Gene Expression Responses to Sequential Nutrient Deficiency Stresses in Soybean
Abstract
1. Introduction
2. Results
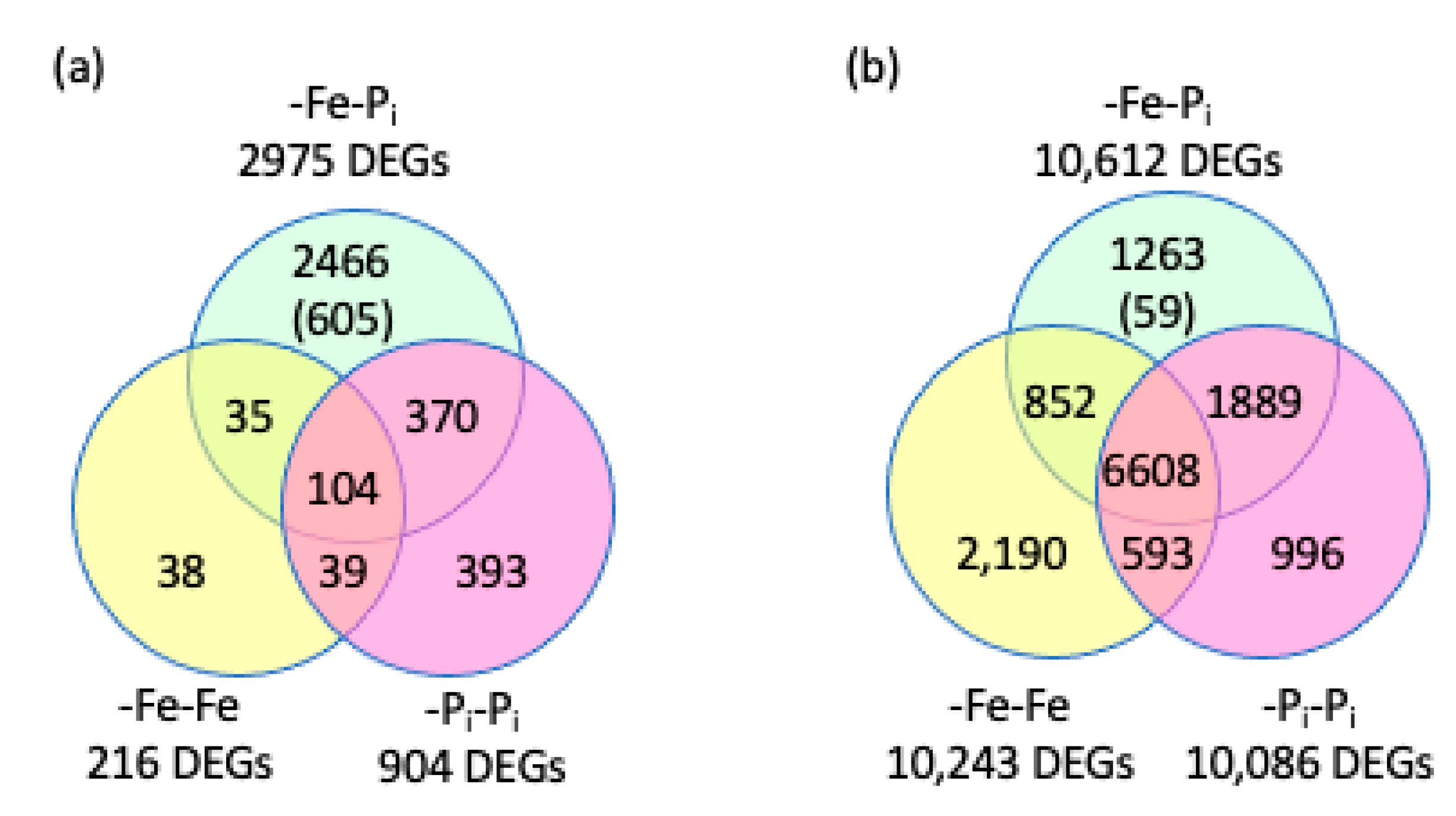
2.1. Comparing Repeated Stress and Sequential Stress
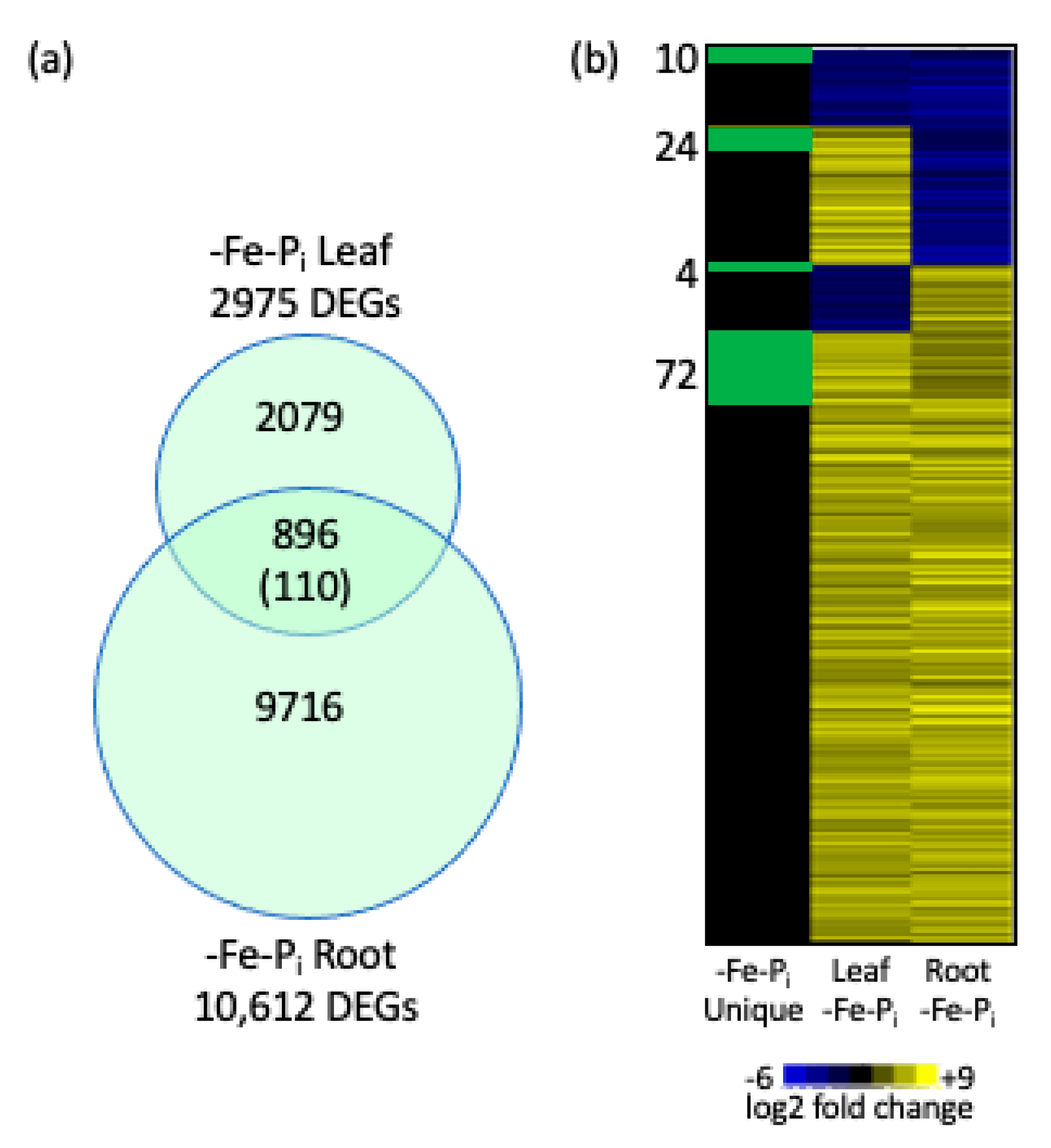
2.2. Identifying Genes Differentially Expressed in Both Leaves and Roots After -Fe-Pi Stress
2.3. Stringent Identification of Sequential Stress Specific Genes
3. Discussion
3.1. A Core Set of Genes Is Differentially Expressed in all Three Stress Profiles
3.2. Genes Required for -Pi Responses
3.3. First Stress Signature Genes
3.4. Sequential Stress Induces More DEGs in Leaves than in Roots
3.5. Genes Unique to Sequential Stress Response
4. Materials and Methods
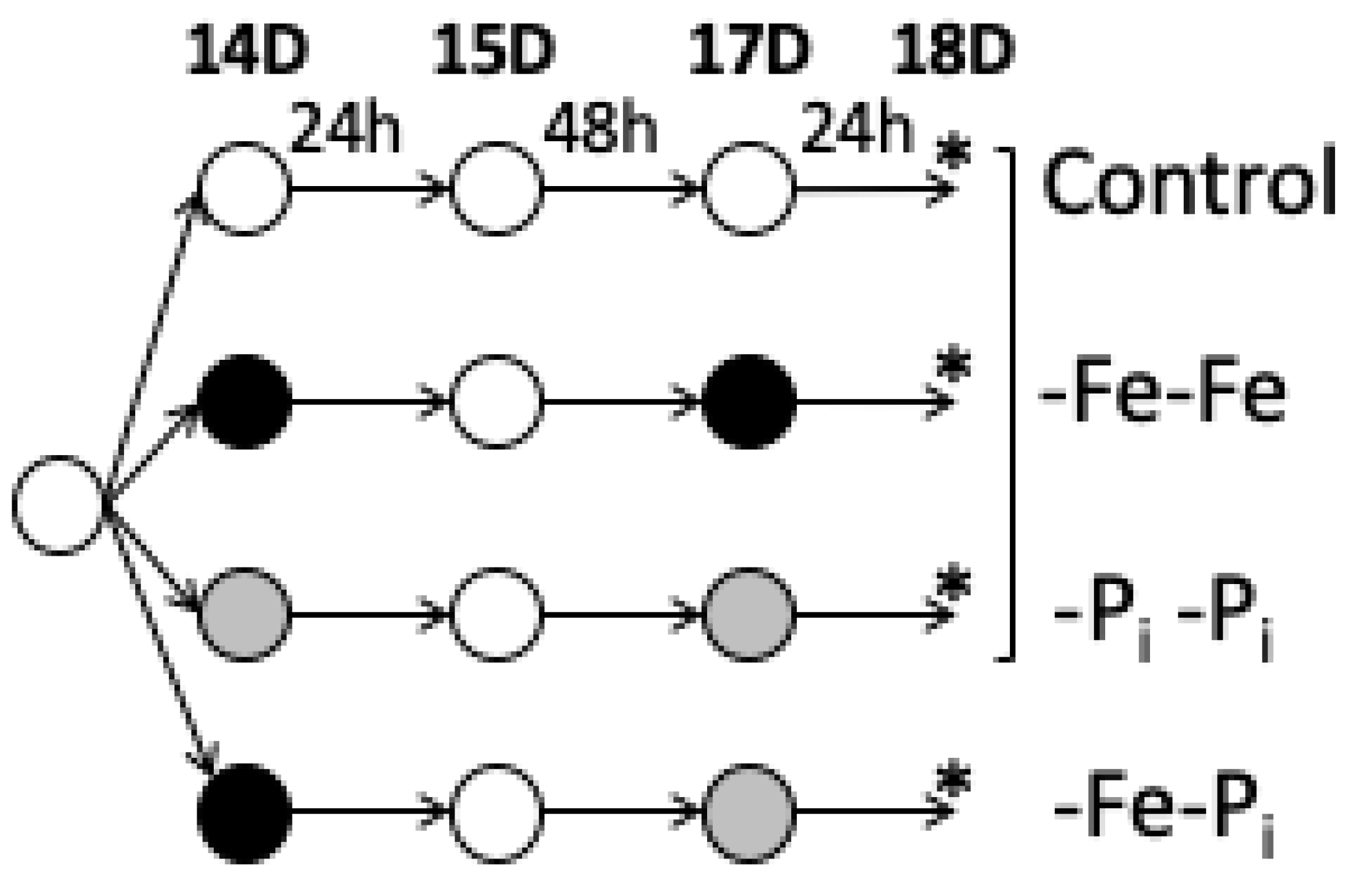
4.1. Growth Conditions
4.2. RNA Isolation
4.3. RNA-Seq and Data Analysis
4.4. Gene Annotation
4.5. Identification of Over-Represented Transcription Factor Binding Sites
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEGs | Differentially expressed genes |
| FDR | False discovery rate |
| -Fe | Iron deficiency stress |
| -Fe-Fe | Iron deficiency stress (repeated stress) |
| -Fe-Pi | Iron deficiency stress followed by phosphate deficiency stress (sequential stress) |
| GO | Gene ontology |
| h | Hours |
| IDC | Iron Deficiency Chlorosis |
| p | Probability |
| -Pi | Phosphate deficiency stress |
| -Pi-Pi | Phosphate deficiency stress (repeated stress) |
| TF | Transcription factor |
| TFBS | Transcription factor binding site |
| TFF | Transcription factor family |
References
- Bonnet, C.; Lassueur, S.; Ponzio, C.; Gols, R.; Dicke, M.; Reymond, P. Combined biotic stresses trigger similar transcriptomic responses but contrasting resistance against a chewing herbivore in Brasica nigra. BMC Plant Biol. 2017, 17, 127. [Google Scholar] [CrossRef]
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J. Transcriptome Responses to Combinations of Stresses in Arabidopsis. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef]
- Coolen, S.; Proietti, S.; Hickman, R.; Davila Olivas, N.H.; Huant, P.-P.; VanVerk, M.C.; VanPelt, J.A.; Wittenberg, A.H.J.; De Voss, M.; Prins, M.; et al. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 2016, 86, 249–267. [Google Scholar] [CrossRef]
- Zhang, H.; Sonnewald, U. Differences and commonalities of plant responses to single and combined stresses. Plant J. 2017, 90, 839–855. [Google Scholar] [CrossRef]
- Marschner, H. Mineral Nutrition of Higher Plants; Academic Press Limited: San Diego, CA, USA, 1995. [Google Scholar]
- Hansen, N.C.; Jolley, V.D.; Naeve, S.L.; Goos, R.J. Iron deficiency of soybean in the north central U.S. and associated soil properties. Soil Sci. Plant Nutr. 2004, 50, 983–987. [Google Scholar] [CrossRef]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, F. Phosphorus dynamics: From soil to plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef]
- MacDonald, G.K.; Bennet, E.M.; Potter, P.A.; Ramankutty, N. Agronomic phosphorus imbalances across the world’s croplands. Proc. Natl. Acad. Sci. USA 2011, 108, 3086–3091. [Google Scholar] [CrossRef]
- Chowdhury, R.B.; Moore, G.A.; Weatherley, A.J.; Arora, M. Key sustainability challenges for the global phosphorus resource, their implications for global food security, and options for mitigation. J. Clean. Prod. 2017, 140, 945–963. [Google Scholar] [CrossRef]
- Blackwell, M.; Darch, T.; Haslam, R. Phosphorus use efficiency and fertilizers: Future opportunities for improvements. Front. Agricult. Sci. Eng. 2019, 6, 332–340. [Google Scholar] [CrossRef]
- Cordell, D.; Drangert, J.-O.; White, S. The story of phosphorus: Global food security and food for thought. Glob. Environ. Change 2009, 19, 292–305. [Google Scholar] [CrossRef]
- Brumbarova, T.; Bauer, P.; Ivanov, R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015, 20, 124–133. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Ann. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef]
- Li, X.; Zhang, H.; Ai, Q.; Liang, G.; Yu, D. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2478–2493. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.-F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.-Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Aparicio-Fabre, R.; Gullien, G.; Loredo, M.; Arellano, J.; Valdes-Lopez, O.; Ramirez, M.; Iniguez, L.P.; Panzeri, D.; Castiglioni, B.; Cremonesi, P. Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol. 2013, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-Y.; Aung, K.; Tseng, C.-Y.; Chang, T.-Y.; Chen, Y.-S.; Chiou, T.-J. Vacuolar Ca 2+/H+ transport activity is required for systemic phosphate homeostasis involving shoot-to-root signaling in Arabidopsis. Plant Physiol. 2011, 156, 1176–1189. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.; Yang, S.S.; Miller, S.S.; Bucciarelli, B.; Liu, J.; Rydeen, A.; Bozsoki, Z.; Uhde-Stone, C.; Tu, Z.J.; Allan, D.; et al. An RNA-Seq transcriptome analysis of orthophosphate deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 2013, 161, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ruan, W.; Shi, J.; Zhang, L.; Xiang, D.; Yang, C.; Li, C.; Wu, Z.; Liu, Y.; Yu, Y. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. PNAS 2014, 111, 14953–14958. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, H.; Moloney, M.; Inźe, D. Translational research: From pot to plot. Plant Biotechnol. J. 2014, 12, 277–285. [Google Scholar] [CrossRef]
- Borrill, P. Blurring the boundaries between cereal crops and model plants. New Phytol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, G.A.; King, K.E.; Severin, A.J.; May, G.D.; Cianzio, S.R.; Lin, S.F.; Lauter, N.C.; Shoemaker, R.C. Identification of candidate genes underlying an iron efficiency quantitative trait locus in soybean. Plant Physiol. 2012, 158, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Chen, C.L.; Cui, M.; Zhou, W.J.; Wu, H.L.; Ling, H.Q. Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol. Plant 2018, 11, P1166–P1183. [Google Scholar] [CrossRef] [PubMed]
- Assefa, T.; Zhang, J.; Chowda-Reddy, R.V.; Lauter, A.N.M.; Singh, A.; O’Rourke, J.A.; Graham, M.A.; Singh, A.K. Deconstrcting the genetic architecture of iron deficiency chlorosis in soybean using genome-wide approaches. BMC Plant Biol. 2020, 20, 42. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; Amundsen, K.; Graef, G. Gene expression profiling of iron deficiency chlorosis sensitive and tolerant soybean indicates key roles for phenylpropanoids under alkalinity stress. Front. Plant Sci. 2018, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Gao, W.; Peng, Q.; Zhou, B.; Kong, Q.; Ying, Y.; Shou, H. Two soybean bHLH factors regulate response to iron deficiency. J. Integr. Plant Biol. 2018, 60, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Moran Lauter, A.N.; Peiffer, G.A.; Yin, T.; Whitham, S.A.; Cook, D.; Shoemaker, R.C.; Graham, M.A. Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef]
- Atwood, S.E.; O’Rourke, J.A.; Peiffer, G.A.; Yin, T.; Majumder, M.; Zhang, C.; Cianzio, S.R.; Hill, J.H.; Cook, D.; Whitham, S.A.; et al. Replication protein A subunit 3 and the iron efficiency response in soybean. Plant Cell Environ. 2014, 37, 213–234. [Google Scholar] [CrossRef]
- Rogers, E.E.; Wu, X.; Stacey, G.; Nguyen, H.T. Two MATE proteins play a role in iron efficiency in soybean. J. Plant Physiol. 2009, 166, 1453–1459. [Google Scholar] [CrossRef]
- Santos, C.S.; Roriz, M.; Carvalho, S.M.; Vasconcelos, M.W. Iron partitioning at an early growth stage impacts iron deficiency responses in soybean plants (Glycine max L.). Front. Plant Sci. 2015, 6, 325. [Google Scholar] [CrossRef]
- Moran Lauter, A.N.; Rutter, L.; Cook, D.; O’Rourke, J.A.; Graham, M.A. Examining short-term responses to a long-term problem: RNA-seq analyses of iron deficiency chlorosis tolerant soybean. Int. J. Mol. Sci. 2020, 21, 3591. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; McCabe, C.E.; Graham, M.A. Dynamic gene expression changes in response to micronutrient, macronutrient, and multiple stress exposure in soybean. Funct. Integr. Genom. 2020, 20, 321–341. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, A.; Arpat, A.B.; Bligny, R.; Gout, E.; Vidoudez, C.; Bensimon, M.; Poirier, Y. Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 2011, 66, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Rouached, H.; Stefanovic, A.; Secco, D.; Arpat, A.B.; Gout, E.; Bligny, R.; Poirier, Y. Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J. 2011, 65, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-F.; Liang, H.-M.; Yang, S.-Y.; Boch, A.; Clemens, S.; Chen, C.-C.; Wu, J.-F.; Huang, J.-L.; Yeh, K.-C. Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 2009, 182, 392–404. [Google Scholar] [CrossRef]
- Wu, H.; Li, L.; Du, J.; Yuan, Y.; Cheng, X.; Ling, H.-Q. Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 1505–1514. [Google Scholar] [CrossRef]
- Gollhofer, J.; Timofeev, R.; Lan, P.; Schmidt, W.; Buckhout, T.J. Vacuolar-iron-transporter1-like proteins mediate iron homeostasis in Arabidopsis. PLoS ONE 2014, 9, e110468. [Google Scholar] [CrossRef]
- Yu, H.; Yang, J.; Shi, Y.; Donelson, J.; Thompson, S.M.; Sprague, S.; Roshan, T.; Wang, D.-L.; Liu, J.; Park, S.; et al. Arabidopsis glutaredoxin S17 contributes to vegetative growth, mineral accumulation, and redox balance during iron deficiency. Front. Plant Sci. 2017, 19. [Google Scholar] [CrossRef]
- Magio, A.; Bressan, R.A.; Zhao, Y.; Park, J.; Yun, D.-J. It’s hard to avoid avoidance: Uncoupling the evolutionary connection between plant growth, productivity and stress “tolerance”. Int. J. Mol. Sci. 2018, 20, 3671. [Google Scholar] [CrossRef]
- McCabe, C.E.; Cianzio, S.R.; O’Rourke, J.A.; Graham, M.A. Leveraging RNA-seq to characterize resistance to brown stem rot and the Rbs3 locus in soybean. Mol. Plant Microbe Interact. 2018, 31, 1083–1094. [Google Scholar] [CrossRef]
- McCabe, C.E.; Singh, A.; Leandro, L.F.; Cianzio, S.R.; Graham, M.A. Identifying new sources of resistance to brown stem rot in soybean. Crop Sci. 2016, 56, 2287–2296. [Google Scholar] [CrossRef]
- Gust, A.A.; Felix, G. Receptor like proteins associate with SOBIR1-type of adaptors to form biomolecular receptor kinases. Curr. Opin. Plant Biol. 2014, 21, 104–111. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; Graham, M.A.; Vodkin, L.; Gonzalez, D.O.; Cianzio, S.R.; Shoemaker, R.C. Recovering from iron deficiency chlorosis in near-isogenic soybeans: A microarray study. Plant Physiol. Biochem. 2007, 45, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W.; Cole, M.; Flier, A.; Werr, W. BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNRÖSCHEN and DORNRÖSCHEN-LIKE. Plant Mol. Biol. 2009, 69, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, H.; Chen, J.; Chang, M.; Palmer, I.A.; Grassmann, W.; Liu, F.; Fu, Z.Q. TCP transcription factors interact with NPR1 and contribute redundantly to systemic acquired resistance. Front. Plant Sci. 2018, 14. [Google Scholar] [CrossRef]
- Gong, S.; Ding, Y.; Hu, S.; Ding, L.; Chen, Z.; Zhu, C. The role of HD-Zip class I transcription factors in plant response to abiotic stress. Physiol. Plant. 2019, 167, 516–525. [Google Scholar] [CrossRef]
- Yang, Y.; Luang, S.; Harris, J.; Riboni, M.; Li, Y.; Bazanova, N.; Hrmova, M. Overexpression of the class I homeodomain transcription factor TaHDZipI-5 increases drought and frost tolerance in transgenic wheat. Plant Biotechnol. J. 2017, 16, 1227–1240. [Google Scholar] [CrossRef]
- Gao, S.; Fang, J.; Xu, F.; Wang, W.; Chu, C. Rice HOX12 reglates panicle exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell 2016, 28, 680–695. [Google Scholar] [CrossRef]
- Chew, W.; Hrmova, M.; Lopato, S. Role of homeodomain leucine zipper (HD-ZIP) IV transcription factors in plant development and plant protection from deleterious environmental factors. Int. J. Mol. Sci. 2013, 14, 8122–8147. [Google Scholar] [CrossRef]
- Pavlū, J.; Novák, J.; Koukalová, V.; Luklová, M.; Brzobohaty, B.; Černy, M. Cytokinin at the crossroads of abiotic stress signalling pathways. Int. J. Mol. Sci. 2018, 19, 2450. [Google Scholar] [CrossRef]
- Franco-Zorilla, J.M.; Martin, A.C.; Leyva, A.; Paz-Ares, J. Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 2005, 138, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Séguéla, M.; Briat, J.F.; Vert, G.; Curie, C. Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant J. 2008, 55, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Zubo, Y.O.; Blakley, I.C.; Yamburenko, M.V.; Worthen, J.M.; Street, I.H.; Franco-Zorilla, J.M.; Zhang, W.; Hill, K.; Raines, T.; Solano, R.; et al. Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. PNAS 2017, 114, E5995–E6004. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Divya, K.; Bhatnagar-Mathur, P.; Sharma, K.K.; Sudhakar Reddy, P. Heat shock proteins (Hsps) mediated signalling pathways during abiotic stress conditions. In Plant Signaling Molecules: Role and Regulation Under Stressful Environments; Ferrante, A., Khan, M.I.R., Khan, N.A., Sudhakar Reddy, P., Eds.; Woodhead Publishing: Sawston, UK, 2019; pp. 499–516. [Google Scholar]
- Mishra, D.; Shekhar, S.; Singh, D.; Chakraborty, S.; Chakraborty, N. Heat Shock Proteins and Abiotic Stress Tolerance in Plants. In Regulation of Heat Shock Protein Responses; Asea, A.A.A., Kaur, P., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 41–69. [Google Scholar] [CrossRef]
- Anderson, M.X.; Stridh, M.H.; Larsson, K.E.; Liljenberg, C.; Sandelius, A.S. Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett. 2003, 537, 128–132. [Google Scholar] [CrossRef]
- Mehra, P.; Pandey, B.K.; Verma, L.; Giri, J. A novel glycerophosphodiester phosphodiesterase improves phosphate deficiency tolerance in rice. Plant Cell Environ. 2018, 42, 1167–1179. [Google Scholar] [CrossRef]
- McCabe, C.E.; Graham, M.A. New tools for characterizing early brown stem rot disease resistance signaling in soybean. Plant Genome 2020, e20037. [Google Scholar] [CrossRef]
- Wang, G.; Ellendorff, U.; Kemp, B.; Mansfield, J.W.; Forsyth, A.; Mitchell, K.; Bastas, K.; Liu, C.-M.; Woods-Tör, A.; Zipfel, C.; et al. A genome-wide functional investigation into the roles of receptor like proteins in Arabidopsis. Plant Physiol. 2008, 147, 503–517. [Google Scholar] [CrossRef]
- Kang, W.H.; Yeom, S.I. Genome-wide identification, classification, and expression analysis of the receptor-like protein family in tomato. Plant Pathol. J. 2018, 34, 435–444. [Google Scholar] [PubMed]
- Kim, S.A.; LaCroix, I.S.; Gerber, S.A.; Guerinot, M.L. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. PNAS 2019, 116, 24933–24942. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.L.; Yamaguchi, K.; Shigenobu, S.; Takahashi, H.; Murase, M.; Fukuyoshi, S.; Hara-Nishimura, I. Exess sterols disrupt plant cellular activity by inducing stress-responsive gene expression. J. Plant Res. 2020, 133, 383–392. [Google Scholar] [CrossRef]
- Saema, S.; ur Rahman, L.; Singh, R.; Niranjan, A.; Zareen Ahmad, I.; Misra, P. Ectopic overexpression of WsSGTL1, a sterol glucosyltransferase gene in Withania somnifera, promotes growth, enhances glycowithanolide and provides tolerance to abiotic and biotic stresses. Plant Cell Rep. 2016, 35, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Kuczyńska, A.; Cardenia, V.; Ogrodowicz, P.; Kempa, M.; Rodriguez-Estrada, M.T.; Mikołajczak, K. Effects of multiple abiotic stresses on lipids and sterols profile in barley leaves (Hordeum vulgare L.). Plant Physiol. Biochem. 2019, 141, 215–224. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Cifuentes-Esquivel, N.; Bou-Torrent, J.; Galstyan, A.; Gallemí, M.; Sessa, G.; Martret, M.S.; Roig-Villanova, I.; Martínez-García, I.R.J.F. The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 2013, 75, 989–1002. [Google Scholar] [CrossRef]
- Xing, S.; Quodt, V.; Chandler, J.; Höhmann, S.; Berndtgen, R.; Huijser, P. SPL8 acts together with brassinosteroid-signaling component BIM1 in controlling Arabidopsis thaliana male fertility. Plants 2013, 2, 416–428. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Li, X.; Liu, A.; Chen, S. Brassinosteroids in plant tolerance to abiotic stress. J. Plant Growth Regulat. 2020, 39, 1451–1464. [Google Scholar] [CrossRef]
- Vardhini, B.V.; Anjum, N.A. Brassinosteroids make plant life easier under abiotic stresses mainly by modulating major components of antioxidant defense system. Front. Environ. Sci. 2015, 2, 67. [Google Scholar] [CrossRef]
- Kim, Y.; Gilmour, S.J.; Chao, L.; Park, S.; Thomashow, M.F. Arabidopsis CAMTA transcription factors regulate pipecolic acid biosynthesis and priming of immunity genes. Mol. Plant 2020, 13, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Mu, R.-L.; Cao, Y.-R.; Liu, Y.-F.; Lei, G.; Zhou, H.-F.; Liao, Y.; Wang, H.-W.; Zhang, W.-K.; Ma, B.; Du, J.-Z.; et al. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 2009, 19, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Fasini, E.; DalCorso, G.; Costa, A.; Zenoni, S.; Furini, A. The Araibidopsis thaliana transcription factor MYB59 regulates calcium signaling during plant growth and stress response. Plant. Mol. Biol. 2019, 99, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Hickman, R.; van Verk, M.C.; van Dijken, A.J.H.; Mendes, M.P.; Vroegop-Vos, I.A.; Caarls, L.; Steenbergen, M.; van der Nagel, I.; Wesselink, G.J.; Jironkin, A.; et al. Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell 2017, 29, 2086–2105. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Engelsdorf, T.; Gigli-Bisceglia, N.; Veerabagu, M.; McKenna, J.F.; Vaahtera, L.; Augstein, F.; Van der Does, D.; Zipfel, C.; Hamann, T. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Pattathil, S.; Hahn, M.G.; Molina, A.; Miedes, E. Characterization of plant cell wall damage-associated molecular patterns regulating immune responses. Methods Mol. Biol. 2017, 1578, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Kurt, F.; Filiz, E. Genome-wide and comparative analysis of bHLH38, bHLH39, bHLH100, and bHLH101 genes in Arabidopsis, tomato, rice, soybean and maize: Insights into iron (Fe) homeostasis. BioMetals 2018, 31, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Li, L.; Wang, L.; Wang, S.; Li, S.; Du, J.; Zhang, S.; Shou, H. MPK3/MPK6 are involved in iron deficiency-induced ethylene production in Arabidopsis. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Romera, F.J.; Alcantara, E.; de la Guardia, M.D. Ethylene production by Fe-deficient roots and its involvement in the regulation of Fe-deficiency stress responses by strategy I plants. Ann. Botany 1999, 83, 51–55. [Google Scholar] [CrossRef]
- Waters, B.M.; Blevins, D.G. Ethylene production, cluster root formation, and location of iron(III) reducing capacity in Fe deficient squash roots. Plant Soil 2000, 225, 21–31. [Google Scholar] [CrossRef]
- Lingam, S.; Mohrbacher, J.; Brumbarova, T.; Potuschak, T.; Fink-Straube, C.; Blondet, E.; Genschik, P.; Bauer, P. Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 2011, 23, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Retzer, K.; Akhmanova, M.; Konstantinova, N.; Malínská, K.; Leitner, J.; Petrášek, J.; Luschnig, C. Brassinosteroid signaling delimits root gravitropism via sorting of the Arabidopsis PIN2 auxin transporter. Nat. Commun. 2019, 10, 5516. [Google Scholar] [CrossRef]
- Xia, Y.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988. [Google Scholar] [CrossRef]
- Stegmann, M.; Anderson, R.G.; Ichimura, K.; Pecenkova, T.; Reuter, P.; Zársky, V.; McDowell, J.M.; Shirasu, K.; Trujillo, M. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 2012, 24, 4703–4716. [Google Scholar] [CrossRef]
- Maathuis, F.J. cGMP modulates gene transcription and cation transport in Arabidopsis roots. Plant J. 2006, 45, 700–711. [Google Scholar] [CrossRef]
- Wang, Y.F.; Munemasa, S.; Nishimura, N.; Ren, H.M.; Robert, N.; Han, M.; Puzõrjova, I.; Kollist, H.; Lee, S.; Mori, I.; et al. Identification of cyclic GMP-activated nonselective Ca2+-permeable cation channels and associated CNGC5 and CNGC6 genes in Arabidopsis guard cells. Plant Physiol. 2013, 163, 578–590. [Google Scholar] [CrossRef]
- Li, J.Y.; Fu, Y.L.; Pike, S.M.; Bao, J.; Tian, W.; Zhang, Y.; Chen, C.Z.; Zhang, Y.; Li, H.M.; Huang, J.; et al. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 2010, 22, 1633–1646. [Google Scholar] [CrossRef]
- He, L.; Jing, Y.; Shen, J.; Li, X.; Liu, H.; Geng, Z.; Wang, M.; Li, Y.; Chen, D.; Gao, J.; et al. Mitochondrial pyruvate carriers prevent cadmium toxicity by sustaining the TCA cycle and glutathione synthesis. Plant Physiol. 2019, 180, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Chaney, R.L.; Coulombe, B.A.; Bell, P.F.; Angle, J.S. Detailed method to screen dicot cultivars for resistance to Fe-chlorosis using FeDTPA and bicarbonate in nutrient solutions. J. Plant Nutr. 1992, 15, 2063–2083. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Team, R. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA,, 2015. [Google Scholar]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level–The DESeq Package; European Molecular Biology Laboratory (EMBL): Heidelberg, Germany, 2012. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer Nature: Houston, TX, USA, 2016. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R. The Design of Experiments, 8th ed.; London Oliver and Boyd: Edinburgh, UK, 1966. [Google Scholar]
- Bonferroni, C. III Calcolo delle assicurazioni su gruppi di teste. Studi Onore Prof. Salvatore Ortu Carboni 1935, 13–60. [Google Scholar]
- Morales, A.M.A.P.; O’Rourke, J.A.; van de Mortel, M.; Scheider, K.T.; Bancroft, T.J.; Borem, A.; Nelson, R.T.; Nettleton, D.; Baum, T.J.; Shoemaker, R.C.; et al. Transcriptome analyses and virus induced gene silencing identify genes in the Rpp4-mediated Asian soybean rust resistance pathway. Funct. Plant Biol. 2013, 40, 1029–1047. [Google Scholar] [CrossRef]
- Wang, Z.; Libault, M.; Joshi, T.; Valliyodan, B.; Nguyen, H.T.; Xu, D.; Stacey, G.; Cheng, J. SoyDB: A knowledge database of soybean transcription factors. BMC Plant Biol. 2010, 10, 14. [Google Scholar] [CrossRef]
- Frith, M.C.; Fu, Y.; Yu, L.; Chen, J.F.; Hansen, U.; Weng, Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004, 32, 1372–1381. [Google Scholar] [CrossRef]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]
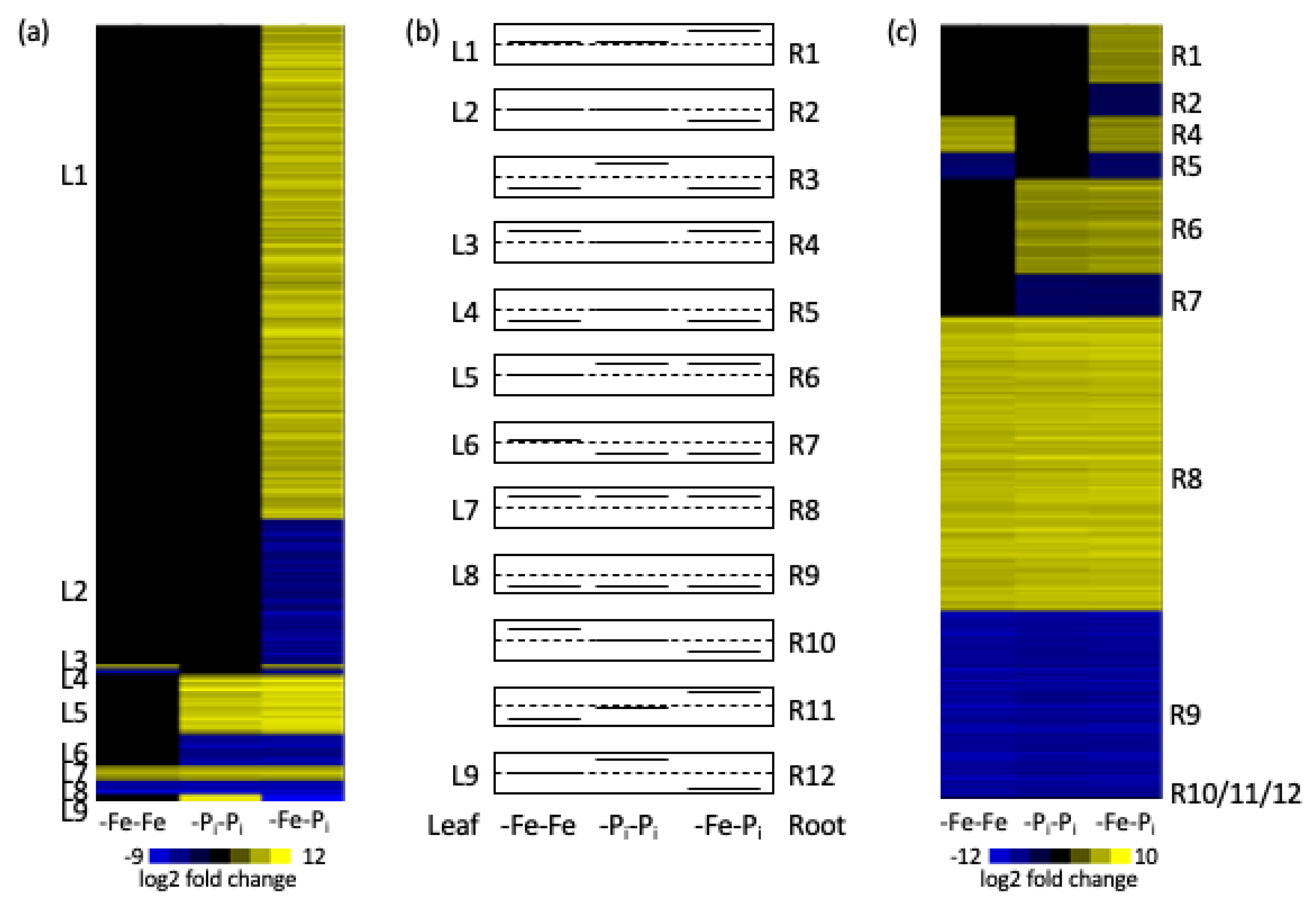

| Cluster | Pattern | # DEGs | Corrected p-Value | GO ID | GO Description |
|---|---|---|---|---|---|
| L1 | 0,0,+ | 1908 | 1.69 × 10−8 | GO:0009733 | Response to auxin stimulus |
| 2.25 × 10−7 | GO:0043481 | Anthocyanin accumulation, UV response | |||
| 9.91 × 10−5 | GO:0010817 | Regulation of hormone levels | |||
| 1.26 × 10−4 | GO:0009611 | Response to wounding | |||
| 5.40 × 10−4 | GO:0055114 | Oxidation-reduction process | |||
| L2 | 0,0,− | 558 | 0.016700 | GO:0042754 | Negative regulation of circadian rhythm |
| L3 | +,0,+ | 15 | No stat sig GO | ||
| L4 | −,0,− | 20 | No stat sig GO | ||
| L5 | 0,+,+ | 227 | 4.06 × 10−4 | GO:0016036 | Response to phosphate starvation |
| 4.83 × 10−3 | GO:0019375 | Galactolipid biosynthetic process | |||
| 4.05 × 10−3 | GO:0030643 | Cellular phosphate ion homeostasis | |||
| L6 | 0,−,− | 121 | 3.93 × 10−5 | GO:0000103 | Sulfate assimilation |
| 0.017285 | GO:0006792 | Regulation of sulfur utilization | |||
| 0.028727 | GO:0010438 | Cellular response to sulfur starvation | |||
| 0.028727 | GO:0019419 | Sulfate reduction | |||
| L7 | +,+,+ | 52 | 0.002435 | GO:0000160 | Phosphorelay signal transduction |
| 0.003631 | GO:2000121 | Regulating superoxide radical removal | |||
| 0.012052 | GO:0034052 | Positive regulation of plant hypersensitive response | |||
| L8 | −,−,− | 52 | 2.58 × 10−9 | GO:0009408 | Response to heat |
| 0.001374 | GO:0042542 | Response to hydrogen peroxide | |||
| 0.004119 | GO:0009644 | Response to high light intensity | |||
| 0.016569 | GO:0006110 | Regulation of glycolysis | |||
| 0.039347 | GO:0006979 | Response to oxidative stress | |||
| L9 | 0,+,− | 21 | No stat sig GO |
| Cluster | Pattern | # DEGs | Corrected p-Value | GO ID | GO Description |
|---|---|---|---|---|---|
| R1 | 0,0,+ | 813 | 2.96 × 10−15 | GO:0009834 | Secondary cell wall biogenesis |
| 1.97 × 10−7 | GO:0010413 | Glucuronoxylan metabolism | |||
| 2.19 × 10−7 | GO:0045492 | Xylan biosynthetic process | |||
| 4.33 × 10−6 | GO:0044036 | Cell wall macromolecule metabolism | |||
| 0.000167 | GO:0046274 | Lignin catabolic process | |||
| R2 | 0,0,− | 450 | 0.002135 | GO:0009863 | Salicylic acid mediated signaling |
| 0.002228 | GO:0002679 | Respiratory burst in defense | |||
| R3 | −,+,− | 1 | No stat sig GO | ||
| R4 | +,0,+ | 487 | No stat Sig GO | ||
| R5 | −,0,− | 361 | 5.30 × 10−5 | GO:0009715 | Chalcone biosynthetic process |
| 9.67 × 10−5 | GO:0009629 | Response to gravity | |||
| 5.91 × 10−4 | GO:0010224 | Response to UV-B | |||
| 0.011275 | GO:0006979 | Response to oxidative stress | |||
| 0.016313 | GO:0031540 | Regulation of anthocyanin biosynthesis | |||
| R6 | 0,+,+ | 1305 | 4.32 × 10−12 | GO:0009832 | Plant-type cell wall biogenesis |
| 1.13 × 10−9 | GO:0007018 | Microtubule-based movement | |||
| 2.39 × 10−9 | GO:0030243 | Cellulose metabolic process | |||
| 4.26 × 10−8 | GO:0016126 | Sterol biosynthetic process | |||
| 1.95 × 10−7 | GO:0010075 | Regulation of meristem growth | |||
| R7 | 0,−,− | 583 | No stat sig GO | ||
| R8 | +,+,+ | 4044 | 6.25 × 10−40 | GO:0000911 | Cytokinesis by cell plate formation |
| 1.28 × 10−32 | GO:0008283 | Cell proliferation | |||
| 3.40 × 10−32 | GO:0010075 | Regulation of meristem growth | |||
| 1.40 × 10−29 | GO:0006275 | Regulation of DNA replication | |||
| 1.28 × 10−26 | GO:0010389 | Regulation of G2/M transition of mitosis | |||
| R9 | −,−,− | 2563 | 2.02 × 10−25 | GO:0006606 | Protein import into nucleus |
| 2.32 × 10−18 | GO:0006626 | Protein targeting to mitochondrion | |||
| 2.46 × 10−18 | GO:0001510 | RNA methylation | |||
| 4.35 × 10−15 | GO:0034976 | Response to ER stress | |||
| 3.01 × 10−12 | GO:0009220 | Pyrimidine ribonucleotide biosynthesis | |||
| R10 | +,0,− | 1 | No stat sig GO | ||
| R11 | −,0,+ | 3 | No stat sig GO | ||
| R12 | 0,+,− | 1 | No stat sig GO |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Rourke, J.A.; Graham, M.A. Gene Expression Responses to Sequential Nutrient Deficiency Stresses in Soybean. Int. J. Mol. Sci. 2021, 22, 1252. https://doi.org/10.3390/ijms22031252
O’Rourke JA, Graham MA. Gene Expression Responses to Sequential Nutrient Deficiency Stresses in Soybean. International Journal of Molecular Sciences. 2021; 22(3):1252. https://doi.org/10.3390/ijms22031252
Chicago/Turabian StyleO’Rourke, Jamie A., and Michelle A. Graham. 2021. "Gene Expression Responses to Sequential Nutrient Deficiency Stresses in Soybean" International Journal of Molecular Sciences 22, no. 3: 1252. https://doi.org/10.3390/ijms22031252
APA StyleO’Rourke, J. A., & Graham, M. A. (2021). Gene Expression Responses to Sequential Nutrient Deficiency Stresses in Soybean. International Journal of Molecular Sciences, 22(3), 1252. https://doi.org/10.3390/ijms22031252





