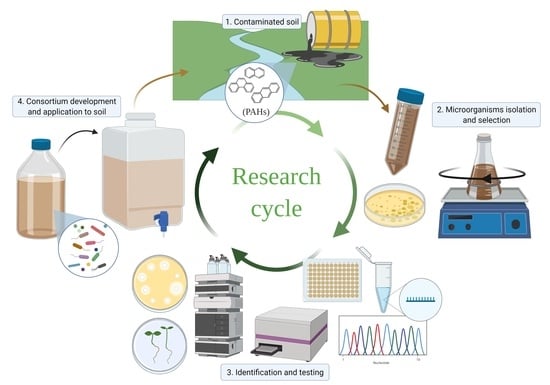
Development of an Autochthonous Microbial Consortium for Enhanced Bioremediation of PAH-Contaminated Soil
Abstract
1. Introduction
2. Results
2.1. Bacteria Isolation and Identification
2.2. Detection of PAH-Degradation-Related Gene
2.3. Effectiveness of In Vitro Hydrocarbon Degradation
2.4. In Situ Application of Microbiological Consortium
3. Discussion
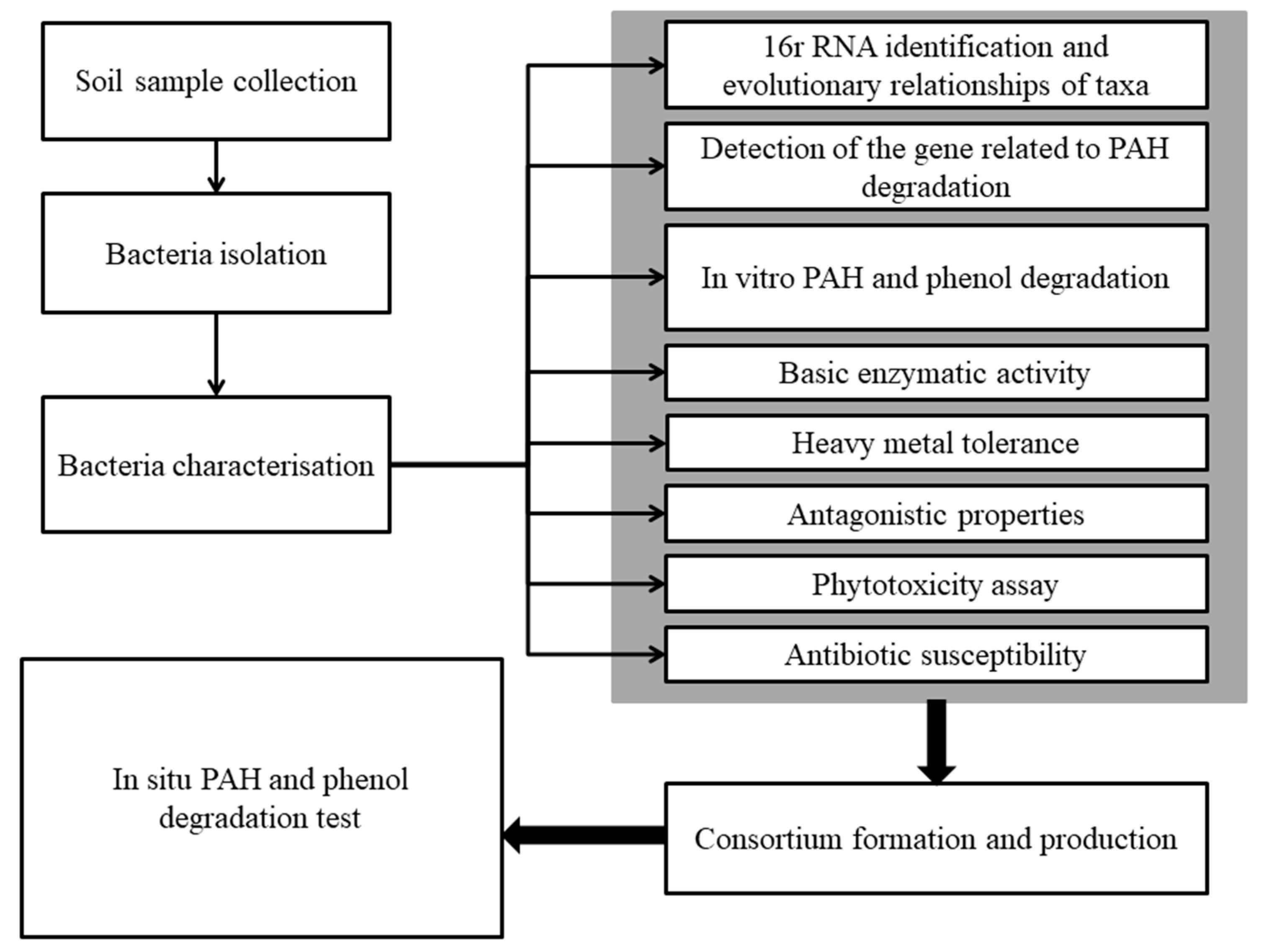
4. Materials and Methods
4.1. Soil Samples and Contamination Measurements
4.2. Bacteria Isolation and Characterization
4.3. Detection of PAH-Degradation-Related Gene
4.4. Hydrocarbons Degradation In Vitro Test
4.5. Consortium Development and Application to Soil
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tang, L.; Tang, X.-Y.; Zhu, Y.-G.; Zheng, M.-H.; Miao, Q.-L. Contamination of polycyclic aromatic hydrocarbons (PAHs) in urban soils in Beijing, China. Environ. Int. 2005, 31, 822–828. [Google Scholar] [CrossRef]
- Jacques, R.J.S.; Okeke, B.C.; Bento, F.M.; Teixeira, A.S.; Peralba, M.C.R.; Camargo, F.A.O. Microbial consortium bioaugmentation of a polycyclic aromatic hydrocarbons contaminated soil. Bioresour. Technol. 2008, 99, 2637–2643. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Thavamani, P.; Venkateswarlu, K.; Lee, Y.B.; Naidu, R.; Megharaj, M. Remediation approaches for polycyclic aromatic hydrocarbons (PAHs) contaminated soils: Technological constraints, emerging trends and future directions. Chemosphere 2017, 168, 944–968. [Google Scholar] [CrossRef]
- Canet, R.; Birnstingl, J.G.; Malcolm, D.G.; Lopez-Real, J.M.; Beck, A.J. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by native microflora and combinations of white-rot fungi in a coal-tar contaminated soil. Bioresour. Technol. 2001, 76, 113–117. [Google Scholar] [CrossRef]
- Smith, M.J.; Flowers, T.H.; Duncan, H.J.; Alder, J. Effects of polycyclic aromatic hydrocarbons on germination and subsequent growth of grasses and legumes in freshly contaminated soil and soil with aged PAHs residues. Environ. Pollut. 2006, 141, 519–525. [Google Scholar] [CrossRef]
- Li, X.; Qu, C.; Bian, Y.; Gu, C.; Jiang, X.; Song, Y. New insights into the responses of soil microorganisms to polycyclic aromatic hydrocarbon stress by combining enzyme activity and sequencing analysis with metabolomics. Environ. Pollut. 2019, 255, 113312. [Google Scholar] [CrossRef]
- Thavamani, P.; Malik, S.; Beer, M.; Megharaj, M.; Naidu, R. Microbial activity and diversity in long-term mixed contaminated soils with respect to polyaromatic hydrocarbons and heavy metals. J. Environ. Manag. 2012, 99, 10–17. [Google Scholar] [CrossRef]
- Samsøe-Petersen, L.; Larsen, E.H.; Larsen, P.B.; Bruun, P. Uptake of Trace Elements and PAHs by Fruit and Vegetables from Contaminated Soils. Environ. Sci. Technol. 2002, 36, 3057–3063. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Lau, E.V.; Ng, H.K. Remediation of soils contaminated with polycyclic aromatic hydrocarbons (PAHs). J. Hazard. Mater. 2009, 172, 532–549. [Google Scholar] [CrossRef]
- Fernández-Luqueño, F.; Valenzuela-Encinas, C.; Marsch, R.; Martínez-Suárez, C.; Vázquez-Núñez, E.; Dendooven, L. Microbial communities to mitigate contamination of PAHs in soil—Possibilities and challenges: A review. Environ. Sci. Pollut. Res. 2011, 18, 12–30. [Google Scholar] [CrossRef]
- Wong, J.W.C.; Lai, K.M.; Wan, C.K.; Ma, K.K.; Fang, M. Isolation and Optimization of PAH-Degradative Bacteria from Contaminated Soil for PAHs Bioremediation. Water Air Soil Pollut. 2002, 139, 1–13. [Google Scholar] [CrossRef]
- Hamdi, H.; Benzarti, S.; Manusadžianas, L.; Aoyama, I.; Jedidi, N. Bioaugmentation and biostimulation effects on PAH dissipation and soil ecotoxicity under controlled conditions. Soil Biol. Biochem. 2007, 39, 1926–1935. [Google Scholar] [CrossRef]
- Madueño, L.; Coppotelli, B.M.; Alvarez, H.M.; Morelli, I.S. Isolation and characterization of indigenous soil bacteria for bioaugmentation of PAH contaminated soil of semiarid Patagonia, Argentina. Int. Biodeterior. Biodegrad. 2011, 65, 345–351. [Google Scholar] [CrossRef]
- Trzesicka-Mlynarz, D.; Ward, O.P. Degradation of polycyclic aromatic hydrocarbons (PAHs) by a mixed culture and its component pure cultures, obtained from PAH-contaminated soil. Can. J. Microbiol. 1995, 41, 470–476. [Google Scholar] [CrossRef]
- Molina, M.C.; González, N.; Bautista, L.F.; Sanz, R.; Simarro, R.; Sánchez, I.; Sanz, J.L. Isolation and genetic identification of PAH degrading bacteria from a microbial consortium. Biodegradation 2009, 20, 789–800. [Google Scholar] [CrossRef]
- Haritash, A.K.; Kaushik, C.P. Biodegradation aspects of Polycyclic Aromatic Hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef]
- Lu, X.-Y.; Zhang, T.; Fang, H.H.-P. Bacteria-mediated PAH degradation in soil and sediment. Appl. Microbiol. Biotechnol. 2011, 89, 1357–1371. [Google Scholar] [CrossRef]
- Lin, Y.; Cai, L.X. PAH-degrading microbial consortium and its pyrene-degrading plasmids from mangrove sediment samples in Huian, China. Mar. Pollut. Bull. 2008, 57, 703–706. [Google Scholar] [CrossRef]
- Isaac, P.; Martínez, F.L.; Bourguignon, N.; Sánchez, L.A.; Ferrero, M.A. Improved PAHs removal performance by a defined bacterial consortium of indigenous Pseudomonas and actinobacteria from Patagonia, Argentina. Int. Biodeterior. Biodegrad. 2015, 101, 23–31. [Google Scholar] [CrossRef]
- Zafra, G.; Absalón, Á.E.; Cuevas, M.D.C.; Cortés-Espinosa, D.V. Isolation and Selection of a Highly Tolerant Microbial Consortium with Potential for PAH Biodegradation from Heavy Crude Oil-Contaminated Soils. Water Air Soil Pollut. 2014, 225, 1–18. [Google Scholar] [CrossRef]
- Laothamteep, N.; Kawano, H.; Vejarano, F.; Suzuki-Minakuchi, C.; Shintani, M.; Nojiri, H.; Pinyakong, O. Effects of environmental factors and coexisting substrates on PAH degradation and transcriptomic responses of the defined bacterial consortium OPK. Environ. Pollut. 2021, 277, 116769. [Google Scholar] [CrossRef]
- Kumari, S.; Regar, R.K.; Manickam, N. Improved polycyclic aromatic hydrocarbon degradation in a crude oil by individual and a consortium of bacteria. Bioresour. Technol. 2018, 254, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, F.; Teijiz, I.; Deive, F.J.; Sanromán, M.A. Efficient PAHs biodegradation by a bacterial consortium at flask and bioreactor scale. Bioresour. Technol. 2012, 119, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Tauler, M.; Grifoll, M. Bacterial PAH degradation in marine and terrestrial habitats. Curr. Opin. Biotechnol. 2015, 33, 95–102. [Google Scholar] [CrossRef]
- Liang, C.; Huang, Y.; Wang, H. pahE, a Functional Marker Gene for Polycyclic Aromatic Hydrocarbon-Degrading Bacteria. Appl. Environ. Microbiol. 2019, 85, 1–17. [Google Scholar] [CrossRef]
- John, R.C.; Essien, J.P.; Akpan, S.B.; Okpokwasili, G.C. Polycyclic aromatic hydrocarbon-degrading bacteria from aviation fuel spill site at Ibeno, Nigeria. Bull. Environ. Contam. Toxicol. 2012, 88, 1014–1019. [Google Scholar] [CrossRef]
- Khalid, A.H.; Jin, H.J. Heavy metal resistance of bacteria and its impact on the production of antioxidant enzymes. African J. Microbiol. Res. 2013, 7, 2288–2296. [Google Scholar] [CrossRef]
- Balachandran, C.; Duraipandiyan, V.; Balakrishna, K.; Ignacimuthu, S. Bioresource Technology Petroleum and polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalene metabolism in Streptomyces sp. (ERI-CPDA-1) isolated from oil contaminated soil. Bioresour. Technol. 2012, 112, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Deeb, B.E.; Altalhi, A.D. Degradative Plasmid and Heavy Metal Resistance Plasmid Naturally Coexist in Phenol and Cyanide Assimilating Bacteria. Am. J. Biochem. Biotechnol. 2009, 5, 84–93. [Google Scholar] [CrossRef]
- Song, J.; Guo, Z.; Xiao, X.; Miao, X.; Wang, F. Environmental availability and profile characteristics of arsenic, cadmium, lead and zinc in metal-contaminated vegetable soils. Trans. Nonferrous Met. Soc. China 2009, 19, 765–772. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Wang, Y.; Luo, G.; Chen, X.; Yang, X.; Hall, M.H.P.; Guo, R.; Wang, H.; Cui, J.; et al. Heavy metal contamination of urban soil in an old industrial city (Shenyang) in Northeast China. Geoderma 2013, 192, 50–58. [Google Scholar] [CrossRef]
- Pages, D.; Rose, J.; Conrod, S.; Cuine, S.; Carrier, P.; Heulin, T.; Achouak, W. Heavy metal tolerance in Stenotrophomonas maltophilia. PLoS ONE 2008, 3, e1539. [Google Scholar] [CrossRef]
- Bhojiya, A.A.; Joshi, H. Heavy Metal Tolerance Pattern of Pseudomonas putida Isolated from Heavy Metal Contaminated Soil of Zawar, Udaipur (India). Int. J. Innov. Knowl. Concepts 2015, 1, 17–21. [Google Scholar]
- Tomova, I.; Stoilova-Disheva, M.; Lazarkevich, I.; Vasileva-Tonkova, E. Antimicrobial activity and resistance to heavy metals and antibiotics of heterotrophic bacteria isolated from sediment and soil samples collected from two Antarctic islands. Front. Life Sci. 2015, 8, 348–357. [Google Scholar] [CrossRef]
- Khan, Z.; Rehman, A.; Hussain, S.Z.; Nisar, M.A.; Zulfiqar, S.; Shakoori, A.R. Cadmium resistance and uptake by bacterium, Salmonella enterica 43C, isolated from industrial effluent. AMB Express 2016, 6, 54. [Google Scholar] [CrossRef]
- Hassen, A.; Saidi, N.; Cherif, M.; Boudabous, A. Resistance of environmental bacteria to heavy metals. Bioresour. Technol. 1998, 64, 7–15. [Google Scholar] [CrossRef]
- Yong, X.; Chen, Y.; Liu, W.; Xu, L.; Zhou, J.; Wang, S.; Chen, P.; Ouyang, P.; Zheng, T. Enhanced cadmium resistance and accumulation in Pseudomonas putida KT2440 expressing the phytochelatin synthase gene of Schizosaccharomyces pombe. Lett. Appl. Microbiol. 2014, 58, 255–261. [Google Scholar] [CrossRef]
- Haroun, A.; KK, K.; Alhaji, I.; Magaji, Y.; Oaikhena, E. Evaluation of Heavy Metal Tolerance Level (MIC) and Bioremediation Potentials of Pseudomonas aeruginosa Isolated from Makera-Kakuri Industrial Drain in Kaduna, Nigeria. Eur. J. Exp. Biol. 2017, 7, 3–6. [Google Scholar] [CrossRef]
- Mosbah, R.; Sahmoune, M.N. Biosorption of heavy metals by Streptomyces species—An overview. Cent. Eur. J. Chem. 2013, 11, 1412–1422. [Google Scholar] [CrossRef]
- Hamedi, J.; Dehhaghi, M.; Mohammdipanah, F. Isolation of extremely heavy metal resistant strains of rare actinomycetes from high metal content soils in Iran. Int. J. Environ. Res. 2015, 9, 475–480. [Google Scholar]
- Baltazar, M.D.P.G.; Gracioso, L.H.; Avanzi, I.R.; Karolski, B.; Tenório, J.A.S.; Do Nascimento, C.A.O.; Perpetuo, E.A. Copper biosorption by Rhodococcus erythropolis isolated from the Sossego Mine—PA—Brazil. J. Mater. Res. Technol. 2019, 8, 475–483. [Google Scholar] [CrossRef]
- Learman, D.R.; Ahmad, Z.; Brookshier, A.; Henson, M.W.; Hewitt, V.; Lis, A.; Morrison, C.; Robinson, A.; Todaro, E.; Wologo, E.; et al. Comparative genomics of 16 Microbacterium spp. that tolerate multiple heavy metals and antibiotics. PeerJ 2019, 6, e6258. [Google Scholar] [CrossRef]
- Augustyniak, A.; Cendrowski, K.; Nawrotek, P.; Barylak, M.; Mijowska, E. Investigating the Interaction Between Streptomyces sp. and Titania/Silica Nanospheres. Water Air Soil Pollut. 2016, 227, 230. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Wojciuk, B.; Roszak, M.; Łubowska, N.; Błażejczak, P.; Jursa-Kulesza, J.; Rakoczy, R.; Masiuk, H.; Dołęgowska, B. Environmental Phage-Based Cocktail and Antibiotic Combination Effects on Acinetobacter baumannii Biofilm in a Human Urine Model. Microb. Drug Resist. 2020, 27, 25–35. [Google Scholar] [CrossRef]
- Berg, J.; Tom-Petersen, A.; Nybroe, O. Copper amendment of agricultural soil selects for bacterial antibiotic resistance in the field. Lett. Appl. Microbiol. 2005, 40, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]
- Walsh, F.; Duffy, B. The Culturable Soil Antibiotic Resistome: A Community of Multi-Drug Resistant Bacteria. PLoS ONE 2013, 8, e65567. [Google Scholar] [CrossRef]
- Máthé, I.; Benedek, T.; Táncsics, A.; Palatinszky, M.; Lányi, S.; Márialigeti, K. Diversity, activity, antibiotic and heavy metal resistance of bacteria from petroleum hydrocarbon contaminated soils located in Harghita County (Romania). Int. Biodeterior. Biodegradation 2012, 73, 41–49. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Zhao, Z.; Chen, J.; Lu, H.; Liu, G.; Zhou, J.; Guan, X. PAHs accelerate the propagation of antibiotic resistance genes in coastal water microbial community. Environ. Pollut. 2017, 231, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; He, R.; Yuan, K.; Chen, E.; Lin, L.; Chen, X.; Sha, S.; Zhong, J.; Lin, L.; Yang, L.; et al. Polycyclic aromatic hydrocarbons (PAHs) enriching antibiotic resistance genes (ARGs) in the soils. Environ. Pollut. 2017, 220, 1005–1013. [Google Scholar] [CrossRef]
- Augustyniak, A.; Grygorcewicz, B.; Nawrotek, P. Isolation of multidrug resistant coliforms and their bacteriophages from swine slurry. Turkish J. Vet. Anim. Sci. 2018, 42, 319–325. [Google Scholar] [CrossRef]
- Maurya, A.P.; Rajkumari, J.; Pandey, P. Enrichment of antibiotic resistance genes (ARGs) in polyaromatic hydrocarbon–contaminated soils: A major challenge for environmental health. Environ. Sci. Pollut. Res. 2021, 28, 12178–12189. [Google Scholar] [CrossRef]
- Palleroni, N.J. Pseudomonas. Bergey’s Man. Syst. Archaea Bact. 2015, 1. [Google Scholar]
- Marion-Sanchez, K.; Pailla, K.; Olive, C.; Le Coutour, X.; Derancourt, C. Achromobacter spp. healthcare associated infections in the French West Indies: A longitudinal study from 2006 to 2016. BMC Infect. Dis. 2019, 19, 795. [Google Scholar] [CrossRef]
- Kämpfer, P. Streptomyces. Bergey’s Man. Syst. Archaea Bact. 2015, 1–414. [Google Scholar]
- Zafra, G.; Absalón, Á.E.; Anducho-Reyes, M.Á.; Fernandez, F.J.; Cortés-Espinosa, D.V. Construction of PAH-degrading mixed microbial consortia by induced selection in soil. Chemosphere 2017, 172, 120–126. [Google Scholar] [CrossRef]
- Polianskaia, L.M.; Kozhevin, P.A.; Zviagintsev, D.G. Dynamics of populations of microbial antagonists in nonsterile soil. Mikrobiologiia 1983, 52, 145–148. [Google Scholar]
- Kerr, J.R. Bacterial inhibition of fungal growth and pathogenicity. Microb. Ecol. Health Dis. 1999, 11, 129–142. [Google Scholar] [CrossRef]
- Sivasakthi, S.; Usharani, G.; Saranraj, P. Biocontrol potentiality of plant growth promoting bacteria (PGPR)—Pseudomonas fluorescens and Bacillus subtilis: A review. Afr. J. Agric. Res. 2014, 9, 1265–1277. [Google Scholar]
- Song, X.; Xu, Y.; Li, G.; Zhang, Y.; Huang, T.; Hu, Z. Isolation, characterization of Rhodococcus sp. P14 capable of degrading high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons. Mar. Pollut. Bull. 2011, 62, 2122–2128. [Google Scholar] [CrossRef]
- Leneva, N.A.; Kolomytseva, M.P.; Baskunov, B.P.; Golovleva, L.A. Phenanthrene and anthracene degradation by microorganisms of the genus Rhodococcus. Appl. Biochem. Microbiol. 2009, 45, 169–175. [Google Scholar] [CrossRef]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nešvera, J.; Křen, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef]
- Favier, L.; Rusu, L.; Simion, A.I.; Hlihor, R.M.; Pacala, M.L.; Augustyniak, A. Efficient degradation of clofibric acid by heterogeneous photocatalytic oxidation process. Environ. Eng. Manag. J. 2019, 18, 1683–1692. [Google Scholar] [CrossRef]
- Mikesková, H.; Novotný, Č.; Svobodová, K. Interspecific interactions in mixed microbial cultures in a biodegradation perspective. Appl. Microbiol. Biotechnol. 2012, 95, 861–870. [Google Scholar] [CrossRef]
- Eaton, R.W. Trans-O-Hydroxybenzylidenepyruvate Hydratase-Aldolase As a Biocatalyst. Appl. Environ. Microbiol. 2000, 66, 2668–2672. [Google Scholar] [CrossRef][Green Version]
- Fulekar, M. Microbial degradation of petrochemical waste-polycyclic aromatic hydrocarbons. Bioresour. Bioprocess. 2017, 4. [Google Scholar] [CrossRef]
- Suja, F.; Rahim, F.; Taha, M.R.; Hambali, N.; Rizal Razali, M.; Khalid, A.; Hamzah, A. Effects of local microbial bioaugmentation and biostimulation on the bioremediation of total petroleum hydrocarbons (TPH) in crude oil contaminated soil based on laboratory and field observations. Int. Biodeterior. Biodegrad. 2014, 90, 115–122. [Google Scholar] [CrossRef]
- Vaidya, S.; Devpura, N.; Jain, K.; Madamwar, D. Degradation of chrysene by enriched bacterial consortium. Front. Microbiol. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Jiménez, B.; Pesciaroli, C.; Maza-Márquez, P.; López-Martínez, S.; Vílchez-Quero, J.L.; Zafra-Gómez, A. Biodegradation of methyl and butylparaben by bacterial strains isolated from amended and non-amended agricultural soil. Identification, behavior and enzyme activities of microorganisms. J. Environ. Manag. 2019, 245, 245–254. [Google Scholar] [CrossRef]
- Briceño, G.; Lamilla, C.; Leiva, B.; Levio, M.; Donoso-Piñol, P.; Schalchli, H.; Gallardo, F.; Diez, M.C. Pesticide-tolerant bacteria isolated from a biopurification system to remove commonly used pesticides to protect water resources. PLoS ONE 2020, 15, e0234865. [Google Scholar] [CrossRef]
- You, J.; Lin, S.; Jiang, T. Origins and Evolution of the α-L-Fucosidases: From Bacteria to Metazoans. Front. Microbiol. 2019, 10, 1756. [Google Scholar] [CrossRef] [PubMed]
- Saltiene, Z.; Brukstiene, D.; Ruzgyte, A. Contamination of Soil by Polycyclic Aromatic Hydrocarbons in Some Urban Areas. Polycycl. Aromat. Compd. 2002, 22, 23–35. [Google Scholar] [CrossRef]
- Motelay-Massei, A.; Ollivon, D.; Garban, B.; Teil, M.J.; Blanchard, M.; Chevreuil, M. Distribution and spatial trends of PAHs and PCBs in soils in the Seine River basin, France. Chemosphere 2004, 55, 555–565. [Google Scholar] [CrossRef]
- Maliszewska-Kordybach, B.; Smreczak, B.; Klimkowicz-Pawlas, A. Concentrations, sources, and spatial distribution of individual polycyclic aromatic hydrocarbons (PAHs) in agricultural soils in the Eastern part of the EU: Poland as a case study. Sci. Total Environ. 2009, 407, 3746–3753. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Wang, R.; Hou, H.; Du, X.; Fan, S.; Liu, J.; Dai, J. Effects of pollution sources and soil properties on distribution of polycyclic aromatic hydrocarbons and risk assessment. Sci. Total Environ. 2013, 463–464, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Wang, M.; Chen, W. Spatial Analysis of PAHs in Soils along an Urban–Suburban–Rural Gradient: Scale effect, distribution patterns, diffusion and influencing factors. Sci. Rep. 2016, 6, 37185. [Google Scholar] [CrossRef]
- Li, Y.T.; Li, F.B.; Chen, J.J.; Yang, G.Y.; Wan, H.F.; Zhang, T.B.; Zeng, X.D.; Liu, J.M. The concentrations, distribution and sources of PAHs in agricultural soils and vegetables from Shunde, Guangdong, China. Environ. Monit. Assess. 2008, 139, 61–76. [Google Scholar] [CrossRef]
- Sivasubramanian, S.; Namasivayam, S.K.R. Phenol degradation studies using microbial consortium isolated from environmental sources. J. Environ. Chem. Eng. 2015, 3, 243–252. [Google Scholar] [CrossRef]
- Breedveld, G.D.; Sparrevik, M. Nutrient-limited biodegradation of PAH in various soil strata at a creosote contaminated site. Biodegradation 2000, 11, 391–399. [Google Scholar] [CrossRef]
- Correa-García, S.; Pande, P.; Séguin, A.; St-Arnaud, M.; Yergeau, E. Rhizoremediation of petroleum hydrocarbons: A model system for plant microbiome manipulation. Microb. Biotechnol. 2018, 11, 819–832. [Google Scholar] [CrossRef] [PubMed]
| Strain | Strain Genera | Gene | |||
|---|---|---|---|---|---|
| pahE1 | pahE2 | pahE3 | pahE4 | ||
| 11 | Microbacterium sp. | - | - | - | + |
| 17 | Pseudomonas sp. | + | - | + | + |
| 19 | Streptomyces sp. | + | - | - | - |
| 20 | Achromobacter sp. | - | - | - | + |
| 22 | Cupriavidus sp. | + | + | + | + |
| 24 | Rhodococcus sp. | - | - | + | + |
| 26 | Arthrobacter sp. | - | - | + | + |
| 27 | Arthrobacter sp. | - | - | + | + |
| 29 | Arthrobacter sp. | + | + | + | + |
| 36 | Pseudomonas sp. | + | + | + | + |
| 38 | Pseudomonas sp. | + | + | + | + |
| 40 | Rhodococcus sp. | + | - | + | + |
| 41 | Pseudomonas sp. | + | - | + | + |
| 44 | Rhodococcus sp. | + | - | + | + |
| Hydrocarbon | Substance Content in Comparison to the Initial Calculated Content [%] | |||
|---|---|---|---|---|
| 1 Day of Incubation | 8 Days of Incubation | |||
| Control Sample (without Consortium) | Tested Sample (Added Consortium) | Control Sample (without Consortium) | Tested Sample (Added Consortium) | |
| Phenol | 73.06 | 62.82 | 65.29 | 0.29 |
| Naphthalene | 19.23 | 15.00 | 10.00 | 0.04 |
| Phenanthrene | 21.36 | 8.64 | 17.27 | 1.73 |
| Anthracene | 8.33 | 2.33 | 4.89 | 0.78 |
| Fluoranthene | 27.65 | 2.53 | 13.53 | 1.82 |
| Chrysene | 4.44 | 0.52 | 1.33 | 0.20 |
| Total PAHs | 18.39 | 7.47 | 10.69 | 0.90 |
| Hydrocarbon | Acceptable Concentration * [mg/kg] | Concentration [mg/kg] in Soil (0–2 m below Ground Level) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 05/2019 | 10/2019 | 06/2020 | ||||||||
| Spot 1 | Spot 2 | Spot 3 | Spot 1 | Spot 2 | Spot 3 | Spot 1 | Spot 2 | Spot 3 | ||
| Phenol index | 50 | 23.2 | 17.8 | 12.5 | 1.7 | 3.5 | 0.8 | 3.8 | 0.28 | 0.17 |
| Naphthalene | 50 | 407.0 | 108.0 | 135.0 | 353.0 | 129.0 | 5.5 | 74.7 | 13.6 | 17.0 |
| Phenanthrene | 50 | 736.0 | 144.0 | 1073.0 | 500.0 | 121.0 | 39.0 | 698.1 | 154.2 | 212.3 |
| Anthracene | 50 | 692.0 | 144.0 | 675.0 | 670.0 | 119.0 | 33.7 | 431.2 | 102.2 | 110.4 |
| Fluoranthene | 50 | 503.0 | 175.0 | 76.0 | 375.0 | 500.0 | 43.5 | 455.1 | 18.5 | 141.6 |
| Chrysene | 50 | 148.0 | 68.0 | 136.0 | 127.0 | 40.7 | 16.6 | 138.4 | 58.1 | 27.3 |
| Total PAHs | 250 | 2635.0 | 724.0 | 2289.0 | 2344.0 | 1265.0 | 160.0 | 977.4 | 349.0 | 242.0 |
| Hydrocarbon | Percentage of the 05/2019 Value [%] | |||||
|---|---|---|---|---|---|---|
| 10/2019 | 06/2020 | |||||
| Spot 1 | Spot 2 | Spot 3 | Spot 1 | Spot 2 | Spot 3 | |
| Phenol index | 7.3 | 19.7 | 6.7 | 16.3 | 1.57 | 1.33 |
| Naphthalene | 86.7 | 119.4 | 4.1 | 18.4 | 12.57 | 12.6 |
| Phenanthrene | 67.9 | 84.0 | 3.6 | 94.9 | 107.1 | 19.8 |
| Anthracene | 96.8 | 82.6 | 5.0 | 62.3 | 71.0 | 16.4 |
| Fluoranthene | 74.6 | 285.7 | 57.2 | 90.5 | 10.6 | 186.4 |
| Chrysene | 85.8 | 59.9 | 12.2 | 93.5 | 85.48 | 20.1 |
| Total PAHs | 88.9 | 174.7 | 7.0 | 37.1 | 48.21 | 10.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roszak, M.; Jabłońska, J.; Stachurska, X.; Dubrowska, K.; Kajdanowicz, J.; Gołębiewska, M.; Kiepas-Kokot, A.; Osińska, B.; Augustyniak, A.; Karakulska, J. Development of an Autochthonous Microbial Consortium for Enhanced Bioremediation of PAH-Contaminated Soil. Int. J. Mol. Sci. 2021, 22, 13469. https://doi.org/10.3390/ijms222413469
Roszak M, Jabłońska J, Stachurska X, Dubrowska K, Kajdanowicz J, Gołębiewska M, Kiepas-Kokot A, Osińska B, Augustyniak A, Karakulska J. Development of an Autochthonous Microbial Consortium for Enhanced Bioremediation of PAH-Contaminated Soil. International Journal of Molecular Sciences. 2021; 22(24):13469. https://doi.org/10.3390/ijms222413469
Chicago/Turabian StyleRoszak, Marta, Joanna Jabłońska, Xymena Stachurska, Kamila Dubrowska, Justyna Kajdanowicz, Marta Gołębiewska, Anna Kiepas-Kokot, Beata Osińska, Adrian Augustyniak, and Jolanta Karakulska. 2021. "Development of an Autochthonous Microbial Consortium for Enhanced Bioremediation of PAH-Contaminated Soil" International Journal of Molecular Sciences 22, no. 24: 13469. https://doi.org/10.3390/ijms222413469
APA StyleRoszak, M., Jabłońska, J., Stachurska, X., Dubrowska, K., Kajdanowicz, J., Gołębiewska, M., Kiepas-Kokot, A., Osińska, B., Augustyniak, A., & Karakulska, J. (2021). Development of an Autochthonous Microbial Consortium for Enhanced Bioremediation of PAH-Contaminated Soil. International Journal of Molecular Sciences, 22(24), 13469. https://doi.org/10.3390/ijms222413469