HIF1A: A Putative Modifier of Hemochromatosis
Abstract
1. Introduction
2. Results
2.1. Iron and Clinical Status
2.2. Analysis of NGS Data
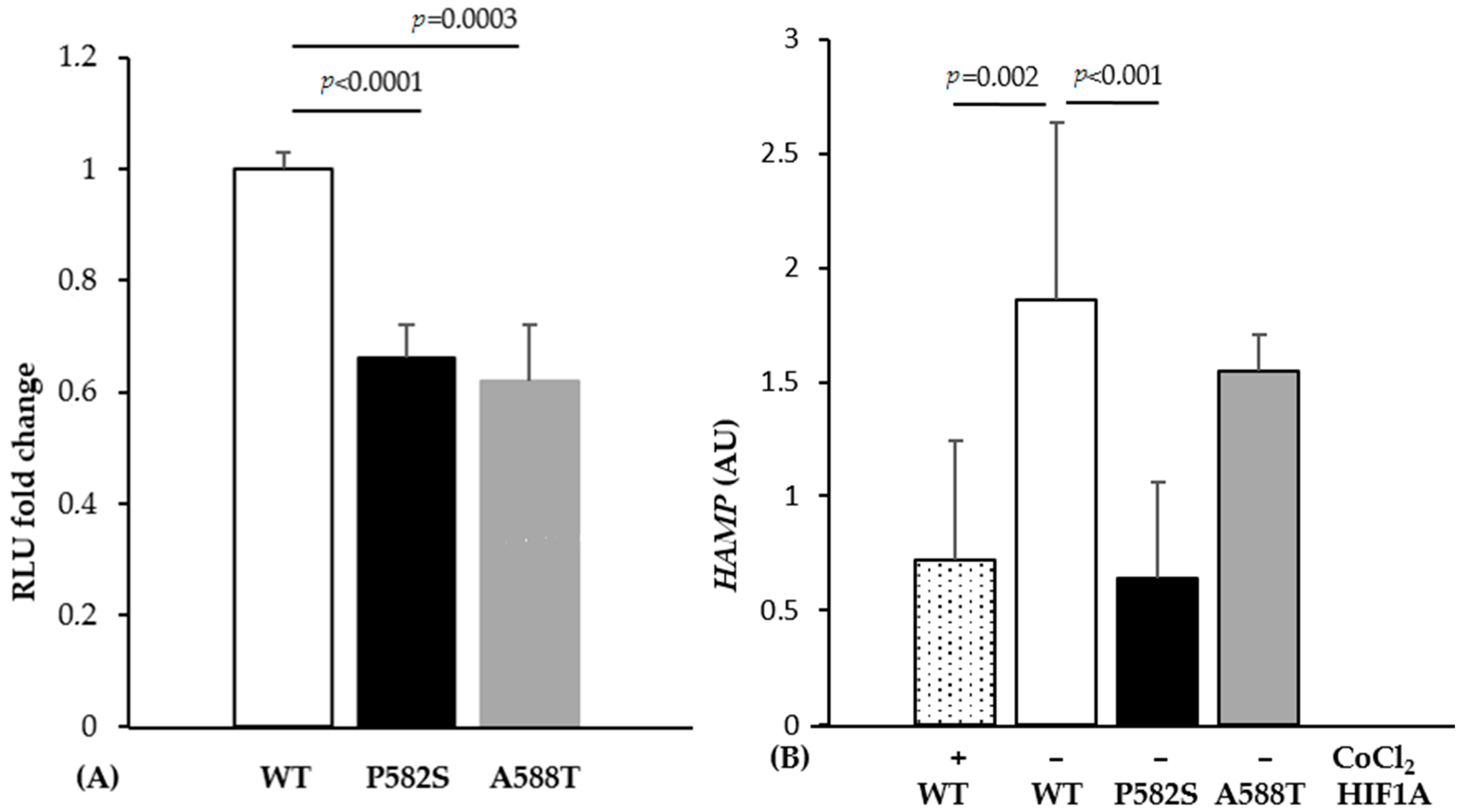
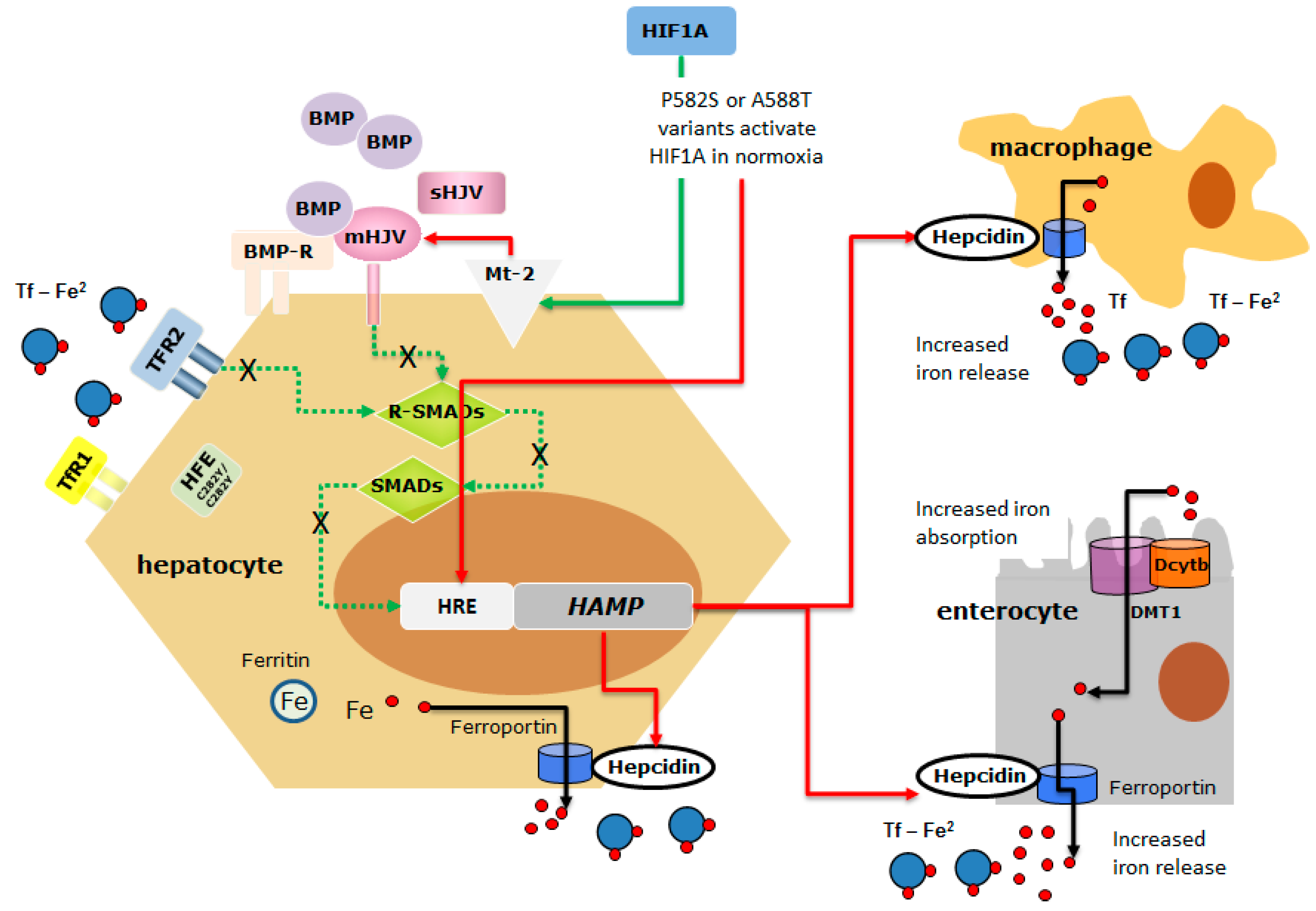
2.3. In Vitro Study
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. NGS Panel
4.3. Library Preparation and Sequencing
4.4. Data Analysis
4.5. PCR and Sequencing
4.6. Mutagenesis
4.7. Cell Culture, Transfection and Hypoxia Treatment
4.8. Dual-Luciferase Reporter Assay
4.9. RNA Extraction and cDNA Synthesis
4.10. Real-Time Quantitative-PCR
4.11. Western Blot
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A1AT | Alpha 1 Anti-Trypsin |
| ALT | Alanine Amino Transferase |
| AST | Aspartate Amino Transferase |
| AU | Arbitrary Unit |
| BMI | Body Mass Index |
| DMEM | Dulbecco’s Modified Eagle Medium |
| EPS | Extreme Phenotype Sampling |
| FBS | Fetal Bovine Serum |
| gnomAD | Genome Aggregation Database |
| HAS | Haute Autorité de Santé |
| HIF1A-WT | Vector Carrying cDNA of HIF1A Wild Type |
| HIF1A-P582S | Vector Carrying cDNA of HIF1A with p.Phe582Ser Variant |
| HIF1A-A588T | Vector Carrying cDNA of HIF1A with p.Ala588Thr Variant |
| HRE | Hypoxia Responsive Element |
| HDL | High-Density Lipoprotein |
| HH | Hereditary Hemochromatosis |
| IR | Iron Removed |
| NGS | Next Generation Sequencing |
| qRT-PCR | Quantitative-Real Time Polymerase Chain Reaction |
| RLU | Relative Luminescence Unit |
| sFerr | Serum Ferritin |
| SNP | Single Nucleotide Polymorphism |
| Try | Triglicerydes |
| TSAT | Transferrin Saturation |
| VUS | Variant of Uncertain Significance |
References
- Hanson, E.H.; Imperatore, G.; Burke, W. HFE gene and hereditary hemochromatosis: A HuGE review. Human Genome Epidemiology. Am. J. Epidemiol. 2001, 154, 193–206. [Google Scholar] [CrossRef]
- Fracanzani, A.L.; Piperno, A.; Valenti, L.; Fraquelli, M.; Coletti, S.; Maraschi, A.; Consonni, D.; Coviello, E.; Conte, D.; Fargion, S. Hemochromatosis in Italy in the last 30 years: Role of genetic and acquired factors. Hepatology 2010, 51, 501–510. [Google Scholar] [CrossRef]
- Piperno, A.; Vergani, A.; Malosio, I.; Parma, L.; Fossati, L.; Ricci, A.; Bovo, G.; Boari, G.; Mancia, G. Hepatic iron overload in patients with chronic viral hepatitis: Role of HFE gene mutations. Hepatology 1998, 28, 1105–1109. [Google Scholar] [CrossRef]
- Milet, J.; Le Gac, G.; Scotet, V.; Gourlaouen, I.; Thèze, C.; Mosser, J.; Bourgain, C.; Deugnier, Y.; Ferec, C. A common SNP near BMP2 is associated with severity of the iron burden in HFE p.C282Y homozygous patients: A follow-up study. Blood Cells Mol. Dis. 2010, 44, 34–37. [Google Scholar] [CrossRef]
- Constantine, C.C.; Anderson, G.J.; Vulpe, C.D.; McLaren, C.E.; Bahlo, M.; Yeap, H.L.; Gertig, D.M.; Osborne, N.J.; Bertalli, N.A.; Beckman, K.B.; et al. A novel association between a SNP in CYBRD1 and serum ferritin levels in a cohort study of HFE hereditary haemochromatosis. Br. J. Haematol. 2009, 147, 140–149. [Google Scholar] [CrossRef]
- Pelucchi, S.; Mariani, R.; Calza, S.; Fracanzani, A.L.; Modignani, G.L.; Bertola, F.; Busti, F.; Trombini, P.; Fraquelli, M.; Forni, G.L.; et al. CYBRD1 as a modifier gene that modulates iron phenotype in HFE p.C282Y homozygous patients. Haematologica 2012, 97, 1818–1825. [Google Scholar] [CrossRef]
- Stickel, F.; Buch, S.; Zoller, H.; Hultcrantz, R.; Gallati, S.; Österreicher, C.; Finkenstedt, A.; Stadlmayr, A.; Aigner, E.; Sahinbegovic, E.; et al. Evaluation of genome-wide loci of iron metabolism in hereditary hemochromatosis identifies PCSK7 as a host risk factor of liver cirrhosis. Hum. Mol. Genet. 2014, 23, 3883–3890. [Google Scholar] [CrossRef]
- Pelucchi, S.; Galimberti, S.; Greni, F.; Rametta, R.; Mariani, R.; Pelloni, I.; Girelli, D.; Busti, F.; Ravasi, G.; Valsecchi, M.G.; et al. Proprotein convertase 7 rs236918 associated with liver fibrosis in Italian patients with HFE-related hemochromatosis. J. Gastroenterol. Hepatol. 2016, 31, 1342–1348. [Google Scholar] [CrossRef]
- Greni, F.; Valenti, L.; Mariani, R.; Pelloni, I.; Rametta, R.; Busti, F.; Ravasi, G.; Girelli, D.; Fargion, S.; Galimberti, S.; et al. GNPAT rs11558492 is not a Major Modifier of Iron Status: Study of Italian Hemochromatosis Patients and Blood Donors. Ann. Hepatol. 2017, 16, 451–456. [Google Scholar] [CrossRef]
- McLaren, C.E.; Emond, M.J.; Subramaniam, V.N.; Phatak, P.D.; Barton, J.C.; Adams, P.C.; Goh Justin, B.; McDonald Cameron, J.; Powell Lawrie, W.; Gurrin Lyle, C.; et al. Exome sequencing in HFE C282Y homozygous men with extreme phenotypes identifies a GNPAT variant associated with severe iron overload. Hepatology 2015, 62, 429–439. [Google Scholar] [CrossRef]
- Angelucci, E.; Brittenham, G.M.; McLaren, C.E.; Ripalti, M.; Baronciani, D.; Giardini, C.; Galimberti, M.; Polchi, P.; Lucarelli, G. Hepatic iron concentration and total body iron stores in thalassemia major. N. Engl. J. Med. 2000, 343, 327–331. [Google Scholar] [CrossRef]
- Powell, E.E.; Ali, A.; Clouston, A.D.; Dixon, J.L.; Lincoln, D.J.; Purdie, D.M.; Fletcher, L.M.; Powell, L.W.; Jonsson, J.R. Steatosis is a cofactor in liver injury in hemochromatosis. Gastroenterology 2005, 129, 1937–1943. [Google Scholar] [CrossRef]
- Allen, K.J.; Bertalli, N.A.; Osborne, N.J.; Constantine, C.C.; Delatycki, M.B.; Nisselle, A.E.; Nicoll, A.J.; Gertig, D.M.; McLaren, C.E.; Giles, G.G.; et al. HFE Cys282Tyr homozygotes with serum ferritin concentrations below 1000 microg/L are at low risk of hemochromatosis. Hepatology 2010, 52, 925–933. [Google Scholar] [CrossRef]
- Adams, P.C.; Agnew, S. Alcoholism in hereditary hemochromatosis revisited: Prevalence and clinical consequences among homozygous siblings. Hepatology 1996, 23, 724–727. [Google Scholar] [CrossRef]
- Murphy, C.J.; Oudit, G.Y. Iron-overload cardiomyopathy: Pathophysiology, diagnosis, and treatment. J. Card. Fail. 2010, 16, 888–900. [Google Scholar] [CrossRef]
- Batar, B.; Bavunoglu, I.; Hacioglu, Y.; Cengiz, M.; Mutlu, T.; Yavuzer, S.; Yavuzer, H.; Ercelebi, D.C.; Erhan, D.; Unal, S.; et al. The role of TMPRSS6 gene variants in iron-related hematological parameters in Turkish patients with iron deficiency anemia. Gene 2018, 673, 201–205. [Google Scholar] [CrossRef]
- Lee, P.L.; Barton, J.C.; Khaw, P.L.; Bhattacharjee, S.Y. Common TMPRSS6 mutations and iron, erythrocyte, and pica phenotypes in 48 women with iron deficiency or depletion. Blood Cells Mol. Dis. 2012, 48, 124–127. [Google Scholar] [CrossRef]
- Tanimoto, K.; Yoshiga, K.; Eguchi, H.; Kaneyasu, M.; Ukon, K.; Kumazaki, T.; Oue, N.; Yasui, W.; Imai, K.; Nakachi, K.; et al. Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis 2003, 24, 1779–1783. [Google Scholar] [CrossRef]
- Daher, R.; Kannengiesser, C.; Houamel, D.; Lefebvre, T.; Bardou-Jacquet, E.; Ducrot, N.; de Kerguenec, C.; Jouanolle, A.; Robreau, A.; Oudin, C.; et al. Heterozygous Mutations in BMP6 Pro-peptide Lead to Inappropriate Hepcidin Synthesis and Moderate Iron Overload in Humans. Gastroenterology 2016, 150, 672–683. [Google Scholar] [CrossRef]
- Zhao, L.; Hadziahmetovic, M.; Wang, C.; Xu, X.; Song, Y.; Jinnah, H.A.; Wodzinska, J.; Iacovelli, J.; Wolkow, N.; Krajacic, P.; et al. Cp/Heph mutant mice have iron-induced neurodegeneration diminished by deferiprone. J. Neurochem. 2015, 135, 958–974. [Google Scholar] [CrossRef]
- Tolosano, E.; Fagoonee, S.; Garuti, C.; Valli, L.; Andrews, N.C.; Altruda, F.; Pietrangelo, A. Haptoglobin modifies the hemochromatosis phenotype in mice. Blood 2005, 105, 3353–3355. [Google Scholar] [CrossRef]
- Hochstrasser, H.; Bauer, P.; Walter, U.; Behnke, S.; Spiegel, J.; Csoti, I.; Zeiler, B.; Bornemann, A.; Pahnke, J.; Becker, G.; et al. Ceruloplasmin gene variations and substantia nigra hyperechogenicity in Parkinson disease. Neurology 2004, 63, 1912–1917. [Google Scholar] [CrossRef]
- Zorzetto, M.; Russi, E.; Senn, O.; Imboden, M.; Ferrarotti, I.; Tinelli, C.; Campo, I.; Ottaviani, S.; Scabini, R.; von Eckardstein, A.; et al. SERPINA1 gene variants in individuals from the general population with reduced alpha1-antitrypsin concentrations. Clin. Chem. 2008, 54, 1331–1338. [Google Scholar] [CrossRef]
- Hamesch, K.; Mandorfer, M.; Pereira, V.M.; Moeller, L.S.; Pons, M.; Dolman, G.E.; Reichert, M.C.; Schneider, C.V.; Woditsch, V.; Voss, J.; et al. Liver Fibrosis and Metabolic Alterations in Adults With α1-Antitrypsin Deficiency Caused by the Pi*ZZ Mutation. Gastroenterology 2019, 157, 705–719. [Google Scholar] [CrossRef]
- Guo, S.; Jiang, S.; Epperla, N.; Ma, Y.; Maadooliat, M.; Ye, Z.; Olson, B.; Wang, M.; Kitchner, T.; Joyce, J.; et al. A gene-based recessive diplotype exome scan discovers. Blood 2019, 133, 1888–1898. [Google Scholar] [CrossRef]
- Barton, J.C.; Chen, W.P.; Emond, M.J.; Phatak, P.D.; Subramaniam, V.N.; Adams, P.C.; Gurrin, L.C.; Anderson, G.J.; Ramm, G.A.; Powell, L.W.; et al. GNPAT p.D519G is independently associated with markedly increased iron stores in HFE p.C282Y homozygotes. Blood Cells Mol. Dis. 2017, 63, 15–20. [Google Scholar] [CrossRef]
- Henninger, B.; Alustiza, J.; Garbowski, M.; Gandon, Y. Practical guide to quantification of hepatic iron with MRI. Eur. Radiol. 2020, 30, 383–393. [Google Scholar] [CrossRef]
- Barnett, I.J.; Lee, S.; Lin, X. Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet. Epidemiol. 2013, 37, 142–151. [Google Scholar] [CrossRef]
- Panarella, M.; Burkett, K.M. A Cautionary Note on the Effects of Population Stratification Under an Extreme Phenotype Sampling Design. Front. Genet. 2019, 10, 398. [Google Scholar] [CrossRef]
- Worwood, M. Laboratory determination of iron status. In Iron Metabolism in Health and Disease; Brock, J.H., Pippard, M.J., Powell, L.W., Eds.; Saunders: London, UK, 1994; pp. 449–476. [Google Scholar]
- Altès, A.; Bach, V.; Ruiz, A.; Esteve, A.; Felez, J.; Remacha, A.F.; Sarda, M.P.; Baiget, M. Mutations in HAMP and HJV genes and their impact on expression of clinical hemochromatosis in a cohort of 100 Spanish patients homozygous for the C282Y mutation of HFE gene. Ann. Hematol. 2009, 88, 951–955. [Google Scholar] [CrossRef][Green Version]
- Donker, A.E.; Brons, P.P.; Swinkels, D.W. Microcytic anaemia with low transferrin saturation, increased serum hepcidin and non-synonymous TMPRSS6 variants: Not always iron-refractory iron deficiency anaemia. Br. J. Haematol. 2015, 169, 150–151. [Google Scholar] [CrossRef][Green Version]
- Nie, N.; Shi, J.; Shao, Y.; Li, X.; Ge, M.; Huang, J.; Zhang, J.; Huang, Z.; Li, D.; Zheng, Y. A novel tri-allelic mutation of TMPRSS6 in iron-refractory iron deficiency anaemia with response to glucocorticoid. Br. J. Haematol. 2014, 166, 300–303. [Google Scholar] [CrossRef]
- Sato, T.; Iyama, S.; Murase, K.; Kamihara, Y.; Ono, K.; Kikuchi, S.; Takada, K.; Miyanishi, K.; Sato, Y.; Takimoto, R.; et al. Novel missense mutation in the TMPRSS6 gene in a Japanese female with iron-refractory iron deficiency anemia. Int. J. Hematol. 2011, 94, 101–103. [Google Scholar] [CrossRef]
- Fu, X.S.; Choi, E.; Bubley, G.J.; Balk, S.P. Identification of hypoxia-inducible factor-1alpha (HIF-1alpha) polymorphism as a mutation in prostate cancer that prevents normoxia-induced degradation. Prostate 2005, 63, 215–221. [Google Scholar] [CrossRef]
- Nicolas, G.; Chauvet, C.; Viatte, L.; Danan, J.L.; Bigard, X.; Devaux, I.; Beaumont, C.; Kahn, A.; Vaulont, S. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J. Clin. Investig. 2002, 110, 1037–1044. [Google Scholar] [CrossRef]
- Ravasi, G.; Pelucchi, S.; Greni, F.; Mariani, R.; Giuliano, A.; Parati, G.; Silvestri, L.; Piperno, A. Circulating factors are involved in hypoxia-induced hepcidin suppression. Blood Cells Mol. Dis. 2014, 53, 204–210. [Google Scholar] [CrossRef]
- Maurer, E.; Gütschow, M.; Stirnberg, M. Matriptase-2 (TMPRSS6) is directly up-regulated by hypoxia inducible factor-1: Identification of a hypoxia-responsive element in the TMPRSS6 promoter region. Biol. Chem. 2012, 393, 535–540. [Google Scholar] [CrossRef]
- McMahon, S.; Grondin, F.; McDonald, P.P.; Richard, D.E.; Dubois, C.M. Hypoxia-enhanced expression of the proprotein convertase furin is mediated by hypoxia-inducible factor-1: Impact on the bioactivation of proproteins. J. Biol. Chem. 2005, 280, 6561–6569. [Google Scholar] [CrossRef]
- Silvestri, L.; Pagani, A.; Nai, A.; De Domenico, I.; Kaplan, J.; Camaschella, C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008, 8, 502–511. [Google Scholar] [CrossRef]
- Volke, M.; Gale, D.P.; Maegdefrau, U.; Schley, G.; Klanke, B.; Bosserhoff, A.K.; Maxwell, P.H.; Eckardt, K.; Warnecke, C. Evidence for a lack of a direct transcriptional suppression of the iron regulatory peptide hepcidin by hypoxia-inducible factors. PLoS ONE 2009, 4, e7875. [Google Scholar] [CrossRef]
- Vecchi, C.; Montosi, G.; Pietrangelo, A. Huh-7: A human “hemochromatotic” cell line. Hepatology 2010, 51, 654–659. [Google Scholar] [CrossRef]
- Peyssonnaux, C.; Zinkernagel, A.S.; Schuepbach, R.A.; Rankin, E.; Vaulont, S.; Haase, V.H.; Nizet, V.; Johnson, R.S. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J. Clin. Investig. 2007, 117, 1926–1932. [Google Scholar] [CrossRef]
- Lakhal, S.; Schödel, J.; Townsend, A.R.; Pugh, C.W.; Ratcliffe, P.J.; Mole, D.R. Regulation of type II transmembrane serine proteinase TMPRSS6 by hypoxia-inducible factors: New link between hypoxia signaling and iron homeostasis. J. Biol. Chem. 2011, 286, 4090–4097. [Google Scholar] [CrossRef]
- Schaefer, B.; Haschka, D.; Finkenstedt, A.; Petersen, B.S.; Theurl, I.; Henninger, B.; Janecke, A.R.; Wang, C.; Lin, H.Y.; Veits, L.; et al. Impaired hepcidin expression in alpha-1-antitrypsin deficiency associated with iron overload and progressive liver disease. Hum. Mol. Genet. 2015, 24, 6254–6263. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Hochstrasser, H.; Tomiuk, J.; Walter, U.; Behnke, S.; Spiegel, J.; Krüger, R.; Becker, G.; Riess, O.; Berg, D. Functional relevance of ceruloplasmin mutations in Parkinson’s disease. FASEB J. 2005, 19, 1851–1853. [Google Scholar] [CrossRef]
- Henrie, A.; Hemphill, S.E.; Ruiz-Schultz, N.; Cushman, B.; DiStefano, M.T.; Azzariti, D.; Harrison, S.M.; Rehm, H.L.; Eilbeck, K. ClinVar Miner: Demonstrating utility of a Web-based tool for viewing and filtering ClinVar data. Hum. Mutat. 2018, 39, 1051–1060. [Google Scholar] [CrossRef]
- Muckenthaler, M.U. How mutant HFE causes hereditary hemochromatosis. Blood 2014, 124, 1212–1213. [Google Scholar] [CrossRef][Green Version]
- Riva, A.; Trombini, P.; Mariani, R.; Salvioni, A.; Coletti, S.; Bonfadini, S.; Paolini, V.; Pozzi, M.; Facchetti, R.; Bovo, G.; et al. Revaluation of clinical and histological criteria for diagnosis of dysmetabolic iron overload syndrome. World J. Gastroenterol. 2008, 14, 4745–4752. [Google Scholar] [CrossRef]
- Adams, P.C.; Barton, J.C. How I treat hemochromatosis. Blood 2010, 116, 317–325. [Google Scholar] [CrossRef]
- Adams, P.C.; Deugnier, Y.; Moirand, R.; Brissot, P. The relationship between iron overload, clinical symptoms, and age in 410 patients with genetic hemochromatosis. Hepatology 1997, 25, 162–166. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Brissot, P.; Troadec, M.B.; Bardou-Jacquet, E.; Le Lan, C.; Jouanolle, A.M.; Deugnier, Y.; Loreal, O. Current approach to hemochromatosis. Blood Rev. 2008, 22, 195–210. [Google Scholar] [CrossRef]
- Piret, J.P.; Mottet, D.; Raes, M.; Michiels, C. CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann. N. Y. Acad. Sci. 2002, 973, 443–447. [Google Scholar] [CrossRef]

| All | HAS-Low | HAS-High | p | |
|---|---|---|---|---|
| N | 99 | 41 | 58 | // |
| Age (years) | 43 (36–52) | 37 (29–45) | 45.5 (38.0–58.0) | <0.0001 |
| Hb (g/dL) | 15.0 (14.5–15.8) | 15.0 (14.7–15.8) | 14.9 (13.9–15.6) | ns |
| sFerritin (µg/L) | 1244 (768–2463.5) | 765 (516–956) | 2164 (1386–3427) | <0.0001 |
| TSAT (%) | 84.5 (70.6–90.8) | 78.5 (42.0–87.0) | 86 (78–95) | <0.0001 |
| IR/age (g/years) | 0.15 (0.10–0.24) | 0.10 (0.08–0.14) | 0.22 (0.14–0.33) | <0.0001 |
| BMI (kg/m2) | 24.5 (23.5–26.4) | 24.4 (23.8–26.6) | 24.5 (23.0–26.0) | ns |
| AST (U/L) | 31 (23–44) | 25.0 (19.3–30.0) | 41 (30–58) | <0.0001 |
| ALT (U/L) | 43.0 (26.3–64.8) | 27.0 (20.5–45.3) | 52.5 (40.0–79.3) | <0.0001 |
| γGT (U/L) | 28.5 (18.0–45.3) | 18.5 (16.0–30.0) | 38.5 (25.0–53.5) | <0.0001 |
| Total cholesterol (mg/dL) | 182 (155–212) | 172.5 (157.8–207.5) | 189 (153–213) | ns |
| HDL (mg/dL) | 49 (42–55) | 49.0 (42.5–54.0) | 49.0 (42.0–56.5) | ns |
| Try (mg/dL) | 104 (79–144) | 92.5 (78.3–127.5) | 118 (82–156) | ns |
| Glu (mg/dL) | 93 (85–102) | 87.5 (81.8–93.3) | 98.5 (89.5–111.3) | 0.0001 |
| All | L-Iron | H-Iron | p | |
|---|---|---|---|---|
| N | 26 | 15 | 11 | // |
| Age (years) | 43 (38–50) | 41.0 (37.0–49.5) | 43.0 (38.0–48.5) | ns |
| Hb (g/dL) | 14.8 (14.4–15.1) | 14.9 (14.6–15.3) | 13.9 (12.15–15.75) | ns |
| sFerritin (µg/L) | 1158 (602–3680) | 698 (509–794) | 4000 (2700–4800) | <0.0001 |
| TSAT (%) | 84.5 (64.5–90.0) | 70 (59–86) | 90 (84–95) | 0.01 |
| IR/age (g/years) | 0.12 (0.08–0.43) | 0.08 (0.07–0.11) | 0.46 (0.40–0.63) | <0.0001 |
| BMI (kg/m2) | 24.2 (23.6–25.3) | 24.5 (24.2–26.3) | 22.2 (18.8–24.0) | 0.008 |
| AST (U/L) | 27.0 (21.5–36.0) | 23.0 (18.1–27.0) | 47.5 (36.0–61.0) | 0.0002 |
| ALT (U/L) | 38.0 (26.0–50.5) | 26 (24–39) | 58 (43–86) | 0.001 |
| γGT (U/L) | 25.0 (16.8–41.5) | 19.5 (14.5–28.5) | 51.0 (30.5–206.0) | 0.009 |
| Total cholesterol (mg/dL) | 176.5 (160.3–203.8) | 169 (157–183) | 205 (189–253) | 0.04 |
| HDL (mg/dL) | 51.0 (42.0–63.8) | 47.5 (42.0–62.5) | 57.5 (52.0–65.5) | ns |
| Try (mg/dL) | 98.5 (75.3–145.8) | 90 (75–124) | 130 (68–170) | ns |
| Glu (mg/dL) | 93 (88–98) | 92 (87–95) | 105.5 (92.5–151.5) | 0.042 |
| Genes | Amino Acid Change | dbSNP | Allelic Frequencies (all) | Allelic Frequencies (L-Iron) | Allelic Frequencies (H-Iron) | Allelic Frequencies (gnomAD) | p |
|---|---|---|---|---|---|---|---|
| ACO1 | p.Lys580Gln | rs73477393 | 0.019 | 0.000 | 0.045 | 0.000 | ns |
| BMP6 | p.Gln114Arg | rs377443730 | 0.019 | 0.033 | 0.000 | 0.000 | ns |
| CP | p.Arg793His | rs115552500 | 0.019 | 0.033 | 0.000 | 0.009 | ns |
| CP | p.Glu544Asp | rs701753 | 0.865 | 0.900 | 0.818 | 0.941 | ns |
| CP | p.Pro477Leu | rs35331711 | 0.019 | 0.000 | 0.045 | 0.004 | ns |
| CP | p.Thr551Ile | rs61733458 | 0.019 | 0.033 | 0.000 | 0.031 | ns |
| CYBRD1 | p.Ser266Asn | rs10455 | 0.481 | 0.333 | 0.682 | 0.672 | <0.0001 * |
| FAM132B | p.Ala260Ser | rs111241405 | 0.019 | 0.000 | 0.045 | 0.021 | ns |
| FURIN | p.Arg81Cys | rs148110342 | 0.019 | 0.000 | 0.045 | 0.002 | ns |
| GNPAT | p.Asp519Gly | rs11558492 | 0.327 | 0.300 | 0.364 | 0.205 | ns |
| HEPH | p.Val39Ala | rs5919015 | 0.692 | 0.733 | 0.636 | 0.679 | ns |
| HIF1A | p.Ala588Thr | rs11549467 | 0.038 | 0.000 | 0.091 | 0.010 | 0.0001 § |
| HIF1A | p.Pro582Ser | rs11549465 | 0.231 | 0.167 | 0.318 | 0.105 | 0.001 § |
| HP | p.Ala371Ser | rs376612221 | 0.019 | 0.033 | 0.000 | 0.000 | ns |
| HP | p.Val258Leu | rs746679401 | 0.019 | 0.033 | 0.000 | 0.000 | ns |
| IREB2 | p.Ile580Thr | rs2230940 | 0.846 | 0.733 | 1.000 | 0.9998 | ns |
| IREB2 | p.Val159Leu | rs2958720 | 0.500 | 0.467 | 0.545 | 0.9998 | <0.0001 * |
| IREB2 | p.Tyr8fs | rs1456427498 | 0.058 | 0.067 | 0.045 | 0.0001 | <0.0001 * < 0.0001 § |
| SCARA5 | p.Ala304Val | rs118119884 | 0.019 | 0.000 | 0.045 | 0.036 | ns |
| SCARA5 | p.Asp316His | rs17058207 | 0.038 | 0.067 | 0.000 | 0.102 | ns |
| SERPINA1 | p.Arg125His | rs709932 | 0.135 | 0.133 | 0.136 | 0.163 | ns |
| SERPINA1 | p.Glu288Val | rs17580 | 0.038 | 0.067 | 0.000 | 0.037 | ns |
| SERPINA1 | p.Glu366Lys | rs28929474 | 0.038 | 0.000 | 0.091 | 0.018 | ns |
| SERPINA1 | p.Glu400Asp | rs1303 | 0.212 | 0.167 | 0.273 | 0.255 | ns |
| SERPINA1 | p.Ile116Met | rs759135389 | 0.019 | 0.033 | 0.000 | 0.0000 | ns |
| SERPINA1 | p.Val237Ala | rs6647 | 0.173 | 0.100 | 0.273 | 0.217 | ns |
| TF | p.Ile448Val | rs2692696 | 1.000 | 1.000 | 1.000 | 1.000 | ns |
| TF | p.Pro589Ser | rs1049296 | 0.135 | 0.167 | 0.091 | 0.160 | ns |
| TFRC | p.Gly142Ser | rs3817672 | 0.635 | 0.633 | 0.636 | 0.551 | ns |
| TMPRSS6 | p.Ala31Val | rs200558933 | 0.019 | 0.033 | 0.000 | 0.000 | ns |
| TMPRSS6 | p.Lys253Glu | rs2235324 | 0.096 | 0.167 | 0.000 | 0.385 | <0.0001 § |
| TMPRSS6 | p.Val736Ala | rs855791 | 0.442 | 0.367 | 0.545 | 0.564 | ns |
| FURIN | p.Leu163Leu | // | 0.019 | 0.000 | 0.045 | ns | |
| HIF1A | p.Tyr659Tyr | // | 0.019 | 0.000 | 0.045 | ns | |
| IREB2 | p.Pro145Ser | // | 0.019 | 0.000 | 0.045 | ns | |
| TMPRSS6 | p.Leu746Leu | // | 0.019 | 0.033 | 0.000 | ns |
| WT + Co | WT | P582S | A588T | |
|---|---|---|---|---|
| HIF1A | 911 ± 21 * | 600 ± 113 | 20,653 ± 2505 * | 287 ± 82 |
| VEGF | 2.93 ± 0.86 * | 0.76 ± 0.05 | 89 ± 5 * | 0.20 ± 0.07 |
| HMOX | 69 ± 9 * | 1.44 ± 0.11 | 6.28 ± 1.66 * | 0.42 ± 0.06 |
| FUR | 0.63 ± 0.15 | 0.43 ± 0.10 | 55 ± 2 * | 0.35 ± 0.07 |
| TMPRSS6 | 0.38 ± 0.04 | 1.06 ± 0.11 | 77 ± 12 * | 0.06 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pelucchi, S.; Ravasi, G.; Arosio, C.; Mauri, M.; Piazza, R.; Mariani, R.; Piperno, A. HIF1A: A Putative Modifier of Hemochromatosis. Int. J. Mol. Sci. 2021, 22, 1245. https://doi.org/10.3390/ijms22031245
Pelucchi S, Ravasi G, Arosio C, Mauri M, Piazza R, Mariani R, Piperno A. HIF1A: A Putative Modifier of Hemochromatosis. International Journal of Molecular Sciences. 2021; 22(3):1245. https://doi.org/10.3390/ijms22031245
Chicago/Turabian StylePelucchi, Sara, Giulia Ravasi, Cristina Arosio, Mario Mauri, Rocco Piazza, Raffaella Mariani, and Alberto Piperno. 2021. "HIF1A: A Putative Modifier of Hemochromatosis" International Journal of Molecular Sciences 22, no. 3: 1245. https://doi.org/10.3390/ijms22031245
APA StylePelucchi, S., Ravasi, G., Arosio, C., Mauri, M., Piazza, R., Mariani, R., & Piperno, A. (2021). HIF1A: A Putative Modifier of Hemochromatosis. International Journal of Molecular Sciences, 22(3), 1245. https://doi.org/10.3390/ijms22031245






