Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes
Abstract
1. Introduction
2. Results
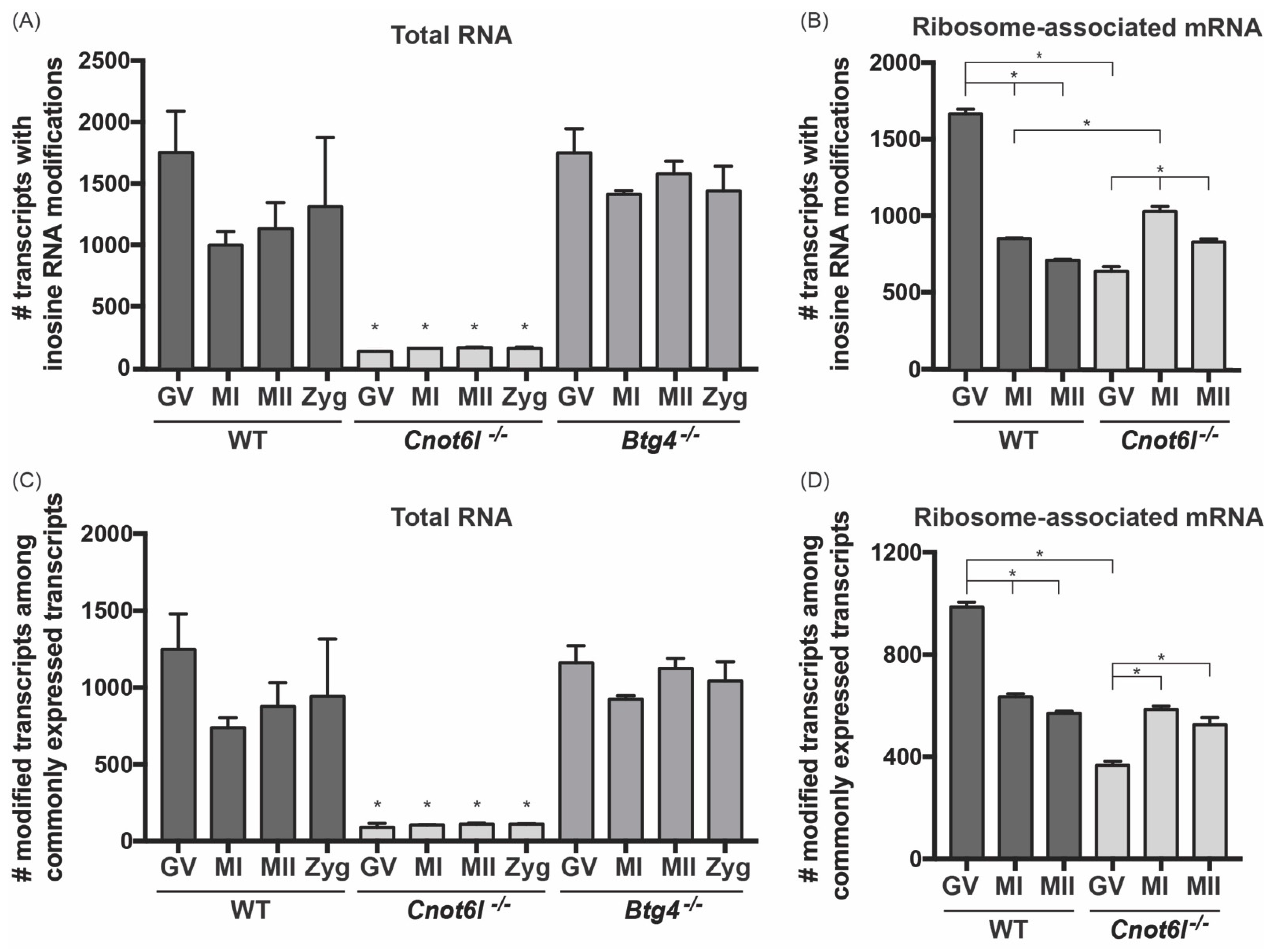
2.1. Inosine RNA Modifications Are Lost in Oocytes from Cnot6l-/- Mice
2.2. Inosine RNA Modifications Are Enriched in Ribosome-Associated mRNA
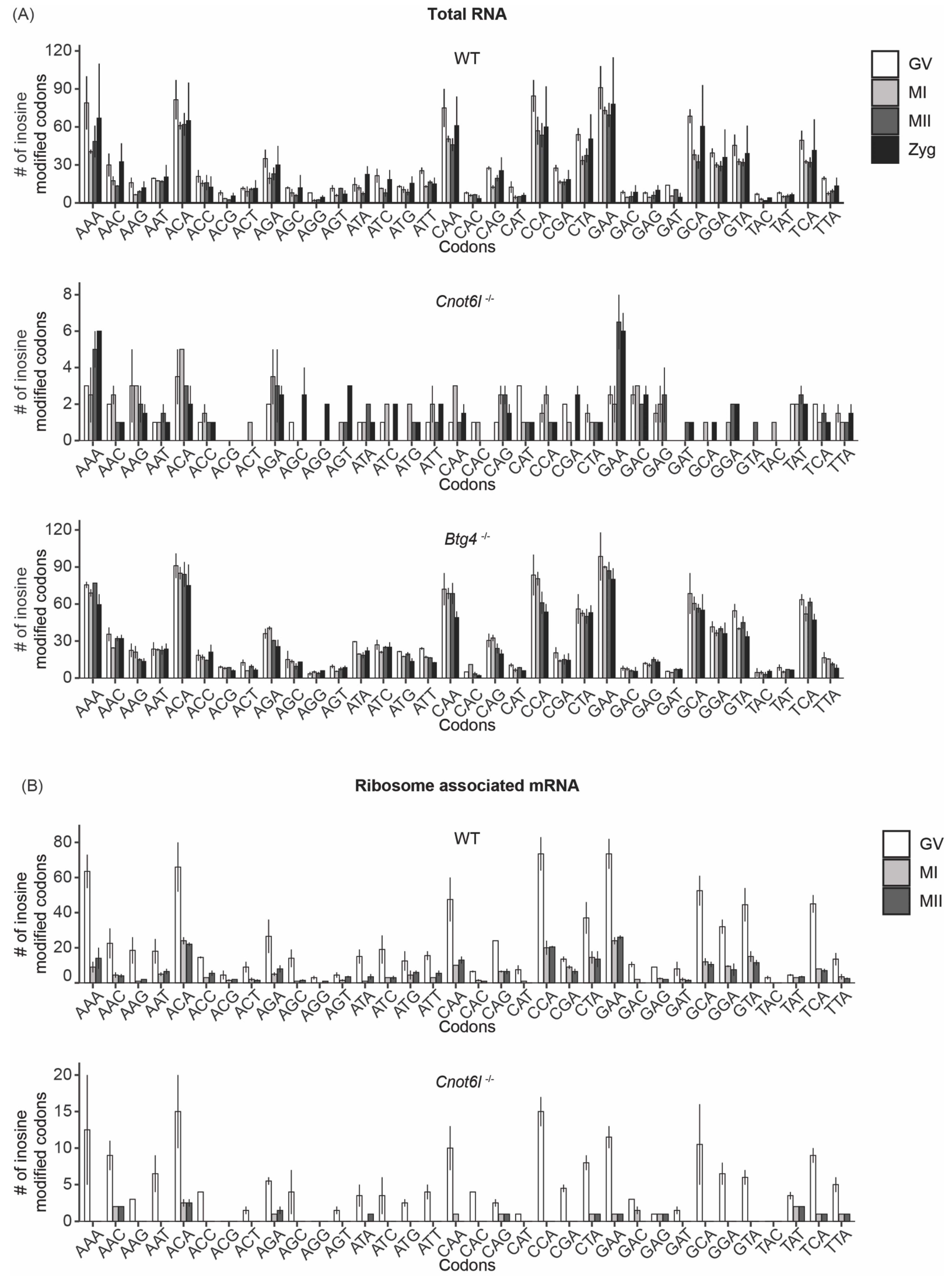
2.3. Pattern of Inosine RNA Modifications in Total and Ribosome-Associated mRNA
2.4. Consequences of Coding Sequence Inosine RNA Modifications in Mouse Oocytes, Eggs, and Embryos
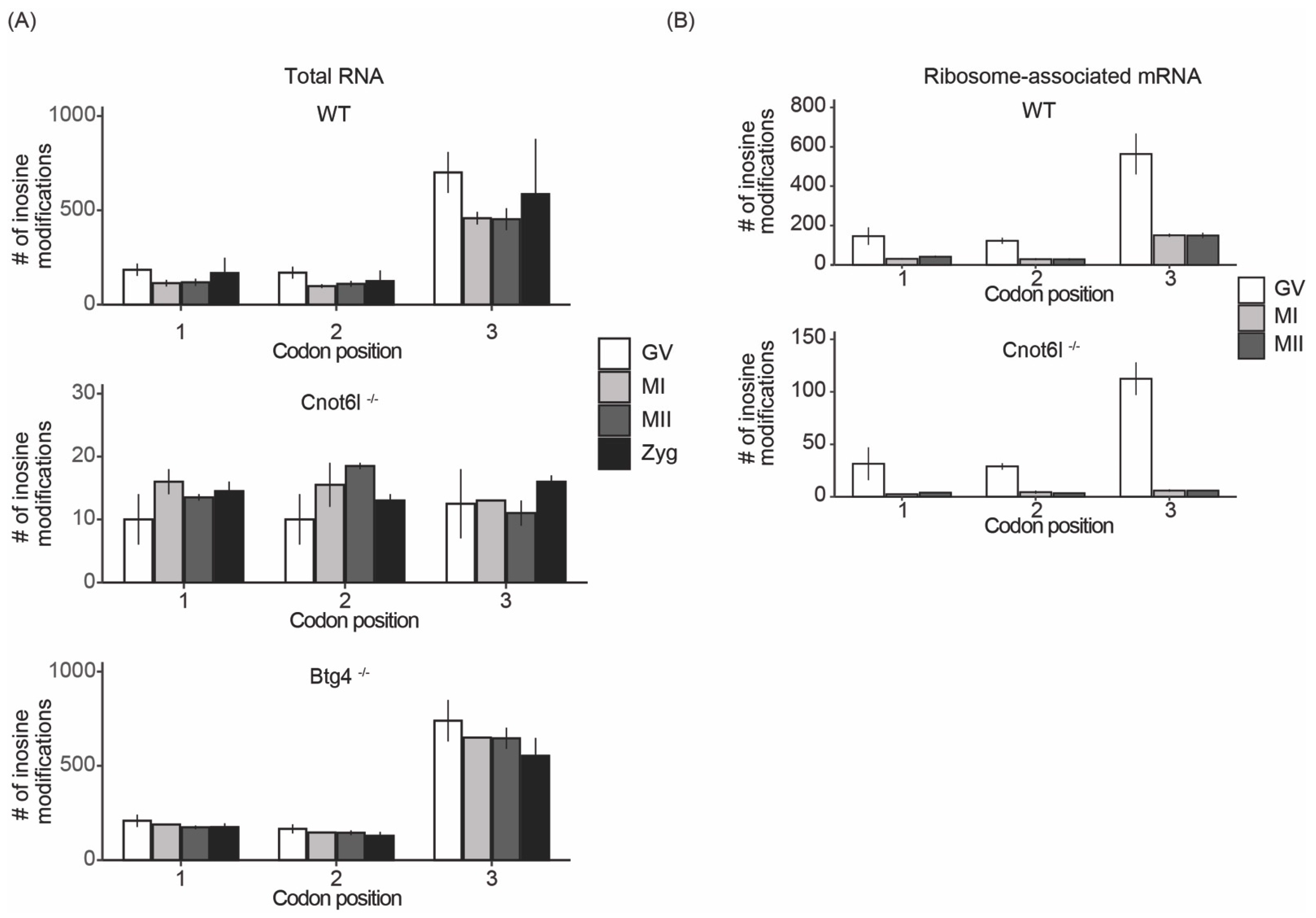
2.5. Inosine RNA Modifications Are Enriched at the Wobble Position in Ribosome-Associated RNA
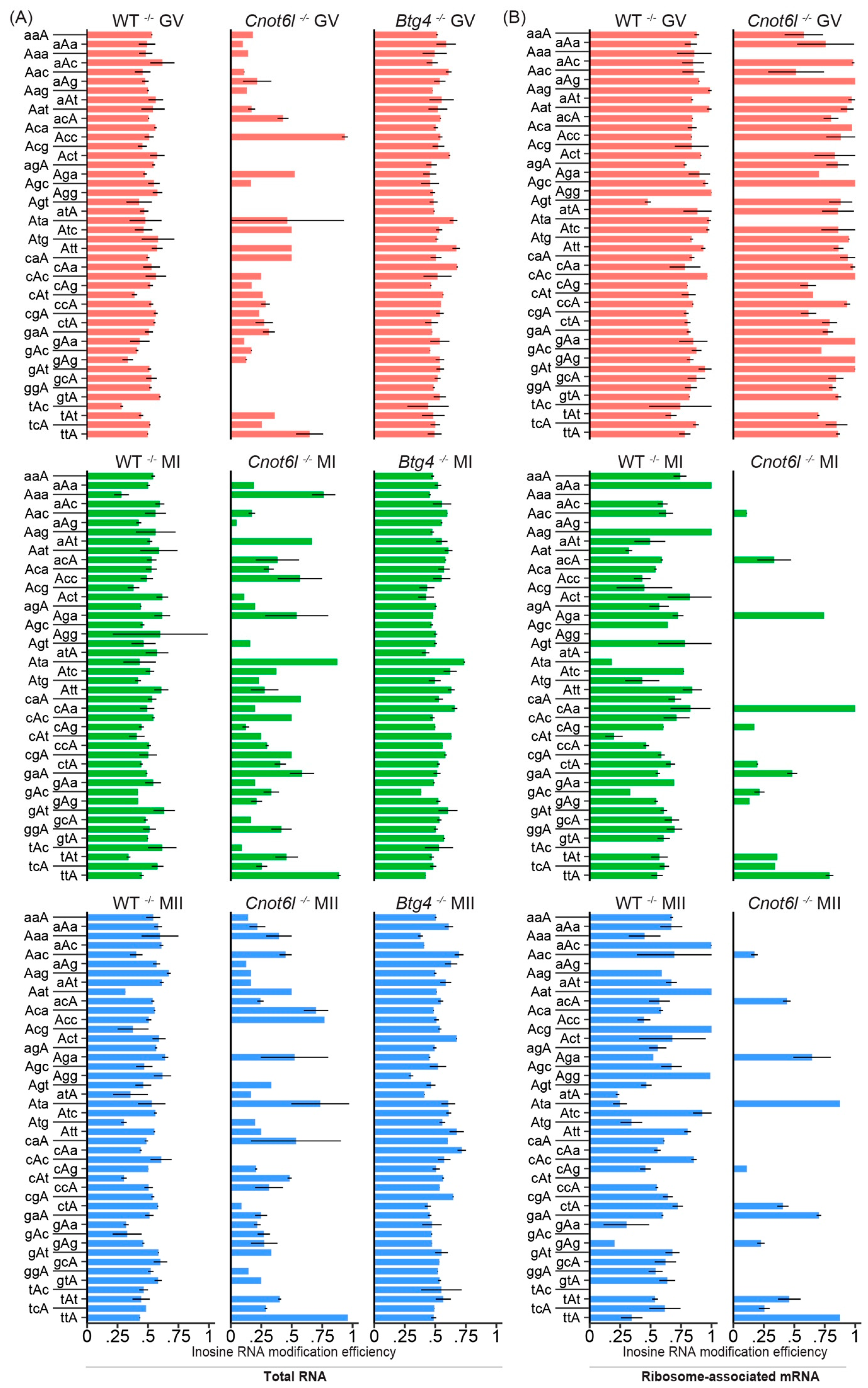
2.6. Efficiency of Inosine RNA Modifications is Highest in Ribosome-Associated mRNA
3. Discussion
4. Materials and Methods
4.1. Sources of GV and MII RNA-Seq Datasets
4.2. Identification and Consequence Analysis of Inosine RNA Modifications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GATK | Genome Analysis Toolkit |
| GV | germinal vesicle oocyte |
| MI | metaphase I stage oocyte |
| MII | metaphase II stage oocyte |
| MZT | maternal to zygotic transition |
| SIFT | Sorting Intolerant From Tolerant |
| RNA | ribonucleic acid |
| UTR | untranslated region |
| VEP | Ensembl Variant Effect Predictor |
| WT | wild-type |
| Zyg | one-cell zygote |
References
- Bachvarova, R.; De Leon, V. Polyadenylated RNA of mouse ova and loss of maternal RNA in early development. Dev. Biol. 1980, 74, 1–8. [Google Scholar] [CrossRef]
- Bachvarova, R. Gene Expression During Oogenesis and Oocyte Development in Mammals. Dev. Biol. 1985, 1, 453–524. [Google Scholar] [CrossRef]
- De La Fuente, R.; Viveiros, M.M.; Burns, K.H.; Adashi, E.Y.; Matzuk, M.M.; Eppig, J.J. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev. Biol. 2004, 275, 447–458. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente, R.; Eppig, J.J. Transcriptional Activity of the Mouse Oocyte Genome: Companion Granulosa Cells Modulate Transcription and Chromatin Remodeling. Dev. Biol. 2001, 229, 224–236. [Google Scholar] [CrossRef]
- Pikó, L.; Clegg, K.B. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev. Biol. 1982, 89, 362–378. [Google Scholar] [CrossRef]
- Su, Y.-Q.; Sugiura, K.; Woo, Y.; Wigglesworth, K.; Kamdar, S.; Affourtit, J.; Eppig, J.J. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev. Biol. 2007, 302, 104–117. [Google Scholar] [CrossRef]
- Svoboda, P.; Franke, V.; Schultz, R.M. Sculpting the Transcriptome During the Oocyte-to-Embryo Transition in Mouse. Curr. Top. Dev. Biol. 2015, 113, 305–349. [Google Scholar] [CrossRef]
- Sha, Q.-Q.; Zhang, J.; Fan, H.-Y. A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals†. Biol. Reprod. 2019, 101, 579–590. [Google Scholar] [CrossRef]
- Sha, Q.-Q.; Dai, X.-X.; Dang, Y.; Tang, F.; Liu, J.; Zhang, Y.-L.; Fan, H.-Y. A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocytes. Development 2016, 144, 452–463. [Google Scholar] [CrossRef]
- Ozturk, S.; Uysal, F. Poly(A)-binding proteins are required for translational regulation in vertebrate oocytes and early embryos. Reprod. Fertil. Dev. 2017, 29, 1890–1901. [Google Scholar] [CrossRef]
- Chen, J.; Melton, C.; Suh, N.; Oh, J.S.; Horner, K.; Xie, F.; Sette, C.; Blelloch, R.; Conti, M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011, 25, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.-X.; Jiang, J.-C.; Sha, Q.-Q.; Jiang, Y.; Ou, X.-H.; Fan, H.-Y. A combinatorial code for mRNA 3′-UTR-mediated translational control in the mouse oocyte. Nucleic Acids Res. 2019, 47, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A.; Panasenko, O.O. The Ccr4–Not complex. Gene 2012, 492, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Sha, Q.-Q.; Yu, J.; Guo, J.; Dai, X.; Jiang, J.; Zhang, Y.; Yu, C.; Ji, S.; Jiang, Y.; Zhang, S.; et al. CNOT 6L couples the selective degradation of maternal transcripts to meiotic cell cycle progression in mouse oocyte. EMBO J. 2018, 37, e99333. [Google Scholar] [CrossRef]
- Horvat, F.; Fulka, H.; Jankele, R.; Malik, R.; Jun, M.; Solcova, K.; Sedlacek, R.; Vlahoviček, K.; Schultz, R.M.; Svoboda, P. Role of Cnot6l in maternal mRNA turnover. Life Sci. Alliance 2018, 1, e201800084. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Ji, S.-Y.; Sha, Q.-Q.; Dang, Y.; Zhou, J.-J.; Zhang, Y.-L.; Liu, Y.; Wang, Z.-W.; Hu, B.; Sun, Q.-Y.; et al. BTG4 is a meiotic cell cycle–coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 2016, 23, 387–394. [Google Scholar] [CrossRef]
- Dumdie, J.N.; Cho, K.; Ramaiah, M.; Skarbrevik, D.; Mora-Castilla, S.; Stumpo, D.J.; Lykke-Andersen, J.; Laurent, L.C.; Blackshear, P.J.; Wilkinson, M.F.; et al. Chromatin Modification and Global Transcriptional Silencing in the Oocyte Mediated by the mRNA Decay Activator ZFP36L2. Dev. Cell 2018, 44, 392–402.e7. [Google Scholar] [CrossRef] [PubMed]
- Vieux, K.-F.; Clarke, H.J. CNOT6 regulates a novel pattern of mRNA deadenylation during oocyte meiotic maturation. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Ball, C.B.; Rodriguez, K.F.; Stumpo, D.J.; Ribeiro-Neto, F.; Korach, K.S.; Blackshear, P.J.; Birnbaumer, L.; Ramos, S.B.V. The RNA-Binding Protein, ZFP36L2, Influences Ovulation and Oocyte Maturation. PLoS ONE 2014, 9, e97324. [Google Scholar] [CrossRef]
- Doidge, R.; Mittal, S.; Aslam, A.; Winkler, G.S. The Anti-Proliferative Activity of BTG/TOB Proteins Is Mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) Deadenylase Subunits of the Ccr4-Not Complex. PLoS ONE 2012, 7, e51331. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, X.; Shi, J.; Yu, X.; Zhang, X.; Zhu, K.; Yi, Z.; Duan, E.; Li, L. BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis. J. Mol. Cell Biol. 2016, 8, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Dean, J. BTG4, a maternal mRNA cleaner. J. Mol. Cell Biol. 2016, 8, 369–370. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pasternak, M.; Pfender, S.; Santhanam, B.; Schuh, M. The BTG4 and CAF1 complex prevents the spontaneous activation of eggs by deadenylating maternal mRNAs. Open Biol. 2016, 6, 160184. [Google Scholar] [CrossRef]
- Ivanova, I.; Much, C.; Di Giacomo, M.; Azzi, C.; Morgan, M.; Moreira, P.N.; Monahan, J.; Carrieri, C.; Enright, A.J.; O’Carroll, D. The RNA m6A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Com-petence. Mol. Cell 2017, 67, 1059–1067.e4. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K.; Yoo, C.; Kim, U.; Murray, J.M.; Estes, P.A.; Cash, F.E.; Liebhaber, S.A. Substrate specificity of the dsRNA unwind-ing/modifying activity. EMBO J. 1991, 10, 3523–3532. [Google Scholar] [CrossRef]
- Bass, B.L.; Weintraub, H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 1988, 55, 1089–1098. [Google Scholar] [CrossRef]
- Wagner, R.W.; Smith, J.E.; Cooperman, B.S.; Nishikura, K. A double-stranded RNA unwinding activity introduces structural al-terations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. USA 1989, 86, 2647–2651. [Google Scholar] [CrossRef]
- Brachova, P.; Alvarez, N.S.; Hong, X.; Gunewardena, S.; A Vincent, K.; E Latham, K.; Christenson, L. Inosine RNA modifications are enriched at the codon wobble position in mouse oocytes and eggs†. Biol. Reprod. 2019, 101, 938–949. [Google Scholar] [CrossRef]
- Licht, K.; Hartl, M.; Amman, F.; Anrather, D.; Janisiw, M.P.; Jantsch, M.F. Inosine induces context-dependent recoding and trans-lational stalling. Nucleic Acids Res. 2019, 47, 3–14. [Google Scholar] [CrossRef]
- Higuchi, M.; Single, F.N.; Köhler, M.; Sommer, B.; Sprengel, R.; Seeburg, P.H. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell 1993, 75, 1361–1370. [Google Scholar] [CrossRef]
- Brusa, R.; Zimmermann, F.; Koh, D.-S.; Feldmeyer, D.; Gass, P.; Seeburg, P.H.; Sprengel, R. Early-Onset Epilepsy and Postnatal Lethality Associated with an Editing-Deficient GluR-B Allele in Mice. Science 1995, 270, 1677–1680. [Google Scholar] [CrossRef] [PubMed]
- Walkley, C.R.; Li, J.B. Rewriting the transcriptome: Adenosine-to-inosine RNA editing by ADARs. Genome Biol. 2017, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Presnyak, V.; Alhusaini, N.; Chen, Y.-H.; Martin, S.; Morris, N.; Kline, N.; Olson, S.; Weinberg, D.; Baker, K.E.; Graveley, B.R.; et al. Codon Optimality Is a Major Determinant of mRNA Stability. Cell 2015, 160, 1111–1124. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Del Viso, F.; A Moreno-Mateos, M.; Johnstone, T.G.; E Vejnar, C.; Qin, Y.; Yao, J.; Khokha, M.K.; Giraldez, A.J. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016, 35, 2087–2103. [Google Scholar] [CrossRef]
- Carneiro, R.L.; Requião, R.D.; Rossetto, S.; Domitrovic, T.; Palhano, F.L. Codon stabilization coefficient as a metric to gain insights into mRNA stability and codon bias and their relationships with translation. Nucleic Acids Res. 2019, 47, 2216–2228. [Google Scholar] [CrossRef]
- Horstick, E.J.; Jordan, D.C.; Bergeron, S.A.; Tabor, K.M.; Serpe, M.; Feldman, B.; Burgess, H.A. Increased functional protein ex-pression using nucleotide sequence features enriched in highly expressed genes in zebrafish. Nucleic Acids Res. 2015, 43, e48. [Google Scholar] [CrossRef]
- Pop, C.; Rouskin, S.; Ingolia, N.T.; Han, L.; Phizicky, E.M.; Weissman, J.S.; Koller, D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014, 10, 770. [Google Scholar] [CrossRef]
- Bergman, S.; Tuller, T. Widespread non-modular overlapping codes in the coding regions. Phys. Biol. 2020, 17, 031002. [Google Scholar] [CrossRef]
- Freund, E.C.; Sapiro, A.L.; Li, Q.; Linder, S.; Moresco, J.J.; Yates, J.R., 3rd; Li, J.B. Unbiased Identification of trans Regulators of ADAR and A-to-I RNA Editing. Cell Rep. 2020, 31, 107656. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.W.; Chen, Y.-H.; Stowell, J.A.W.; Alhusaini, N.; Sweet, T.; Graveley, B.R.; Coller, J.; Passmore, L.A. mRNA Deadenyl-ation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol. Cell 2018, 70, 1089–1100.e8. [Google Scholar] [CrossRef] [PubMed]
- Buschauer, R.; Matsuo, Y.; Sugiyama, T.; Chen, Y.-H.; Alhusaini, N.; Sweet, T.; Ikeuchi, K.; Cheng, J.; Matsuki, Y.; Nobuta, R.; et al. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 2020, 368, eaay6912. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.B.; Daly, M.J.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Ramaswami, G.; Zhang, R.; Piskol, R.; Keegan, L.P.; Deng, P.; O’Connell, M.A.; Li, J.B. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 2013, 10, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, B.; Wong, K.; Agam, A.; Goodson, M.; Keane, T.M.; Gan, X.; Nellåker, C.; Goodstadt, L.; Nicod, J.; Bhomra, A.; et al. Se-quence-based characterization of structural variation in the mouse genome. Nature 2011, 477, 326–329. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential analysis of RNA-seq incorporating quantification uncer-tainty. Nat. Methods 2017, 14, 687–690. [Google Scholar] [CrossRef]
- R. Core Team. R: A Language and Environment for Statistical Computing. 2013. Available online: https://cran.microsoft.com/snapshot/2014-09-08/web/packages/dplR/vignettes/xdate-dplR.pdf (accessed on 25 January 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. 2009. Available online: https://cran.r-project.org/package= ggplot2 (accessed on 10 January 2016).
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brachova, P.; Alvarez, N.S.; Christenson, L.K. Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes. Int. J. Mol. Sci. 2021, 22, 1191. https://doi.org/10.3390/ijms22031191
Brachova P, Alvarez NS, Christenson LK. Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes. International Journal of Molecular Sciences. 2021; 22(3):1191. https://doi.org/10.3390/ijms22031191
Chicago/Turabian StyleBrachova, Pavla, Nehemiah S. Alvarez, and Lane K. Christenson. 2021. "Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes" International Journal of Molecular Sciences 22, no. 3: 1191. https://doi.org/10.3390/ijms22031191
APA StyleBrachova, P., Alvarez, N. S., & Christenson, L. K. (2021). Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes. International Journal of Molecular Sciences, 22(3), 1191. https://doi.org/10.3390/ijms22031191







