and feoA), and hence strong Fur binding. Unexpectedly, we found that OmpR inhibits fepA and fecA expression, which appears at odds with the inhibition of their repressor Fur mediated by the same regulator. EMSAs revealed that OmpR is able to bind the promoter regions of fecA and fepA, suggesting that it directly influences their expression. Based on these results, we hypothesize that the negative regulation of both receptor genes by OmpR may overcome the modest positive effect of OmpR resulting from its inhibition of Fur. Interestingly, we observed a positive impact of OmpR on fepA expression in the absence of Fur, suggesting that additional regulatory mechanisms involving these two regulators may exist. It may be speculated that as yet unrecognized transcriptional or post-transcriptional regulatory mechanisms could modulate fepA expression in Y. enterocolitica and possibly integrate different environmental cues [131]. Although we did not analyze the influence of environmental signals on the OmpR-dependent fepA and fecA expression, from a physiological point of view, it would be expected that the inhibition of fepA and fecA by OmpR could alter levels of the encoded receptors in response to environmental cues besides transcriptional control by the Fur repressor. The advantage of limiting FepA and FecA synthesis by OmpR is unclear. Since the solubility of iron increases in the acidic conditions known to activate EnvZ/OmpR in E. coli [57,58], the uptake of ferric siderophores by FepA and FecA receptors might be less necessary under these circumstances. In addition, decreasing the level of the outer membrane proteins FepA and FecA, which are known targets of the host immune system [132], may help to limit any host response to Y. enterocolitica infection.
With regard to the function of Fur and OmpR in the regulation of Y. enterocolitica feoA, we revealed that the former slightly represses feoA expression, while the latter weakly activates it. Since our EMSA analysis suggested a poor direct interaction of OmpR with the feoA promoter sequence, the positive effect of OmpR on feoA expression may result from indirect (by lowering the level of Fur repressor) and weak direct control of feoA transcription. Curiously, the positive effect of OmpR on feoA expression was also observed in the absence of Fur, and thus it seems that OmpR might also be involved in other regulatory mechanisms not directly connected with the expression of fur. Recent studies have shown that a constitutively activated EnvZ/OmpR system caused by a specific mutation in envZ (envZR397L) induces feoA/feoB and downregulates fepA and fecA expression in E. coli [133]. In addition, binding of OmpR to the feoABC operon promoter region was indicated. Moreover, these authors hypothesized that the porins OmpC/F and transporter FeoB are involved in the uptake of ferrous ions, leading to an increase in the intracellular Fur-Fe2+ level and hence downregulation of fepA and fecA expression. Thus, although the net regulatory effect of OmpR on fepA, fecA and feoA expression in Y. enterocolitica is consistent with that observed in E. coli, the regulatory mechanisms involved might be more complicated. From a physiological point of view, the upregulation of the FeoABC system by OmpR could be important at low pH and in microaerobic or anaerobic environments where ferrous iron dominates.
The results of this study raise questions concerning the adaptive role of OmpR associated with the dual control of different iron transport systems. Y. enterocolitica can survive and grow outside and inside the host organism, and this localization influences the nature of the iron available as well as its dedicated transport mechanisms. Thus, the OmpR-mediated induction or repression of the iron uptake system appropriate to the local environment may contribute to the fitness of Y. enterocolitica.
Finally, it is noteworthy that studies on pathogenic bacteria have identified Fur as a global regulator that can activate or repress a variety of genes involved in diverse non-iron functions [41,134]. The identification of such direct Fur targets in Y. enterocolitica would help to verify the physiological role of the OmpR-mediated inhibition of fur expression. Taken together, the presented results lay the foundations for future work to discover further targets for the activity of Fur and thus under OmpR regulatory impact.
4. Materials and Methods
4.1. Strains, Media and Growth Conditions
The Y. enterocolitica and E. coli strains and plasmids used in this work are described in Supplementary Table S1. The Y. enterocolitica subsp. palearctica strain Ye9 of bio-serotype 2/O:9 and its derivatives were used (including its isogenic ompR (AR4) and fur deletion mutants). E. coli strain S17-1 λpir was used as a host for recombinant plasmids. LB broth (Lennox, Sigma Aldrich, St. Louis, MO, USA) and LB0 (10 g/L Tryptone, 5 g/L Yeast Extract) were used as growth media. When necessary, growth media were supplemented with the appropriate antibiotics at the following concentrations: 25 μg/mL chloramphenicol (Cm), 40 µg/mL gentamicin (Gm; 10 µg/mL for E. coli), 50 µg/mL kanamycin (Km), 30 µg/mL nalidixic acid (Nal), 12.5 µg/mL tetracycline (Tet). To test the effects of high osmolarity (350 mM NaCl), iron content (10 µM FeCl3 or 150 µM 2,2-dipyridyl), oxidative stress (100 µM hydrogen peroxide or 100 µM paraquat) and novobiocin (25 µg/mL), cells were grown overnight (to OD600 ~1.5), diluted to an OD600 of 0.1 and grown for ~4 h to OD600 0.4 when the studied compounds were added. To investigate the effect of acidic conditions, the pH of LB medium was adjusted to 5.6 with MES buffer. Untreated cells were cultured in parallel in an identical fashion to a control.
4.2. Molecular Biology Techniques
Standard DNA manipulation methods including polymerase chain reactions (PCRs), restriction digests, ligations and DNA electrophoresis were performed as described previously [135]. Plasmid DNA was isolated using a GeneJET Plasmid Miniprep Kit (Thermo Scientific, Waltham, MA, USA). Genomic DNA was isolated using a Bacterial & Yeast Genomic DNA Purification Kit (EurX, Gdańsk, Poland). PCRs were performed with Phusion High-Fidelity DNA polymerase or DreamTaq DNA polymerase (Thermo Scientific). Oligonucleotide primers used for PCR were purchased from Sigma Aldrich and are listed in Supplementary Table S2. DNA fragments amplified by PCR or obtained by restriction digestion were purified using a PCR/DNA Clean-Up Purification Kit (EurX). DNA sequencing was performed by Genomed S.A. (Warsaw, Poland).
4.3. Construction of a Chromosomal Pfur’::lacZYA Fusion
To obtain a Pfur’::lacZYA chromosomal fusion, a 498-bp DNA fragment corresponding to the fur promoter region (−498/−1) was amplified by PCR with the primers LPfurXbaI and PPfurSmaI (Table S2) using Y. enterocolitica Ye9N genomic DNA as the template. The obtained fragment was then purified, digested with restriction endonucleases XbaI and SmaI (FastDigest, Thermo Scientific) and ligated into the mobilizable suicide vector pFUSE, cleaved with the same enzymes. The fur promoter fragment was inserted upstream of the lacZYA operon in pFUSE to form plasmid pFUSEfur. This construct was introduced into E. coli S17-1 λpir by transformation and then transferred into Y. enterocolitica strains (Ye9N and the ΔompR mutant AR4) by conjugation. Recombinants harboring plasmid co-integrates were selected on LB agar plus antibiotics (Cm + Nal for strain Ye9N and Cm + Km for strain AR4). The integrity and chromosomal location of the integrated plasmid were confirmed by PCR using primers LFur695 and lacZH991 (Table S2), followed by sequencing of the amplicons.
4.4. Construction of Pfur::lacZ Fusions in Plasmid pCM132Gm
Five fur promoter fragments of varying length (containing a different number of predicted OmpR-binding motifs) were PCR-amplified using Y. enterocolitica Ye9N genomic DNA as template and primers containing unique restriction sites (Table S2). The purified amplicons were digested with restriction endonucleases EcoRI and KpnI and ligated into vector pCM132Gm (derivative of pCM132 [136]) cleaved with the same enzymes, to fuse the promoter fragments to the lacZ gene. Plasmids pCMF1234 (−443/+87 relative to the fur translational start site), pCMF234 (−276/+88), pCMF34 (−201/+88), pCMF4 (−173/+88) and pCMF0 (−168/−27) containing promoter fragments of diminishing size and number of putative OmpR-binding sites were constructed using the primer pairs FurAEcoRI/FurBKpnI, FurCEcoRI/FurBKpnI, FurDEcoRI/FurBKpnI, FurEEcoRI/FurBKpnI and FurFEcoRI/FurGKpnI (Table S2), respectively. The obtained promoter-lacZ fusions were verified by PCR and DNA sequencing using primers pCM132GmSpr1/pCM132GmSpr2 (Table S2). To obtain an analogous construct carrying the fur promoter region of E. coli, a fragment was PCR-amplified with primers FurEcEcoRI/FurEcKpnI using genomic DNA of E. coli BW25113 as template. This amplicon and vector pCM132Gm were both digested with EcoRI and KpnI and ligated to produce the construct pCMFEc. The resulting fusion was verified by PCR and sequencing with primers pCM132GmSpr1/pCM132GmSpr2 (Table S2).
4.5. Complementation of the ΔompR Mutation
To complement the ΔompR mutation in Y. enterocolitica strains, plasmid pBOmpR was used to transform E. coli DH5α [74]. Next, pBOmpR was mobilized into recipient Y. enterocolitica ΔompR strains by triparental conjugation using E. coli DH5α carrying the helper plasmid pRK2013, which provides tra and mob functions (Table S1). In the case of E. coli ΔompR mutant JW3368-1, pBOmpR was introduced by transformation [135].
4.6. β-Galactosidase Assay
The modified method of Miller [137] was used to measure β-galactosidase activity in bacterial cells. Overnight cultures of Y. enterocolitica or E. coli, incubated at 26 or 37 °C, respectively, were diluted to OD600 0.1 in fresh medium and incubation was continued until OD600 0.4 (exponential phase) or overnight (OD600 ~1, early stationary phase). To examine the influence of environmental factors on the activity of the tested promoters, cultures were supplemented with different reagents as described above. After incubation, 80 µL of culture (early stationary phase cultures were diluted to OD600 ~0.4) was mixed with 20 µL of PopCulture Reagent (Merck, Darmstadt, Germany) to disrupt the cell membranes. After 15-min incubation at room temperature, 20 µL of cell lysate was mixed with 130 µL of Z buffer and 30 µL of ONPG (4 mg/mL). The reaction was stopped after sufficient yellow color had developed by adding 75 µL of 1 M Na2CO3. The absorbance was then measured at both 420 and 550 nm and β-galactosidase activity, expressed in Miller Units, was calculated as described previously [137]. Each assay was performed at least in triplicate.
4.7. RT-qPCR
Total RNA was isolated from cultures of Y. enterocolitica grown at 26 °C to OD600 ~1 using a NucleoSpin RNA purification kit (Macherey-Nagel, Düren, Germany). The purity and quality of the RNA were assessed using an Agilent RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA). The RNAs were DNase-treated using a TURBO DNA-free Kit (Invitrogen, Carlsbad, CA, USA) and 1 µg of each preparation was used for cDNA synthesis with a Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) using random hexamer primers. The cDNAs were then used as the template for real-time qPCR reactions performed with 5× HOT FIREPol EvaGreen qPCR Mix Plus (Solis Biodyne, Tartu, Estonia) and primers listed in Table S2. Reactions were performed using a LightCycler 480 II (Roche, Basel, Switzerland). Relative quantification of transcripts in various genetic backgrounds was performed using the 2−ΔΔCT method with 16S rDNA serving as a reference gene.
4.8. Construction of Strains Expressing Fur Carrying a 3×FLAG Epitope
Y. enterocolitica strains Ye9Fflag and AR4Fflag, expressing a Fur-3×FLAG fusion protein carrying a 3×FLAG epitope peptide at the C-terminal end, were obtained by homologous recombination using suicide vector pDS132 carrying the sucrose sensitivity system [138]. An in-frame fusion of fur with the 3×FLAG epitope coding sequence was constructed by overlap extension PCR using primers listed in Table S2. As the first step, two DNA fragments (A and B) were PCR-amplified using Y. enterocolitica Ye9 genomic DNA as the template. Fragment A comprising the 132-bp sequence upstream of fur plus the fur ORF without a STOP codon (444 bp), with an overhang of 20 bp of the 1×FLAG coding sequence, was amplified using the primer pair 1FurFLAGXba-F/2FurFLAG-R. Fragment B composed of a 3×FLAG coding sequence (69 bp) plus 635 bp downstream of the fur STOP codon was amplified with the second pair of primers 3FLAGFur-F/4FLAGFurXba-R. A mixture of these two amplicons was then used as the template in a PCR with the flanking primers 1FurFLAGXba-F and 4FLAGFurXba-R to generate fragment C (1308 bp). This fragment was purified, digested with XbaI and then cloned into the corresponding site of suicide vector pDS132. The resulting construct pDSFur-FLAG was sequenced to confirm the absence of errors and used to transform E. coli S17-1 λpir. Plasmid pDSFur-FLAG was then introduced into different Y. enterocolitica strains via conjugation. Transconjugants carrying pDSFur-FLAG integrated into the chromosome were selected on LB agar supplemented with Cm (25 µg/mL). To force the second cross-over recombination, strains were plated on LB agar supplemented with 10% sucrose. Sucrose-resistant colonies that had lost chloramphenicol resistance were screened for the 3×FLAG coding sequence using colony PCR with the primer pair FlagSpr1/FlagSpr2. The presence of an intact fur::3×flag fusion in selected strains Ye9Fflag and AR4Fflag was confirmed by PCR and sequencing using primer pair 0FurFLAG-F/5FurFLAG-R (Table S2).
4.9. Western Blotting
For immunological detection of HemR and the FLAG-tagged Fur protein, Y. enterocolitica strains were grown in LB medium under the desired conditions until OD600 ~1. Volumes of 1 mL of each culture were then centrifuged and the cell pellets were resuspended in OD600 ×100 μL of Laemmli buffer [135]. Samples were then heated at 95 °C for 10 min to solubilize cellular proteins and separated by electrophoresis on 12% TGX Stain-Free FastCast Acrylamide gels (Bio-Rad, Hercules, CA, USA). Each gel was then Stain-Free-activated by a 10 min exposure to UV and imaged using a GE Healthcare AI600 Imager to confirm equal protein loading in each lane. Next, the proteins were transferred to a nitrocellulose membrane (Amersham Protran 0.2 µm Western Blotting Membrane; GE Healthcare, Chicago, IL, USA) using a wet electroblotting system (100 V for 1 h; Bio-Rad). The membranes were then blocked with 3.5% non-fat dried milk diluted in TBST and treated with rabbit anti-HemR polyclonal antibody (1:10,000; generous gift of Prof. Jürgen Heesemann) [15] or mouse anti-FLAG monoclonal antibody (1:5000; Sigma Aldrich). Sheep anti-rabbit IgG conjugated with horseradish peroxidase (HRP) was used as the secondary antibody (1:15,000; Promega, Madison, WI, USA) to detect anti-HemR antibody, while sheep anti-mouse IgG conjugated with HRP was used as the secondary antibody (1:8000; Sigma Aldrich) with the anti-FLAG antibody. Positive immunoreaction was visualized using Clarity Western ECL Blotting Substrate (Bio-Rad) for HRP-based chemiluminescent detection with a GE Healthcare AI600 Imager. As a further protein loading control, membranes were also probed with an antiserum specific for the cytoplasmic molecular chaperone DnaK (rabbit anti-DnaK, 1:10,000; kind gift of Prof. Sabina Kędzierska-Mieszkowska, University of Gdansk, Poland) [139]. The immunoreactive band intensities were quantified by densitometric analysis using ImageQuant TL analysis software (GE Healthcare).
4.10. Electrophoretic Mobility Shift Assays (EMSAs)
Overproduction and purification of recombinant OmpR-His6 for in vitro DNA binding studies were performed as described previously [80]. Briefly, Y. enterocolitica OmpR protein with a His6 motif at its N terminus was expressed from plasmid pETOmpR in E. coli BL21(DE3) and purified using Ni-NTA resin (Qiagen, Hilden, Germany). The concentration of the purified OmpR was determined using the RC DC protein assay (Bio-Rad). DNA fragments comprising the regulatory regions upstream of fur, fecA, fepA and feoA of Y. enterocolitica and fur of E. coli were PCR-amplified using primers listed in Table S2, with the respective genomic DNAs as template. The amplicons were purified using a Gene Matrix PCR/DNA Clean-Up Kit (EurX) and the concentration of DNA was determined with a NanoDrop 2000 spectrophotometer (Thermo Scientific). EMSA reactions were carried out in 10 μL of OmpR-binding buffer (50 mM Tris-HCl pH 8.0, 100 mM KCl, 1 mM EDTA, 1 mM DTT, 20 mM MgCl2, 12% glycerol, 100 µg/mL BSA, 0.1% Triton X-100) and contained appropriate DNA fragments of the studied promoters (0.05 or 0.3 pmol) with increasing amounts of OmpR-His6. DNA probes were mixed with OmpR-His6 with the molar ratio of 1:20–910. The purified protein was used in EMSAs without in vitro phosphorylation. To confirm binding specificity, a 304-bp or a 211-bp fragment of Y. enterocolitica Ye9 16S rDNA generated by PCR was included as a non-specific competitor in some binding reactions (Table S2). OmpR binding to the DNA targets was performed at 22 °C for 15 min, then 2 µL of 30% (v/v) glycerol was added and the samples were separated on 4.2% native polyacrylamide gels (19:1 acrylamide/bisacrylamide, 0.2×TBE) containing 2% glycerol. To improve the visualization of slower migrating OmpR/DNA complexes, non-denaturing polyacrylamide gels comprised of two layers were used, i.e., a large pore 4.2% gel on the top and a small pore 8.4% gel on the bottom. All gels were pre-run in 0.2×TBE running buffer for 40 min at 80 V. The samples were then loaded, and electrophoresis continued at 110 V for approximately 3 h. The gels were then stained in 1×SYBRgreen solution (Invitrogen) and visualized using a GE Healthcare AI600 imager.
4.11. Bioinformatic and Statistical Analyses
In silico analyses of the fur locus were performed on a shotgun genome sequence of Y. enterocolitica subsp. palearctica Ye9N (bio-serotype 2/O:9; NCBI/GenBank: JAALCX000000000). BLAST analysis (https://blast.ncbi.nlm.nih.gov/BlastAlign.cgi) and Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) were used for sequence alignment. Promoter prediction was performed using the software BPROM [81]. Statistical analyses were performed using Prism 7 software (v.7.02, GraphPad, San Diego, CA, USA). One-way ANOVA was used to determine statistically significant differences.
5. Conclusions
The human enteropathogen Y. enterocolitica is characterized by various iron transport systems that are important for the pathogenic abilities of this species. This study revealed the importance of the two-component system response regulator OmpR in the regulation of iron homeostasis in Y. enterocolitica by modulation of the level of Fur, the main regulator of iron transport and metabolism. It also established that OmpR can directly influence the expression of ferric and ferrous iron uptake genes and hence the level of iron in Y. enterocolitica cells. This dual control via OmpR probably plays a role in the precise expression of the appropriate iron uptake system, according to the ecological niche of the host, and contributes to the fitness and virulence of Y. enterocolitica. These findings expand the role of the OmpR regulator in the physiology of Y. enterocolitica.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/22/3/1475/s1.
Author Contributions
Conceptualization, A.R., K.J. and K.B.; methodology, K.J., M.L. and E.M.; investigation, K.J., M.L. and E.M.; data curation, K.J.; writing—original draft preparation, A.R. and K.J.; writing—review and editing, A.R., K.J. and K.B.; visualization, K.J.; supervision, A.R. and K.B.; project administration, K.B.; funding acquisition, K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Center, Poland, OPUS grant no. UMO-2016/21/B/NZ6/01009. Publication co-financed from the resources of the University of Warsaw.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Fàbrega, A.; Vila, J. Yersinia enterocolitica: Pathogenesis, virulence and antimicrobial resistance. Enferm. Infecc. Microbiol. Clin. 2012, 30, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.M.; Brandt, K.; Starke, M.; Rattei, T. Shotgun sequencing of Yersinia enterocolitica strain W22703 (biotype 2, serotype O:9): Genomic evidence for oscillation between invertebrates and mammals. BMC Genom. 2011, 12, 168. [Google Scholar] [CrossRef] [PubMed]
- Bottone, E.J. Yersinia enterocolitica: Revisitation of an Enduring Human Pathogen. Clin. Microbiol. Newsl. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- Bottone, E.J. Yersinia enterocolitica: A panoramic view of a charismatic microorganism. CRC Crit. Rev. Microbiol. 1977, 5, 211–241. [Google Scholar] [CrossRef]
- Bialas, N.; Kasperkiewicz, K.; Radziejewska-Lebrecht, J.; Skurnik, M. Bacterial cell surface structures in Yersinia enterocolitica. Arch. Immunol. Ther. Exp. 2012, 60, 199–209. [Google Scholar] [CrossRef]
- Zughaier, S.M.; Cornelis, P. Editorial: Role of Iron in Bacterial Pathogenesis. Front. Cell. Infect. Microbiol. 2018, 8, 344. [Google Scholar] [CrossRef]
- Bullen, J.J. The significance of iron in infection. Rev. Infect. Dis. 1981, 3, 1127–1138. [Google Scholar] [CrossRef]
- Stojiljkovic, I.; Hantke, K. Hemin uptake system of Yersinia enterocolitica: Similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992, 11, 4359–4367. [Google Scholar] [CrossRef]
- Stojiljkovic, I.; BaumLer, A.J.; Hantke, K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 1994, 236, 531–545. [Google Scholar] [CrossRef]
- Straley, S.C.; Perry, R.D. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995, 3, 310–317. [Google Scholar] [CrossRef]
- Carniel, E.; Guilvout, I.; Prentice, M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 1996, 178, 6743–6751. [Google Scholar] [CrossRef] [PubMed]
- Rakin, A.; Noelting, C.; Schubert, S.; Heesemann, J. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 1999, 67, 5265–5274. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.; Fischer, D.; Heesemann, J. Ferric enterochelin transport in Yersinia enterocolitica: Molecular and evolutionary aspects. J. Bacteriol. 1999, 181, 6387–6395. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.D.; Fetherston, J.D. Iron and heme uptake systems. In Yersinia Molecular and Cellular Biology; Carniel, E., Hinnebusch, B.J., Eds.; Bioscience Horizons: Wymondham, UK, 2004; pp. 257–283. [Google Scholar]
- Jaworska, K.; Nieckarz, M.; Ludwiczak, M.; Raczkowska, A.; Brzostek, K. OmpR-Mediated Transcriptional Regulation and Function of Two Heme Receptor Proteins of Yersinia enterocolitica Bio-Serotype 2/O:9. Front. Cell. Infect. Microbiol. 2018, 8, 333. [Google Scholar] [CrossRef] [PubMed]
- Heesemann, J.; Hantke, K.; Vocke, T.; Saken, E.; Rakin, A.; Stojiljkovic, I.; Berner, R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol. Microbiol. 1993, 8, 397–408. [Google Scholar] [CrossRef]
- BäumLer, A.; Koebnik, R.; Stojiljkovic, I.; Heesemann, J.; Braun, V.; Hantke, K. Survey on Newly Characterized Iron Uptake Systems of Yersinia enterocolitica. Zent. Bakteriol. 1993, 278, 416–424. [Google Scholar] [CrossRef]
- KammLer, M.; Schön, C.; Hantke, K. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 1993, 175, 6212–6219. [Google Scholar] [CrossRef]
- Ge, R.; Sun, X. Iron trafficking system in Helicobacter pylori. BioMetals 2012, 25, 247–258. [Google Scholar] [CrossRef]
- O’Connor, L.; Fetherston, J.D.; Perry, R.D. The feoABC Locus of Yersinia pestis Likely Has Two Promoters Causing Unique Iron Regulation. Front. Cell. Infect. Microbiol. 2017, 7, 331. [Google Scholar] [CrossRef]
- Frawley, E.R.; Fang, F.C. The ins and outs of bacterial iron metabolism. Mol. Microbiol. 2014, 93, 609–616. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol. Lett. 1995, 82–83, 969–974. [Google Scholar] [CrossRef]
- Masse, E.; Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 4620–4625. [Google Scholar] [CrossRef] [PubMed]
- McHugh, J.P.; Rodríguez-Quinoñes, F.; Abdul-Tehrani, H.; Svistunenko, D.A.; Poole, R.K.; Cooper, C.E.; Andrews, S.C. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 2003, 278, 29478–29486. [Google Scholar] [CrossRef]
- Escolar, L.; Perez-Martin, J.; de Lorenzo, V. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 1998, 283, 537–547. [Google Scholar] [CrossRef]
- Hantke, K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001, 4, 172–177. [Google Scholar] [CrossRef]
- Escolar, L.; Perez-Martin, J.; de Lorenzo, V. Opening the iron box: Transcriptional metalloregulation by the Fur protein. J. Bacteriol. 1999, 181, 6223–6229. [Google Scholar] [CrossRef] [PubMed]
- Hantke, K. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 1984, 197, 337–341. [Google Scholar] [CrossRef]
- Schaffer, S.; Hantke, K.; Braun, V. Nucleotide sequence of the iron regulatory gene fur. Mol. Gen. Genet. 1985, 200, 110–113. [Google Scholar] [CrossRef]
- Tardat, B.; Touati, D. Iron and oxygen regulation of Escherichia coli MnSOD expression: Competition between the global regulators Fur and ArcA for binding to DNA. Mol. Microbiol. 1993, 9, 53–63. [Google Scholar] [CrossRef]
- Lee, H.S.; Lee, Y.S.; Kim, H.S.; Choi, J.Y.; Hassan, H.M.; Chung, M.H. Mechanism of regulation of 8-hydroxyguanine endonuclease by oxidative stress: Roles of FNR, ArcA, and Fur. Free Radic. Biol. Med. 1998, 24, 1193–1201. [Google Scholar] [CrossRef]
- Hantke, K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K-12: Fur not only affects iron metabolism. Mol. Gen. Genet. 1987, 210, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.K.; Foster, J.W. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 1996, 178, 5683–5691. [Google Scholar] [CrossRef] [PubMed]
- Fillat, M.F. The FUR (ferric uptake regulator) superfamily: Diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 2014, 546, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Ernst, F.D.; Bereswill, S.; Waidner, B.; Stoof, J.; Mäder, U.; Kusters, J.G.; Kuipers, E.J.; Kist, M.; van Vliet, A.H.M.; Homuth, G. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 2005, 151, 533–546. [Google Scholar] [CrossRef]
- Yuhara, S.; Komatsu, H.; Goto, H.; Ohtsubo, Y.; Nagata, Y.; Tsuda, M. Pleiotropic roles of iron-responsive transcriptional regulator Fur in Burkholderia multivorans. Microbiology 2008, 154, 1763–1774. [Google Scholar] [CrossRef]
- Porcheron, G.; Dozois, C.M. Interplay between iron homeostasis and virulence: Fur and RyhB as major regulators of bacterial pathogenicity. Vet. Microbiol. 2015, 179, 2–14. [Google Scholar] [CrossRef]
- Mathieu, S.; Cissé, C.; Vitale, S.; Ahmadova, A.; Degardin, M.; Pérard, J.; Colas, P.; Miras, R.; Boturyn, D.; Covès, J.; et al. From Peptide Aptamers to Inhibitors of FUR, Bacterial Transcriptional Regulator of Iron Homeostasis and Virulence. ACS Chem. Biol. 2016, 11, 2519–2528. [Google Scholar] [CrossRef]
- Staggs, T.M.; Perry, R.D. Identification and cloning of a fur regulatory gene in Yersinia pestis. J. Bacteriol. 1991, 173, 417–425. [Google Scholar] [CrossRef][Green Version]
- Schwiesow, L.; Mettert, E.; Wei, Y.; Miller, H.K.; Herrera, N.G.; Balderas, D.; Kiley, P.J.; Auerbuch, V. Control of hmu Heme Uptake Genes in Yersinia pseudotuberculosis in Response to Iron Sources. Front. Cell. Infect. Microbiol. 2018, 8, 47. [Google Scholar] [CrossRef]
- Gao, H.; Zhou, D.; Li, Y.; Guo, Z.; Han, Y.; Song, Y.; Zhai, J.; Du, Z.; Wang, X.; Lu, J.; et al. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 2008, 190, 3063–3075. [Google Scholar] [CrossRef]
- Zhou, D.; Qin, L.; Han, Y.; Qiu, J.; Chen, Z.; Li, B.; Song, Y.; Wang, J.; Guo, Z.; Zhai, J.; et al. Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol. Lett. 2006, 258, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Lesic, B.; Carniel, E. The high pathogenicity island: A broad-host-range pathogenicity island. In Yersinia: Molecular and Cellular Biology; Bioscience Horizons: Norfolk, UK, 2004; pp. 285–306. [Google Scholar]
- Beauchene, N.A.; Mettert, E.L.; Moore, L.J.; Keles, S.; Willey, E.R.; Kiley, P.J. O2 availability impacts iron homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 12261–12266. [Google Scholar] [CrossRef]
- van Vliet, A.H.; Rock, J.D.; Madeleine, L.N.; Ketley, J.M. The iron-responsive regulator Fur of Campylobacter jejuni is expressed from two separate promoters. FEMS Microbiol. Lett. 2000, 188, 115–118. [Google Scholar] [CrossRef]
- De Lorenzo, V.; Herrero, M.; Giovannini, F.; Neilands, J.B. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur. J. Biochem. 1988, 173, 537–546. [Google Scholar] [CrossRef]
- Zheng, M.; Doan, B.; Schneider, T.D.; Storz, G. OxyR and SoxRS regulation of fur. J. Bacteriol. 1999, 181, 4639–4643. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, J.J.E.; Waidner, B.; van Vliet, A.H.M.; Hughes, N.J.; Hag, S.; Bereswill, S.; Kelly, D.J.; Vandenbroucke-Grauls, C.M.J.E.; Kist, M.; Kusters, J.G. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 2002, 70, 606–611. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Delany, I.; Spohn, G.; Pacheco, A.-B.F.; Ieva, R.; Alaimo, C.; Rappuoli, R.; Scarlato, V. Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol. Microbiol. 2002, 46, 1107–1122. [Google Scholar] [CrossRef]
- Lee, H.-J.; Bang, S.H.; Lee, K.-H.; Park, S.-J. Positive regulation of fur gene expression via direct interaction of fur in a pathogenic bacterium, Vibrio vulnificus. J. Bacteriol. 2007, 189, 2629–2636. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vecerek, B.; Moll, I.; Blasi, U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007, 26, 965–975. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-Component Signal Transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Soncini, F.C.; Groisman, E.A. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 1996, 178, 6796–6801. [Google Scholar] [CrossRef] [PubMed]
- Pratt, L.A.; Hsing, W.; Gibson, K.E.; Silhavy, T.J. From acids to osmZ: Multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 1996, 20, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.J.; Inouye, M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 2002, 277, 24155–24161. [Google Scholar] [CrossRef] [PubMed]
- Kenney, L.J. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 2002, 5, 135–141. [Google Scholar] [CrossRef]
- Stincone, A.; Daudi, N.; Rahman, A.S.; Antczak, P.; Henderson, I.; Cole, J.; Johnson, M.D.; Lund, P.; Falciani, F. A systems biology approach sheds new light on Escherichia coli acid resistance. Nucleic Acids Res. 2011, 39, 7512–7528. [Google Scholar] [CrossRef] [PubMed]
- Quinn, H.J.; Cameron, A.D.S.; Dorman, C.J. Bacterial regulon evolution: Distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet. 2014, 10, e1004215. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Mizusaki, H.; Kenney, L.J. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol. 2015, 13, e1002116. [Google Scholar] [CrossRef]
- Chakraborty, S.; Winardhi, R.S.; Morgan, L.K.; Yan, J.; Kenney, L.J. Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nat. Commun. 2017, 8, 1587. [Google Scholar] [CrossRef]
- Chakraborty, S.; Kenney, L.J. A New Role of OmpR in Acid and Osmotic Stress in Salmonella and E. coli. Front. Microbiol. 2018, 9, 2656. [Google Scholar] [CrossRef]
- Bontemps-Gallo, S.; Fernandez, M.; Dewitte, A.; Raphaël, E.; Gherardini, F.C.; Elizabeth, P.; Koch, L.; Biot, F.; Reboul, A.; Sebbane, F. Nutrient depletion may trigger the Yersinia pestis OmpR-EnvZ regulatory system to promote flea-borne plague transmission. Mol. Microbiol. 2019, 112, 1471–1482. [Google Scholar] [CrossRef]
- Slauch, J.M.; Silhavy, T.J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 1989, 210, 281–292. [Google Scholar] [CrossRef]
- Shin, S.; Park, C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 1995, 177, 4696–4702. [Google Scholar] [CrossRef] [PubMed]
- Vidal, O.; Longin, R.; Prigent-Combaret, C.; Dorel, C.; Hooreman, M.; Lejeune, P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998, 180, 2442–2449. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Nagura, R.; Tanabe, H.; Fujita, N.; Ishihama, A.; Utsumi, R. Negative regulation of the bolA1p of Escherichia coli K-12 by the transcription factor OmpR for osmolarity response genes. FEMS Microbiol. Lett. 2000, 186, 257–262. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bernardini, M.L.; Fontaine, A.; Sansonetti, P.J. The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J. Bacteriol. 1990, 172, 6274–6281. [Google Scholar] [CrossRef]
- Perkins, T.T.; Davies, M.R.; Klemm, E.J.; Rowley, G.; Wileman, T.; James, K.; Keane, T.; Maskell, D.; Hinton, J.C.D.; Dougan, G.; et al. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol. Microbiol. 2013, 87, 526–538. [Google Scholar] [CrossRef]
- Gao, H.; Zhang, Y.; Han, Y.; Yang, L.; Liu, X.; Guo, Z.; Tan, Y.; Huang, X.; Zhou, D.; Yang, R. Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Yersinia pestis. BMC Microbiol. 2011, 11, 39. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, Y.; Ding, L.; Lu, P.; Atkinson, S.; Chen, S. Positive regulation of flhDC expression by OmpR in Yersinia pseudotuberculosis. Microbiology 2009, 155, 3622–3631. [Google Scholar] [CrossRef][Green Version]
- Hu, Y.; Lu, P.; Wang, Y.; Ding, L.; Atkinson, S.; Chen, S. OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 2009, 155, 2522–2531. [Google Scholar] [CrossRef]
- Dorrell, N.; Li, S.R.; Everest, P.H.; Dougan, G.; Wren, B.W. Construction and characterisation of a Yersinia enterocolitica O:8 ompR mutant. FEMS Microbiol. Lett. 1998, 165, 145–151. [Google Scholar] [CrossRef][Green Version]
- Brzostek, K.; Raczkowska, A.; Zasada, A. The osmotic regulator OmpR is involved in the response of Yersinia enterocolitica O:9 to environmental stresses and survival within macrophages. FEMS Microbiol. Lett. 2003, 228, 265–271. [Google Scholar] [CrossRef][Green Version]
- Brzostek, K.; Brzostkowska, M.; Bukowska, I.; Karwicka, E.; Raczkowska, A. OmpR negatively regulates expression of invasin in Yersinia enterocolitica. Microbiology 2007, 153, 2416–2425. [Google Scholar] [CrossRef] [PubMed]
- Brzostkowska, M.; Raczkowska, A.; Brzostek, K. OmpR, a response regulator of the two-component signal transduction pathway, influences inv gene expression in Yersinia enterocolitica O9. Front. Cell. Infect. Microbiol. 2012, 2, 153. [Google Scholar] [CrossRef] [PubMed]
- Skorek, K.; Raczkowska, A.; Dudek, B.; Miętka, K.; Guz-Regner, K.; Pawlak, A.; Klausa, E.; Bugla-Płoskońska, G.; Brzostek, K. Regulatory protein OmpR influences the serum resistance of Yersinia enterocolitica O:9 by modifying the structure of the outer membrane. PLoS ONE 2013, 8, e79525. [Google Scholar] [CrossRef] [PubMed]
- Raczkowska, A.; Trzos, J.; Lewandowska, O.; Nieckarz, M.; Brzostek, K. Expression of the AcrAB Components of the AcrAB-TolC Multidrug Efflux Pump of Yersinia enterocolitica Is Subject to Dual Regulation by OmpR. PLoS ONE 2015, 10, e0124248. [Google Scholar] [CrossRef] [PubMed]
- Nieckarz, M.; Raczkowska, A.; Jaworska, K.; Stefańska, E.; Skorek, K.; Stosio, D.; Brzostek, K. The Role of OmpR in the Expression of Genes of the KdgR Regulon Involved in the Uptake and Depolymerization of Oligogalacturonides in Yersinia enterocolitica. Front. Cell. Infect. Microbiol. 2017, 7, 366. [Google Scholar] [CrossRef]
- Raczkowska, A.; Skorek, K.; Bielecki, J.; Brzostek, K. OmpR controls Yersinia enterocolitica motility by positive regulation of flhDC expression. Antonie Van Leeuwenhoek 2011, 99, 381–394. [Google Scholar] [CrossRef]
- Nieckarz, M.; Raczkowska, A.; Debski, J.; Kistowski, M.; Dadlez, M.; Heesemann, J.; Rossier, O.; Brzostek, K. Impact of OmpR on the membrane proteome of Yersinia enterocolitica in different environments: Repression of major adhesin YadA and heme receptor HemR. Environ. Microbiol. 2016, 18, 997–1021. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies; Nova Science Publishers, Inc.: New York, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Maeda, S.; Takayanagi, K.; Nishimura, Y.; Maruyama, T.; Sato, K.; Mizuno, T. Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: A study involving synthetic OmpR-binding sequences. J. Biochem. 1991, 110, 324–327. [Google Scholar] [CrossRef][Green Version]
- Lucas, R.L.; Lee, C.A. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2001, 183, 2733–2745. [Google Scholar] [CrossRef]
- Cameron, A.D.S.; Dorman, C.J. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet. 2012, 8, e1002615. [Google Scholar] [CrossRef] [PubMed]
- de Lorenzo, V.; Wee, S.; Herrero, M.; Neilands, J.B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 1987, 169, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Keseler, I.M.; Mackie, A.; Santos-Zavaleta, A.; Billington, R.; Bonavides-Martínez, C.; Caspi, R.; Fulcher, C.; Gama-Castro, S.; Kothari, A.; Krummenacker, M.; et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2016, 45, D543–D550. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Aiba, H.; Masuda, Y.; Kanaya, S.; Sugiura, M.; Wanner, B.L.; Mori, H.; Mizuno, T. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 2002, 46, 281–291. [Google Scholar] [CrossRef]
- Nieckarz, M.; Kaczor, P.; Jaworska, K.; Raczkowska, A.; Brzostek, K. Urease Expression in Pathogenic Yersinia enterocolitica Strains of Bio-Serotypes 2/O:9 and 1B/O:8 Is Differentially Regulated by the OmpR Regulator. Front. Microbiol. 2020, 11, 607. [Google Scholar] [CrossRef]
- Andrews, S.C.; Robinson, A.K.; Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Perry, R.D.; Fetherston, J.D. Yersiniabactin iron uptake: Mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011, 13, 808–817. [Google Scholar] [CrossRef]
- Achenbach, L.A.; Yang, W. The fur gene from Klebsiella pneumoniae: Characterization, genomic organization and phylogenetic analysis. Gene 1997, 185, 201–207. [Google Scholar] [CrossRef]
- Delany, I.; Ieva, R.; Alaimo, C.; Rappuoli, R.; Scarlato, V. The iron-responsive regulator fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J. Bacteriol. 2003, 185, 6032–6041. [Google Scholar] [CrossRef]
- Lowe, C.A.; Asghar, A.H.; Shalom, G.; Shaw, J.G.; Thomas, M.S. The Burkholderia cepacia fur gene: Co-localization with omLA and absence of regulation by iron. Microbiology 2001, 147, 1303–1314. [Google Scholar] [CrossRef]
- Russo, F.D.; Silhavy, T.J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 1991, 222, 567–580. [Google Scholar] [CrossRef]
- Head, C.G.; Tardy, A.; Kenney, L.J. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 1998, 281, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Owen-Hughes, T.A.; Pavitt, G.D.; Santos, D.S.; Sidebotham, J.M.; Hulton, C.S.; Hinton, J.C.; Higgins, C.F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 1992, 71, 255–265. [Google Scholar] [CrossRef]
- Lucht, J.M.; Dersch, P.; Kempf, B.; Bremer, E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 1994, 269, 6578. [Google Scholar] [CrossRef]
- Puente, J.L.; Verdugo-Rodríguez, A.; Calva, E. Expression of Salmonella typhi and Escherichia coli OmpC is influenced differently by medium osmolarity; dependence on Escherichia coli OmpR. Mol. Microbiol. 1991, 5, 1205–1210. [Google Scholar] [CrossRef]
- Martínez-Flores, I.; Cano, R.; Bustamante, V.H.; Calva, E.; Puente, J.L. The ompB operon partially determines differential expression of OmpC in Salmonella typhi and Escherichia coli. J. Bacteriol. 1999, 181, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Goh, E.-B.; Siino, D.F.; Igo, M.M. The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes. J. Bacteriol. 2004, 186, 4019–4024. [Google Scholar] [CrossRef][Green Version]
- Argandoña, M.; Nieto, J.J.; Iglesias-Guerra, F.; Calderón, M.I.; García-Estepa, R.; Vargas, C. Interplay between iron homeostasis and the osmotic stress response in the halophilic bacterium Chromohalobacter salexigens. Appl. Environ. Microbiol. 2010, 76, 3575–3589. [Google Scholar] [CrossRef]
- Higashitani, A.; Nishimura, Y.; Hara, H.; Aiba, H.; Mizuno, T.; Horiuchi, K. Osmoregulation of the fatty acid receptor gene fadL in Escherichia coli. Mol. Gen. Genet. 1993, 240, 339–347. [Google Scholar] [CrossRef]
- Bang, I.S.; Audia, J.P.; Park, Y.K.; Foster, J.W. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol. Microbiol. 2002, 44, 1235–1250. [Google Scholar] [CrossRef]
- Rentschler, A.E.; Lovrich, S.D.; Fitton, R.; Enos-Berlage, J.; Schwan, W.R. OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology 2013, 159, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.W.; Kim, D.; Szubin, R.; Palsson, B.O. Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. Cell Rep. 2015, 12, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Loprasert, S.; Sallabhan, R.; Whangsuk, W.; Mongkolsuk, S. Characterization and mutagenesis of fur gene from Burkholderia pseudomallei. Gene 2000, 254, 129–137. [Google Scholar] [CrossRef]
- Wei, Q.; Le Minh, P.N.; Dötsch, A.; Hildebrand, F.; Panmanee, W.; Elfarash, A.; Schulz, S.; Plaisance, S.; Charlier, D.; Hassett, D.; et al. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012, 40, 4320–4333. [Google Scholar] [CrossRef] [PubMed]
- Nouaille, S.; Mondeil, S.; Finoux, A.-L.; Moulis, C.; Girbal, L.; Cocaign-Bousquet, M. The stability of an mRNA is influenced by its concentration: A potential physical mechanism to regulate gene expression. Nucleic Acids Res. 2017, 45, 11711–11724. [Google Scholar] [CrossRef]
- Bernhardt, H.S.; Tate, W.P. Primordial soup or vinaigrette: Did the RNA world evolve at acidic pH? Biol. Direct 2012, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef]
- Pratt, L.A.; Silhavy, T.J. Identification of base pairs important for OmpR-DNA interaction. Mol. Microbiol. 1995, 17, 565–573. [Google Scholar] [CrossRef]
- Shimada, T.; Takada, H.; Yamamoto, K.; Ishihama, A. Expanded roles of two-component response regulator OmpR in Escherichia coli: Genomic SELEX search for novel regulation targets. Genes Cells 2015, 20, 915–931. [Google Scholar] [CrossRef]
- Griffith, K.L.; Wolf, R.E.J. Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: Identifying nucleotides required for DNA binding and transcription activation. Mol. Microbiol. 2001, 40, 1141–1154. [Google Scholar] [CrossRef]
- Dunn, T.M.; Hahn, S.; Ogden, S.; Schleif, R.F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: Addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl. Acad. Sci. USA 1984, 81, 5017–5020. [Google Scholar] [CrossRef] [PubMed]
- Ortuno, M.J.; Lawther, R.P. Effect of the deletion of upstream DNA sequences on expression from the ilvGp2 promoter of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987, 15, 1521–1542. [Google Scholar] [CrossRef]
- Rampersaud, A.; Norioka, S.; Inouye, M. Characterization of OmpR binding sequences in the upstream region of the ompF promoter essential for transcriptional activation. J. Biol. Chem. 1989, 264, 18693–18700. [Google Scholar] [CrossRef]
- Rampersaud, A.; Harlocker, S.L.; Inouye, M. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J. Biol. Chem. 1994, 269, 12559–12566. [Google Scholar] [CrossRef]
- Harlocker, S.L.; Bergstrom, L.; Inouye, M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 1995, 270, 26849–26856. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.J.; Lan, C.Y.; Igo, M.M. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc. Natl. Acad. Sci. USA 1997, 94, 2828–2832. [Google Scholar] [CrossRef] [PubMed]
- Bergstrom, L.C.; Qin, L.; Harlocker, S.L.; Egger, L.A.; Inouye, M. Hierarchical and co-operative binding of OmpR to a fusion construct containing the ompC and ompF upstream regulatory sequences of Escherichia coli. Genes Cells 1998, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Gerstel, U.; Park, C.; RömLing, U. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 2003, 49, 639–654. [Google Scholar] [CrossRef]
- Jubelin, G.; Vianney, A.; Beloin, C.; Ghigo, J.-M.; Lazzaroni, J.-C.; Lejeune, P.; Dorel, C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 2005, 187, 2038–2049. [Google Scholar] [CrossRef]
- Perini, L.T.; Doherty, E.A.; Werner, E.; Senear, D.F. Multiple specific CytR binding sites at the Escherichia coli deoP2 promoter mediate both cooperative and competitive interactions between CytR and cAMP receptor protein. J. Biol. Chem. 1996, 271, 33242–33255. [Google Scholar] [CrossRef]
- Hantke, K. Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol. 2003, 11, 192–195. [Google Scholar] [CrossRef]
- Perry, R.D.; Mier, I.J.; Fetherston, J.D. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. BioMetals 2007, 20, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Kanaujia, P.K.; Bajaj, P.; Virdi, J.S. Analysis of iron acquisition and storage-related genes in clinical and non-clinical strains of Yersinia enterocolitica biovar 1A. APMIS 2015, 123, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.D.; Pettis, G.S.; McIntosh, M.A. Promoter and operator determinants for fur-mediated iron regulation in the bidirectional fepA-fes control region of the Escherichia coli enterobactin gene system. J. Bacteriol. 1994, 176, 3944–3955. [Google Scholar] [CrossRef] [PubMed]
- Angerer, A.; Braun, V. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol. 1998, 169, 483–490. [Google Scholar] [CrossRef]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef]
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef]
- Guillier, M.; Gottesman, S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006, 59, 231–247. [Google Scholar] [CrossRef]
- Fernandez-Beros, M.E.; Gonzalez, C.; McIntosh, M.A.; Cabello, F.C. Immune response to the iron-deprivation-induced proteins of Salmonella typhi in typhoid fever. Infect. Immun. 1989, 57, 1271–1275. [Google Scholar] [CrossRef]
- Gerken, H.; Vuong, P.; Soparkar, K.; Misra, R. Roles of the EnvZ/OmpR Two-Component System and Porins in Iron Acquisition in Escherichia coli. mBio 2020, 11. [Google Scholar] [CrossRef]
- Troxell, B.; Hassan, H.M. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 2013, 3, 59. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Marx, C.J.; Lidstrom, M.E. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 2001, 147, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbour, NY, USA, 1992; ISBN 0-87969-106-9. [Google Scholar]
- Philippe, N.; Alcaraz, J.-P.; Coursange, E.; Geiselmann, J.; Schneider, D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 2004, 51, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Kędzierska, S.; Chesnokova, L.S.; Witt, S.N.; Zolkiewski, M. Interactions within the ClpB/DnaK bi-chaperone system from Escherichia coli. Arch. Biochem. Biophys. 2005, 444, 61–65. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).