A Missing Link: Engagements of Dendritic Cells in the Pathogenesis of SARS-CoV-2 Infections
Abstract
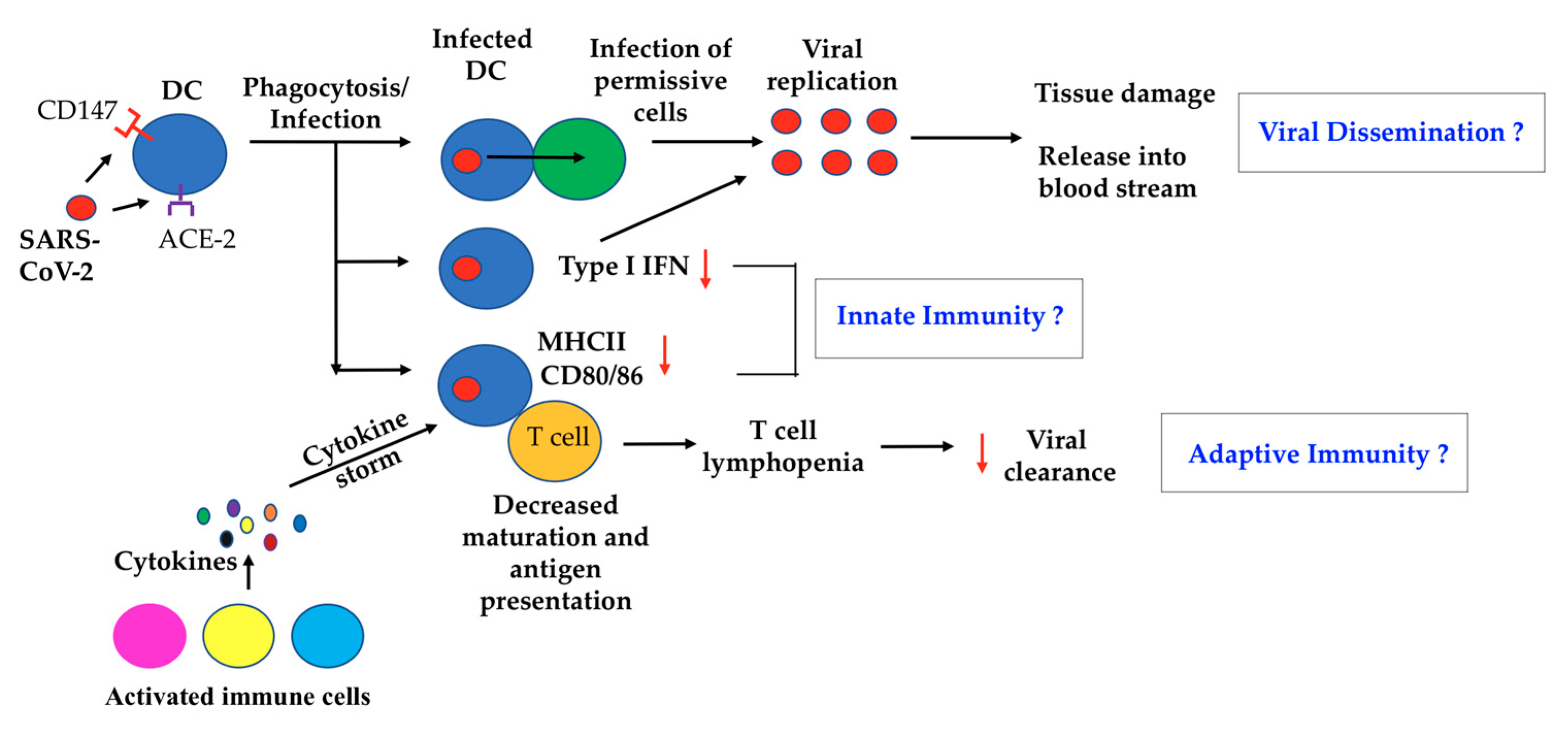
1. Introduction
2. A Possible Role of DC in SARS-CoV-2 Virus Dissemination
3. DC and Innate Immunity to COVID-19
4. DC and Adaptive Immunity toward COVID-19
5. Integrating DC Biology into Potential Treatments for COVID-19
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Jamilloux, Y.; Henry, T.; Belot, A.; Viel, S.; Fauter, M.; Jammal, T.; Walzer, T.; François, B.; Sève, P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020, 19, 102567. [Google Scholar] [CrossRef] [PubMed]
- Nile, S.H.; Nile, A.; Qiu, J.; Li, L.; Jia, X.; Kai, G. COVID-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020, 53, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Yi, Q.; Fan, S.; Lv, J.; Zhang, X.; Guo, L.; Lang, C.; Xiao, Q.; Xiao, K.; Yi, Z.; et al. Relationships among lymphocyte subsets, cytokines, and the pulmonary inflammation index in coronavirus (COVID-19) infected patients. Br. J. Haematol. 2020, 189, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.M.; Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem. Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 762–768. [Google Scholar] [CrossRef]
- Gayo, M.E.; Yu, X.G. Role of Dendritic Cells in Natural Immune Control of HIV-1 Infection. Front. Immunol. 2019, 10, 1306. [Google Scholar] [CrossRef]
- Soto, J.A.; Gálvez, N.M.S.; Andrade, C.A.; Pacheco, G.A.; Bohmwald, K.; Berrios, R.V.; Bueno, S.M.; Kalergis, A.M. The Role of Dendritic Cells During Infections Caused by Highly Prevalent Viruses. Front. Immunol. 2020, 11, 1513. [Google Scholar] [CrossRef]
- Patente, T.A.; Pinho, M.P.; Oliveira, A.A.; Evangelista, G.C.M.; Santos, B.P.C.; Barbuto, J.A.M. Human Dendritic Cells: Their Heterogeneity and Clinical Application Potential in Cancer Immunotherapy. Front. Immunol. 2018, 9, 3176. [Google Scholar] [CrossRef] [PubMed]
- Haeryfar, S.M. The importance of being a pDC in antiviral immunity: The IFN mission versus Ag presentation? Trends Immunol. 2005, 26, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, A.; Lutz, K.; Winheim, E.; Krug, A.B. What Makes a pDC: Recent Advances in Understanding Plasmacytoid DC Development and Heterogeneity. Front. Immunol. 2019, 10, 1222. [Google Scholar] [CrossRef] [PubMed]
- Swiecki, M.; Colonna, M. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol. Rev. 2010, 234, 142–162. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.L.; Abendroth, A.; Jones, C.; Nasr, N.; Turville, S. Viruses and Langerhans cells. Immunol. Cell Biol. 2010, 88, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.L.; Carbone, F.; Geijtenbeek, T.B. Langerhans cells and viral immunity. Eur. J. Immunol. 2008, 38, 2377–2385. [Google Scholar] [CrossRef]
- Huau, T.T.L.; Segura, E. Human in vivo-differentiated monocyte-derived dendritic cells. Semin. Cell Dev. Biol. 2019, 86, 44–49. [Google Scholar] [CrossRef]
- Menter, T.; Haslbauer, J.D.; Nienhold, R.; Savic, S.; Hopfer, H.; Deigendesch, N.; Frank, S.; Turek, D.; Willi, N.; Pargger, H.; et al. Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 2020, 77, 198–209. [Google Scholar] [CrossRef]
- Wichmann, D.; Sperhake, J.P.; Lütgehetmann, M.; Steurer, S.; Edler, C.; Heinemann, A.; Heinrich, F.; Mushumba, H.; Kniep, I.; Schröder, A.S.; et al. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann. Intern. Med. 2020, 173, 268–277. [Google Scholar] [CrossRef]
- Gu, J.; Gong, E.; Zhang, B.; Zheng, J.; Gao, Z.; Zhong, Y.; Zou, W.; Zhan, J.; Wang, S.; Xie, Z.; et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005, 202, 415–424. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Gobeaux, C.C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Donald, D. Dendritic cells and HIV-1 trans-infection. Viruses 2010, 2, 1704–1717. [Google Scholar]
- Zhou, Y.; Lu, K.; Pfefferle, S.; Bertram, S.; Glowacka, I.; Drosten, C.; Pöhlmann, S.; Simmons, G. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms. J. Virol. 2010, 84, 8753–8764. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Huang, Y.; Ganesh, L.; Leung, K.; Kong, W.P.; Schwartz, O.; Subbarao, K.; Nabel, G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004, 78, 5642–5650. [Google Scholar] [CrossRef] [PubMed]
- Amraei, R.; Yin, W.; Napoleon, M.A.; Suder, E.L.; Berrigan, J.; Zhao, Q.; Olejnik, J.; Chandler, K.B.; Xia, C.; Feldman, J.; et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells. bioRxiv 2020, 2020. [Google Scholar] [CrossRef]
- Donald, D.; Wu, L.; Bohks, S.M.; Ramani, K.V.N.; Unutmaz, D.; Hope, T.J. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 2003, 300, 1295–1297. [Google Scholar]
- Yang, D.; Chu, H.; Hou, Y.; Chai, Y.; Shuai, H.; Lee, A.C.; Zhang, X.; Wang, Y.; Hu, B.; Huang, X.; et al. Attenuated Interferon and Proinflammatory Response in SARS-CoV-2-Infected Human Dendritic Cells Is Associated With Viral Antagonism of STAT1 Phosphorylation. J. Infect. Dis. 2020, 222, 734–745. [Google Scholar] [CrossRef]
- Liu, L.; Wei, Q.; Nishiura, K.; Peng, J.; Wang, H.; Midkiff, C.; Alvarez, X.; Qin, C.; Lackner, A.; Chen, Z. Spatiotemporal interplay of severe acute respiratory syndrome coronavirus and respiratory mucosal cells drives viral dissemination in rhesus macaques. Mucosal Immunol. 2016, 9, 1089–1101. [Google Scholar] [CrossRef]
- Ragab, D.; Eldin, S.H.; Taeimah, M.; Khattab, R.; Salem, R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020, 11, 1446. [Google Scholar] [CrossRef]
- Perricone, C.; Triggianese, P.; Bartoloni, E.; Cafaro, G.; Bonifacio, A.F.; Bursi, R.; Perricone, R.; Gerli, R. The anti-viral facet of anti-rheumatic drugs: Lessons from COVID-19. J. Autoimmun. 2020, 111, 102468. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; Ary, M.P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Venet, F.; Huang, X.; Chung, C.S.; Chen, Y.; Ayala, A. Plasmacytoid dendritic cells control lung inflammation and monocyte recruitment in indirect acute lung injury in mice. Am. J. Pathol. 2010, 176, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Hu, W.; Yu, H.; Zhao, L.; Zhao, Y.; Zhao, X.; Xue, H.H.; Zhao, Y. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytom. Part A J. Int. Soc. Anal. Cytol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wang, Y.; Li, K.; Meyerholz, D.K.; Allamargot, C.; Perlman, S. SARS-CoV-2-induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Leitner, J.; Pfistershammer, G.K.; Majdic, O.; Zlabinger, G.; Steinberger, P. Interaction of antithymocyte globulins with dendritic cell antigens. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2011, 11, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Law, H.K.; Cheung, C.Y.; Ng, H.Y.; Sia, S.F.; Chan, Y.O.; Luk, W.; Nicholls, J.M.; Peiris, J.S.; Lau, Y.L. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 2005, 106, 2366–2374. [Google Scholar] [CrossRef]
- Chi, Y.; Ge, Y.; Wu, B.; Zhang, W.; Wu, T.; Wen, T.; Liu, J.; Guo, X.; Huang, C.; Jiao, Y.; et al. Serum Cytokine and Chemokine Profile in Relation to the Severity of Coronavirus Disease 2019 in China. J. Infect. Dis. 2020, 222, 746–754. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, C.; Li, J.; Yuan, J.; Wei, J.; Huang, F.; Wang, F.; Li, G.; Li, Y.; Xing, L.; et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020, 146, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, A.; Kuba, K.; Morita, M.; Chida, S.; Tezuka, H.; Hara, H.; Sasaki, T.; Ohteki, T.; Ranieri, V.M.; Santos, C.C.; et al. CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am. J. Respir. Crit. Care Med. 2013, 187, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Hayney, M.S.; Henriquez, K.M.; Barnet, J.H.; Ewers, T.; Champion, H.M.; Flannery, S.; Barrett, B. Serum IFN-γ-induced protein 10 (IP-10) as a biomarker for severity of acute respiratory infection in healthy adults. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2017, 90, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Hartl, D.; Etschmann, K.S.; Koller, B.; Hordijk, P.L.; Kuijpers, T.W.; Hoffmann, F.; Hector, A.; Eber, E.; Marcos, V.; Bittmann, I.; et al. Infiltrated neutrophils acquire novel chemokine receptor expression and chemokine responsiveness in chronic inflammatory lung diseases. J. Immunol. 2008, 181, 8053–8067. [Google Scholar] [CrossRef] [PubMed]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Colón, M.G.J.; Kechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020, 179, 104811. [Google Scholar] [CrossRef] [PubMed]
- Barragan, C.L.; Züst, R.; Weber, F.; Spiegel, M.; Lang, K.S.; Akira, S.; Thiel, V.; Ludewig, B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 2007, 109, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Scheuplein, V.A.; Seifried, J.; Malczyk, A.H.; Miller, L.; Höcker, L.; Alert, V.J.; Dolnik, O.; Zielecki, F.; Becker, B.; Spreitzer, I.; et al. High Secretion of Interferons by Human Plasmacytoid Dendritic Cells upon Recognition of Middle East Respiratory Syndrome Coronavirus. J. Virol. 2015, 89, 3859–3869. [Google Scholar] [CrossRef]
- Fung, S.Y.; Yuen, K.S.; Ye, Z.W.; Chan, C.P.; Jin, D.Y. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: Lessons from other pathogenic viruses. Emerg. Microbes Infect. 2020, 9, 558–570. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tsang, T.Y.O.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Magro, C.M.; Mulvey, J.J.; Laurence, J.; Sanders, S.; Crowson, A.N.; Grossman, M.; Harp, J.; Nuovo, G. The differing pathophysiologies that underlie COVID-19-associated perniosis and thrombotic retiform purpura: A case series. Br. J. Dermatol. 2021, 184, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Pan, J.; Tao, J.; Guo, D. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 2011, 42, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Xu, J.; Zhang, L.; Shu, T.; Wu, D.; Huang, M.; Ren, Y.; Li, X.; Geng, Q.; Xu, Y.; et al. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 2020, 63, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Broggi, A.; Ghosh, S.; Sposito, B.; Spreafico, R.; Balzarini, F.; Cascio, A.; Clementi, N.; Santis, M.; Mancini, N.; Granucci, F.; et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 2020, 369, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Gorden, K.B.; Gorski, K.S.; Gibson, S.J.; Kedl, R.M.; Kieper, W.C.; Qiu, X.; Tomai, M.A.; Alkan, S.S.; Vasilakos, J.P. Synthetic TLR agonists reveal functional differences between human TLR7 and TLR8. J. Immunol. 2005, 174, 1259–1268. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Cao, H.; Zhu, Y.; Zheng, J.; Zhou, H. Extraordinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response. Microbes Infect. 2013, 15, 88–95. [Google Scholar] [CrossRef]
- Eutimio, M.M.A.; Macías, L.C.; Palacios, P.R. Bioinformatic analysis and identification of single-stranded RNA sequences recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. Microbes Infect. 2020, 22, 226–229. [Google Scholar] [CrossRef]
- Schreibelt, G.; Tel, J.; Sliepen, K.H.; Ribas, B.D.; Figdor, C.G.; Adema, G.J.; Vries, I.J. Toll-like receptor expression and function in human dendritic cell subsets: Implications for dendritic cell-based anti-cancer immunotherapy. Cancer Immunol. Immunother. CII 2010, 59, 1573–1582. [Google Scholar] [CrossRef]
- Gabriele, L.; Fragale, A.; Romagnoli, G.; Parlato, S.; Lapenta, C.; Santini, S.M.; Ozato, K.; Capone, I. Type I IFN-dependent antibody response at the basis of sex dimorphism in the outcome of COVID-19. Cytokine Growth Factor Rev. 2020. [Google Scholar] [CrossRef]
- Cerrillo, S.I.; Landete, P.; Aldave, B.; Alonso, S.S.; Azofra, A.S.; Jiménez, M.A.; Ávalos, E.; Serna, A.A.; Santos, I.; Albero, M.T.; et al. Differential Redistribution of Activated Monocyte and Dendritic Cell Subsets to the Lung Associates with Severity of COVID-19. medRxiv 2020, 2020. [Google Scholar] [CrossRef]
- Mick, E.; Kamm, J.; Pisco, A.O.; Ratnasiri, K.; Babik, J.M.; Calfee, C.S.; Castañeda, G.; Risi, J.L.; Detweiler, A.M.; Hao, S.; et al. Upper airway gene expression differentiates COVID-19 from other acute respiratory illnesses and reveals suppression of innate immune responses by SARS-CoV-2. medRxiv 2020. [Google Scholar] [CrossRef]
- Mauro, G.; Scavone, C.; Rafaniello, C.; Rossi, F.; Capuano, A. SARS-Cov-2 infection: Response of human immune system and possible implications for the rapid test and treatment. Int. Immunopharmacol. 2020, 84, 106519. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Long, W.; Tu, M.; Chen, S.; Huang, Y.; Wang, S.; Zhou, W.; Chen, D.; Zhou, L.; Wang, M.; et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Infect. 2020, 81, 318–356. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wherry, J.E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020, 20, 529–536. [Google Scholar] [CrossRef]
- Law, H.K.; Cheung, C.Y.; Sia, S.F.; Chan, Y.O.; Peiris, J.S.; Lau, Y.L. Toll-like receptors, chemokine receptors and death receptor ligands responses in SARS coronavirus infected human monocyte derived dendritic cells. BMC Immunol. 2009, 10, 35. [Google Scholar] [CrossRef]
- Carter, M.J.; Fish, M.; Jennings, A.; Doores, K.J.; Wellman, P.; Seow, J.; Acors, S.; Graham, C.; Timms, E.; Kenny, J.; et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat. Med. 2020, 26, 1701–1707. [Google Scholar] [CrossRef]
- Parackova, Z.; Zentsova, I.; Bloomfield, M.; Vrabcova, P.; Smetanova, J.; Klocperk, A.; Mesežnikov, G.; Mendez, C.L.F.; Vymazal, T.; Sediva, A. Disharmonic Inflammatory Signatures in COVID-19: Augmented Neutrophils’ but Impaired Monocytes’ and Dendritic Cells’ Responsiveness. Cells 2020, 9, 2206. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Hill, T.; Li, K.; Peters, C.J.; Tseng, C.T. Severe acute respiratory syndrome (SARS) coronavirus-induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte-derived macrophages and dendritic cells. J. Virol. 2009, 83, 3039–3048. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal immunological analyses reveal inflammatory misfiring in severe COVID-19 patients. medRxiv 2020, 2020. [Google Scholar] [CrossRef]
- Wen, W.; Su, W.; Tang, H.; Le, W.; Zhang, X.; Zheng, Y.; Liu, X.; Xie, L.; Li, J.; Ye, J.; et al. Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discov. 2020, 6, 31. [Google Scholar] [CrossRef]
- Hegde, S.; Pahne, J.; Hess, S.S. Novel immunosuppressive properties of interleukin-6 in dendritic cells: Inhibition of NF-kappaB binding activity and CCR7 expression. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2004, 18, 1439–1441. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Xu, X.; Wei, H. Why tocilizumab could be an effective treatment for severe COVID-19? J. Transl. Med. 2020, 18, 164. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Han, M.; Li, T.; Sun, W.; Wang, D.; Fu, B.; Zhou, Y.; Zheng, X.; Yang, Y.; Li, X.; et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA 2020, 117, 10970–10975. [Google Scholar] [CrossRef] [PubMed]
- Strohbehn, G.W.; Heiss, B.L.; Rouhani, S.J.; Trujillo, J.A.; Yu, J.; Kacew, A.J.; Higgs, E.F.; Bloodworth, J.C.; Cabanov, A.; Wright, R.C.; et al. COVIDOSE: Low-dose tocilizumab in the treatment of Covid-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, D.; Li, Y.; Jin, L.; Shi, M.; Wang, M.; Zhou, X.; Wu, H.; Gao, G.F.; Wang, F.S. Longitudinal alteration of circulating dendritic cell subsets and its correlation with steroid treatment in patients with severe acute respiratory syndrome. Clin. Immunol. 2005, 116, 225–235. [Google Scholar] [CrossRef]
- Hamilton, J.A. GM-CSF in inflammation and autoimmunity. Trends Immunol. 2002, 23, 403–408. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, M.; Chen, X.; Montaner, L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020, 108, 17–41. [Google Scholar] [CrossRef]
- Bonaventura, A.; Vecchié, A.; Wang, T.S.; Lee, E.; Cremer, P.C.; Carey, B.; Rajendram, P.; Hudock, K.M.; Korbee, L.; Tassell, B.W.; et al. Targeting GM-CSF in COVID-19 Pneumonia: Rationale and Strategies. Front. Immunol. 2020, 11, 1625. [Google Scholar] [CrossRef]
- Bhattacharya, P.; Budnick, I.; Singh, M.; Thiruppathi, M.; Alharshawi, K.; Elshabrawy, H.; Holterman, M.J.; Prabhakar, B.S. Dual Role of GM-CSF as a Pro-Inflammatory and a Regulatory Cytokine: Implications for Immune Therapy. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2015, 35, 585–599. [Google Scholar] [CrossRef]
- Martinez, O.M.; Bridges, N.D.; Goldmuntz, E.; Pascual, V. The immune roadmap for understanding multi-system inflammatory syndrome in children: Opportunities and challenges. Nat. Med. 2020, 26, 1819–1824. [Google Scholar] [CrossRef]
- Luca, G.; Cavalli, G.; Campochiaro, C.; Torre, D.E.; Angelillo, P.; Tomelleri, A.; Boffini, N.; Tentori, S.; Mette, F.; Farina, N.; et al. GM-CSF blockade with mavrilimumab in severe COVID-19 pneumonia and systemic hyperinflammation: A single-centre, prospective cohort study. Lancet Rheumatol. 2020, 2, e465–e473. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, V.; Shannon, C.P.; Wei, X.S.; Xiang, X.; Wang, X.; Wang, Z.H.; Tebbutt, S.J.; Kollmann, T.R.; Fish, E.N. Interferon-α2b Treatment for COVID-19. Front. Immunol. 2020, 11, 1061. [Google Scholar] [CrossRef] [PubMed]
- Scarfò, L.; Chatzikonstantinou, T.; Rigolin, G.M.; Quaresmini, G.; Motta, M.; Vitale, C.; Marco, G.J.A.; Rivas, H.J.Á.; Mirás, F.; Baile, M.; et al. COVID-19 severity and mortality in patients with chronic lymphocytic leukemia: A joint study by ERIC, the European Research Initiative on CLL, and CLL Campus. Leukemia 2020, 34, 2354–2363. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Agrawal, S.; Gupta, S. Role of Dendritic Cells in Inflammation and Loss of Tolerance in the Elderly. Front. Immunol. 2017, 8, 896. [Google Scholar] [CrossRef]
- Zhao, J.; Lenane, W.C.; Zhao, J.; Fleming, E.; Lane, T.E.; Cray, P.B., Jr.; Perlman, S. Intranasal treatment with poly(I•C) protects aged mice from lethal respiratory virus infections. J. Virol. 2012, 86, 11416–11424. [Google Scholar] [CrossRef]
- Julius, G.A.; Harning, E.K.; Abernathy, L.M.; Yung, R.L. Impaired dendritic cell function in aging leads to defective antitumor immunity. Cancer Res. 2008, 68, 6341–6349. [Google Scholar] [CrossRef]
- Gupta, A.; Chiang, C.K. Prostaglandin D(2) as a mediator of lymphopenia and a therapeutic target in COVID-19 disease. Med. Hypotheses 2020, 143, 110122. [Google Scholar] [CrossRef]
- Yao, Z.; Zheng, Z.; Wu, K.; Junhua, Z. Immune environment modulation in pneumonia patients caused by coronavirus: SARS-CoV, MERS-CoV and SARS-CoV-2. Aging 2020, 12, 7639–7651. [Google Scholar] [CrossRef]
- Borges, R.C.; Hohmann, M.S.; Borghi, S.M. Dendritic cells in COVID-19 immunopathogenesis: Insights for a possible role in determining disease outcome. Int. Rev. Immunol. 2020, 1–18. [Google Scholar] [CrossRef]
- Kielian, M. Enhancing host cell infection by SARS-CoV-2. Science 2020, 370, 765–766. [Google Scholar] [CrossRef]
- Vinson, V. How SARS-CoV-2 binds to human cells. Science 2020, 367, 1438–1439. [Google Scholar] [CrossRef]
- Guo, H.F.; Kooi, V.C.W. Neuropilin Functions as an Essential Cell Surface Receptor. J. Biol. Chem. 2015, 290, 29120–29126. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Plágaro, A.C.; Shoemark, D.K.; Gracia, S.L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Castelvetri, C.L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Alamri, A.; Rahman, R.; Zhang, M.; Alamri, A.; Gounni, A.S.; Kung, S.K.P. Semaphorin-3E Produced by Immature Dendritic Cells Regulates Activated Natural Killer Cells Migration. Front. Immunol. 2018, 9, 1005. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Jiang, X.; Zhao, Y.; Yang, L.; Chen, L.; Dong, M.; Luan, Z.; Yan, C.; Jiao, J.; et al. Preliminary evaluation of the safety and efficacy of oral human antimicrobial peptide LL-37 in the treatment of patients of COVID-19, a small-scale, single-arm, exploratory safety study. medRxiv 2020, 2020. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection results in alterations of the kynurenine pathway and fatty acid metabolism that correlate with IL-6 levels and renal status. medRxiv 2020, 2020. [Google Scholar] [CrossRef]
- Stone, T.W. Inhibitors of the kynurenine pathway. Eur. J. Med. Chem. 2000, 35, 179–186. [Google Scholar] [CrossRef]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of chloroquine on viral infections: An old drug against today’s diseases. Lancet Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef]
- Tarek, M.; Savarino, A. Pharmacokinetic bases of the hydroxychloroquine response in COVID-19: Implications for therapy and prevention. medRxiv 2020, 2020. [Google Scholar] [CrossRef]
| DC Subset | Potential Roles in Viral Infections |
|---|---|
| cDC1 | |
| cDC2 | |
| pDC |
|
| LC | |
| moDC |
| Treatment | Availability | Mechanisms of Action | Potential Target | Evidences |
|---|---|---|---|---|
| Anti-S protein | Available | Prevents viral entry, neutralizes virus | DC & Other cells | Prevents trans-infection of SARS-CoV-1 by DC |
| Anti-IL-6 | Available | IL-6 inhibitor, blocks cytokine storm | DC & Other cells | Increases the capacity of DC to activate T-cell responses against SARS-CoV-1 |
| Tocilizumab/Sarilumab | Available | IL-6 receptor inhibitor, blocks cytokine storm | DC & Other cells | Expected to have same effect as anti-IL-6 on DC |
| Corticosteroids | Available | lower the levels of pDC, cDC, CD4 and CD8+ T cells | DC & Other cells | Limit DC levels, may further delay pDC response |
| Chloroquine/hydroxychloroquine | Available | Not known, change the pH of endosomes, prevent viral entry, transport, and post-entry events | DC & Other cells | Increases antigen presentation by DC |
| Mavrilimumab (anti-GMCSFRα) | Available | Decrease leukocyte activation, ameliorate immunosuppression | DC, macrophages, NK, and T cells | GM-CSF promotes tolerogenic DC. Downstream effects resemble multi-system inflammatory syndrome |
| Type I IFN | Available | Compensate for insufficient type I IFN production by DC | Cells that are permissive to SARS-CoV-2 infection. | Production by pDC is suppressed, is lowest in severest cases. |
| TLR3 Agonist (poly I:C) | Potential | Stimulate type I IFN production by DC | DC & Other cells | Prophylactic intranasal use is 100% protective for SARS-CoV-1-infected mice |
| Anti-NRP | Potential | Inhibit viral entry | DC, respiratory and olfactory epithelia | Five out of 6 autopsy samples are positive for both S protein and NRP1 |
| LL-37 | Potential | Induces inflammasome activation, IL-1β and IL-18 | DC & Other cells | Induces DC maturation and release of type I IFN by APCs |
| Anti- Kynurenine pathway (nicotinylalanine/meta-nitrobenzoylalanine) | Potential | Strengthen adaptive immunity | DC and CD8 T cells | Pathway drives immunosuppressive activity of DC and CD8 T cell |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alamri, A.; Fisk, D.; Upreti, D.; Kung, S.K.P. A Missing Link: Engagements of Dendritic Cells in the Pathogenesis of SARS-CoV-2 Infections. Int. J. Mol. Sci. 2021, 22, 1118. https://doi.org/10.3390/ijms22031118
Alamri A, Fisk D, Upreti D, Kung SKP. A Missing Link: Engagements of Dendritic Cells in the Pathogenesis of SARS-CoV-2 Infections. International Journal of Molecular Sciences. 2021; 22(3):1118. https://doi.org/10.3390/ijms22031118
Chicago/Turabian StyleAlamri, Abdulaziz, Derek Fisk, Deepak Upreti, and Sam K. P. Kung. 2021. "A Missing Link: Engagements of Dendritic Cells in the Pathogenesis of SARS-CoV-2 Infections" International Journal of Molecular Sciences 22, no. 3: 1118. https://doi.org/10.3390/ijms22031118
APA StyleAlamri, A., Fisk, D., Upreti, D., & Kung, S. K. P. (2021). A Missing Link: Engagements of Dendritic Cells in the Pathogenesis of SARS-CoV-2 Infections. International Journal of Molecular Sciences, 22(3), 1118. https://doi.org/10.3390/ijms22031118





