OsFH3 Encodes a Type II Formin Required for Rice Morphogenesis
Abstract
:1. Introduction
2. Results
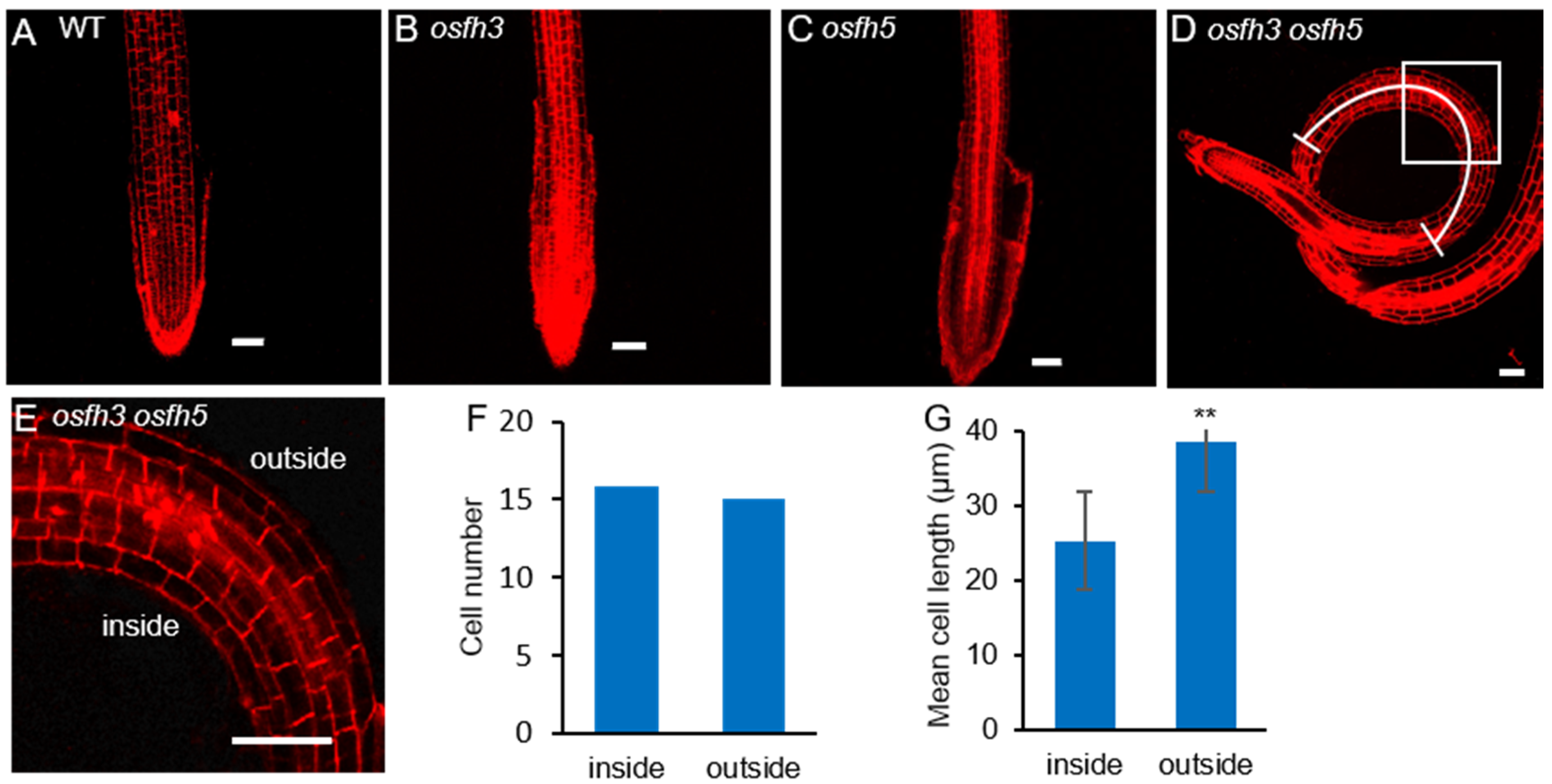
2.1. Phenotypes of osfh3 Mutant
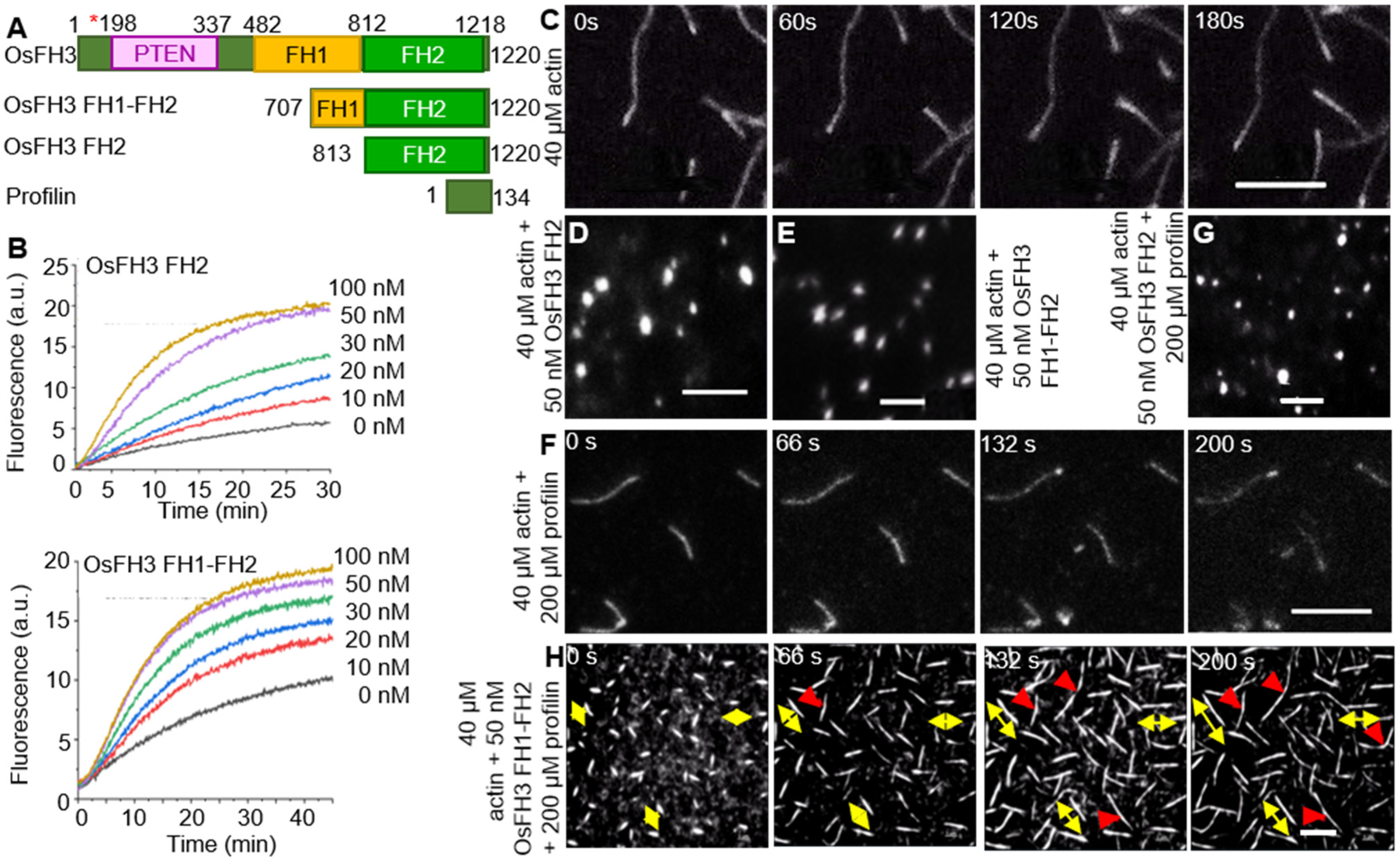
2.2. OsFH3 Nucleates Actin
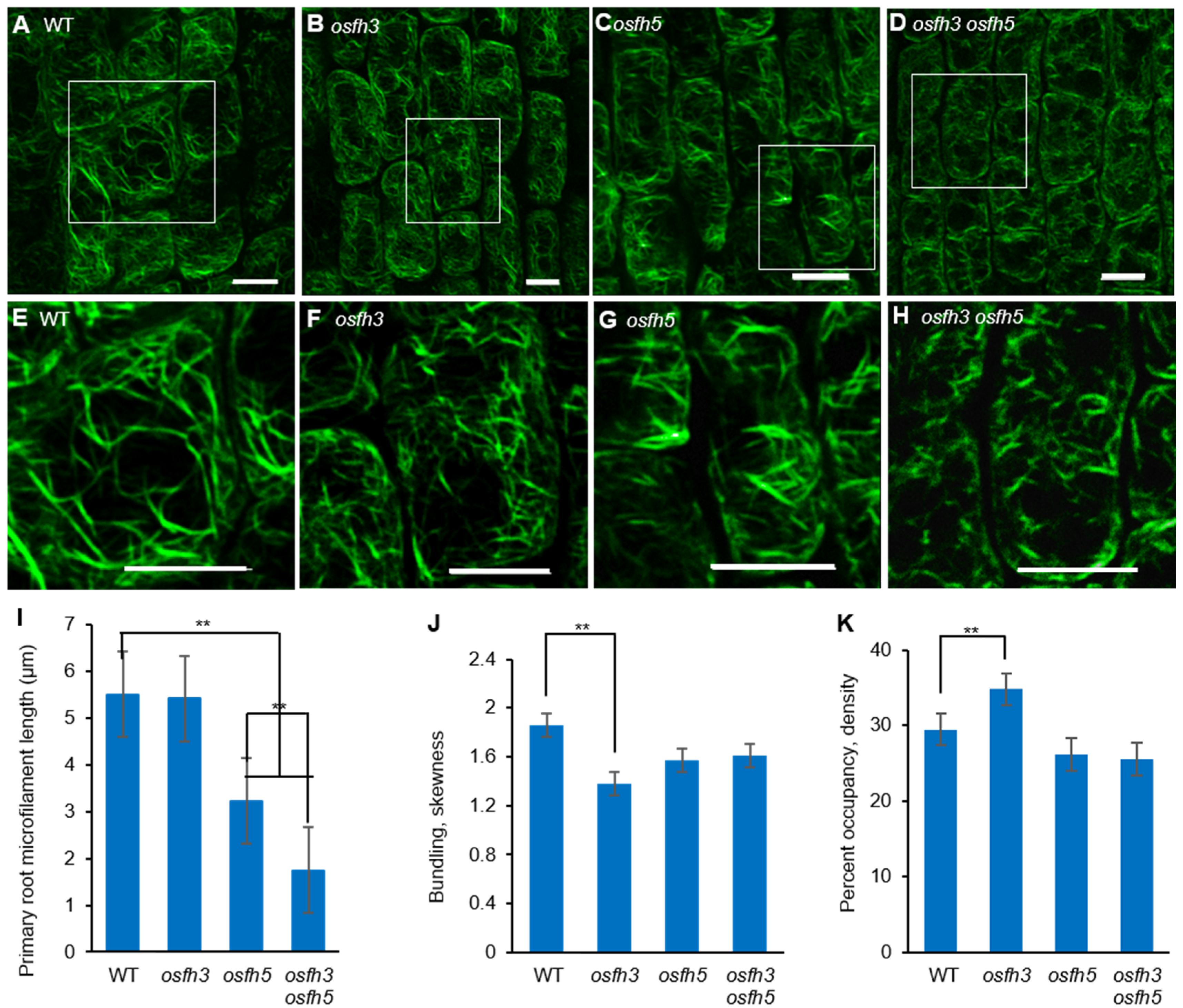
2.3. OsFH3 Bundles and Caps AF
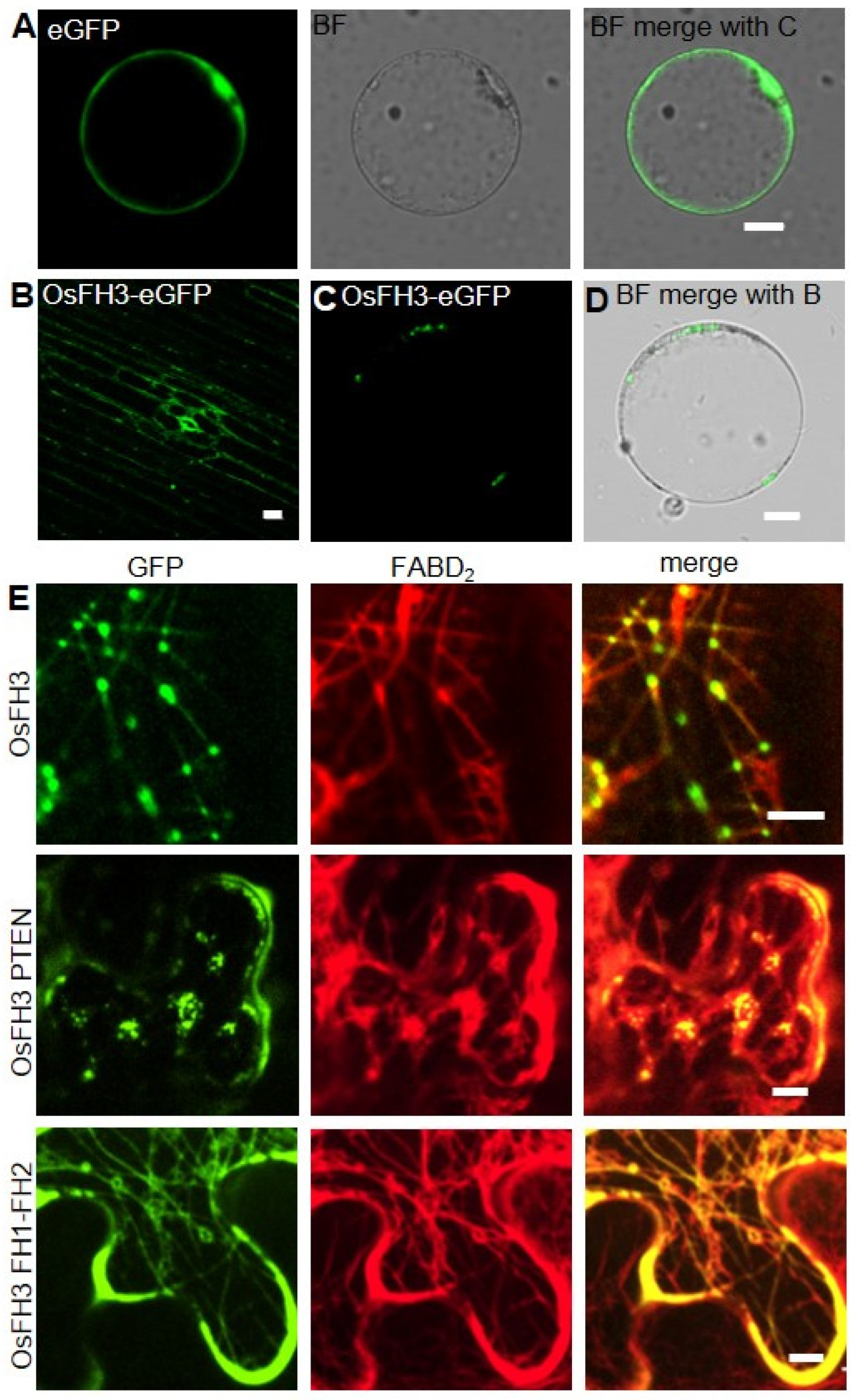
2.4. PTEN Domain Affects the Localization of OsFH3
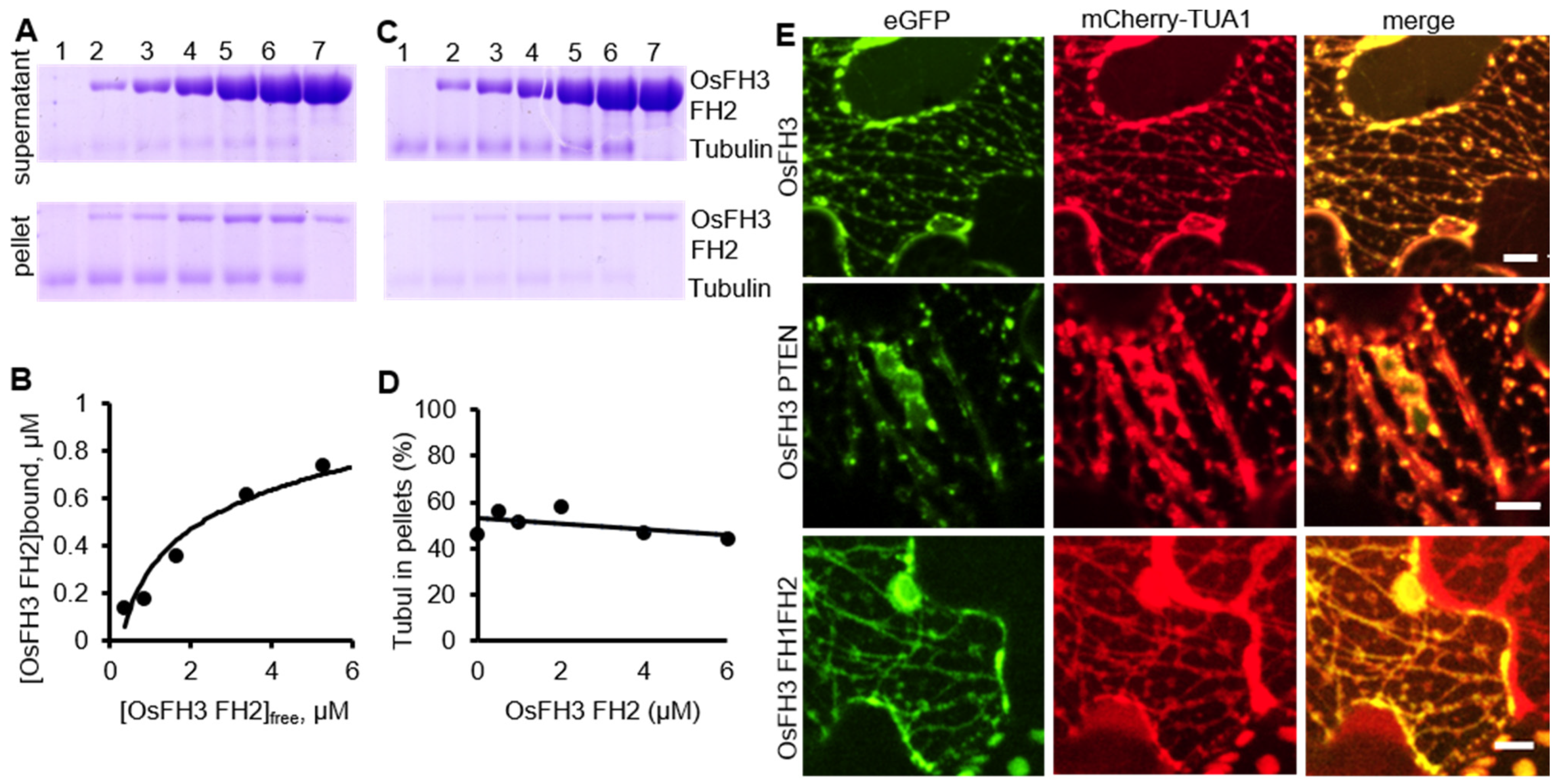
2.5. OsFH3 Binds Microtubules
3. Discussion
3.1. OsFH3 and OsFH5 Synergistically Regulate Rice Morphogenesis
3.2. OsFH3 Acts on AF Organization in Rice
3.3. OsFH3 PTEN-Domain Directs Protein Locating to Actin Cytoskeleton Intersections
3.4. OsFH3 Binds to But Does Not Bundle Microtubules
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. AF Staining of Roots
4.3. OsFH3 In Vivo Expression
4.4. Protein Production
4.5. Actin Nucleation Assays
4.6. Time-Lapse Microscopy of AF Elongation
4.7. Actin Filament Depolymerization Assays
4.8. Co-Sedimentation Assays
4.9. Microtubules Staining of Roots
4.10. Colocalization Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suarez, C.; Carroll, R.T.; Burke, T.A.; Christensen, J.R.; Bestul, A.J.; Sees, J.A.; James, M.L.; Sirotkin, V.; Kovar, D.R. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev. Cell 2015, 32, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Lennarz, W.J.; Lane, M.D. Encyclopedia of Biological Chemistry; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Zhang, Z.; Zhang, Y.; Tan, H.; Wang, Y.; Li, G.; Liang, W.; Yuan, Z.; Hu, J.; Ren, H.; Zhang, D. RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 2011, 23, 681–700. [Google Scholar] [CrossRef] [Green Version]
- Qian, D.; Xiang, Y. Actin cytoskeleton as actor in upstream and downstream of calcium signaling in plant cells. Int. J. Mol. Sci. 2019, 20, 1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Blanchoin, L.; Staiger, C.J. Signaling to actin stochastic dynamics. Annu. Rev. Plant Biol. 2015, 66, 415–440. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Jiang, Y.; Zhao, Y.; Huang, S.; Yuan, M.; Zhao, Y.; Guo, Y. CASEIN KINASE1-LIKE PROTEIN2 regulates actin filament stability and stomatal closure via phosphorylation of actin depolymerizing factor. Plant Cell 2016, 28, 1422–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J. Phospholipase D and phosphatidic acid in plant defence response: From protein-protein and lipid-protein interactions to hormone signalling. J. Exp. Bot. 2015, 66, 1721–1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, N.; Wang, X.; Jing, Y.; Lin, J. Regulation of cytoskeleton-associated protein activities: Linking cellular signals to plant cytoskeletal function. J. Integr. Plant Biol. 2021, 63, 241–250. [Google Scholar] [CrossRef]
- Li, P.; Day, B. Battlefield cytoskeleton: Turning the tide on plant immunity. Mol. Plant Microbe Interact. 2019, 32, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Cao, L.; Staiger, C.J. Capping protein modulates actin remodeling in response to reactive oxygen species during plant innateimmunity. Plant Physiol. 2017, 173, 1125–1136. [Google Scholar] [CrossRef]
- Breuer, D.; Nowak, J.; Ivakov, A.; Somssich, M.; Persson, S.; Nikoloski, Z. System-wide organization of actin cytoskeleton determines organelle transport in hypocotyl plant cells. Proc. Natl. Acad. Sci. USA 2017, 114, E5741–E5749. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhang, Y.; Ren, H. Actin polymerization mediated by AtFH5 directs the polarity establishment and vesicle trafficking for pollen germination in Arabidopsis. Mol. Plant 2018, 11, 1389–1399. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Shen, Y.; Cai, C.; Zhong, C.; Zhu, L.; Yuan, M.; Ren, H. The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 2010, 22, 2710–2726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheuring, D.; Lofke, C.; Kruger, F.; Kittelmann, M.; Eisa, A.; Hughes, L.; Smith, R.S.; Hawes, C.; Schumacher, K.; Kleine-Vehn, J. Actin-dependent vacuolar occupancy of the cell determines auxin-induced growth repression. Proc. Natl. Acad. Sci. USA 2016, 113, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Edwards, M.; Zwolak, A.; Schafer, D.A.; Sept, D.; Dominguez, R.; Cooper, J.A. Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 677–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chereau, D.; Dominguez, R. Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J. Struct. Biol. 2006, 155, 195–201. [Google Scholar] [CrossRef]
- Goley, E.D.; Welch, M.D. The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006, 7, 713–726. [Google Scholar] [CrossRef]
- Ferron, F.O.; Rebowski, G.; Lee, S.H.; Dominguez, R.J.E.J. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2014, 26, 4597–4606. [Google Scholar] [CrossRef] [Green Version]
- García-González, J.; Kebrlová, T.; Semerák, M.; Lacek, J.; Schwarzerová, K. Arp2/3 complex is required for auxin-driven cell expansion through regulation of auxin transporter homeostasis. Front. Plant Sci. 2020, 11, 486. [Google Scholar] [CrossRef] [PubMed]
- Grunt, M.; Žárský, V.; Cvrčková, F. Roots of angiosperm formins: The evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 2008, 8, 115. [Google Scholar] [CrossRef] [Green Version]
- Van Gisbergen, P.A.; Bezanilla, M. Plant formins: Membrane anchors for actin polymerization. Trends Cell Biol. 2013, 23, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Martinière, A.; Gayral, P.; Hawes, C.; Journal, J.R.J.P. Building bridges: Formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J. 2011, 66, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Cvrčková, F.; Novotný, M.; Pícková, D.; Žárský, V. Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genom. 2004, 5, 44. [Google Scholar] [CrossRef] [Green Version]
- Chalkia, D.; Nikolaidis, N.; Makalowski, W.; Klein, J.; Nei, M.J.M.B. Evolution, origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol. Biol. Evol. 2008, 25, 2717–2733. [Google Scholar] [CrossRef] [Green Version]
- Paul, A.S.; Pollard, T.D. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 2008, 18, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Henty-Ridilla, J.L.; Blanchoin, L.; Staiger, C.J. Profilin-dependent nucleation and assembly of actin filaments controls cell elongation in Arabidopsis. Plant Physiol. 2016, 170, 220–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Zheng, Y.; Yan, A.; Chen, N.; Wang, Z.; Huang, S.; Yang, Z. Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 2009, 21, 3868–3884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingouff, M.; Gerald, J.N.F.; Guerin, C.; Robert, H.; Sorensen, M.B.; Van Damme, D.; Geelen, D.; Blanchoin, L.; Berger, F. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 2005, 7, 374–380. [Google Scholar] [CrossRef]
- Van Gisbergen, P.; Wu, S.Z.; Cheng, X.; Pattavina, K.A.; Bezanilla, M. In vivo analysis of formin dynamics in the moss P. patens reveals functional class diversification. J. Cell Sci. 2020, 133, jcs233791. [Google Scholar] [CrossRef]
- Oulehlová, D.; Kollárová, E.; Cifrová, P.; Pejchar, P.; Žárský, V.; Cvrčková, F. Arabidopsis class I formin FH1 relocates between membrane compartments during root cell ontogeny and associates with plasmodesmata. Plant Cell Physiol. 2019, 60, 1855–1870. [Google Scholar] [CrossRef]
- Xue, X.; Guo, C.; Du, F.; Lu, Q.; Zhang, C.; Ren, H. AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol. Plant 2011, 4, 264–278. [Google Scholar] [CrossRef] [Green Version]
- Yi, K.; Guo, C.; Chen, D.; Zhao, B.; Yang, B.; Ren, H. Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis1. Plant Physiol. 2005, 138, 00001071–00001082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Li, S.; Ren, H. OsFH15, a class I formin, interacts with microfilaments and microtubules to regulate grain size via affecting cell expansion in rice. Sci. Rep. 2017, 7, 6538. [Google Scholar] [CrossRef] [PubMed]
- Cvrčková, F.; Grunt, M.; Žárský, V. Expression of GFP-mTalin reveals an actin-related role for the Arabidopsis Class II formin AtFH12. Biol. Plant. 2012, 56, 431–440. [Google Scholar] [CrossRef]
- Vidali, L.; van Gisbergen, P.A.; Guerin, C.; Franco, P.; Li, M.; Burkart, G.M.; Augustine, R.C.; Blanchoin, L.; Bezanilla, M. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA 2009, 106, 13341–13346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, P.; Wang, J.; He, Y.; Zhang, S.; Hu, B.; Xue, X.; Miao, L.; Ren, H. AtFH14 crosslinks actin filaments and microtubules in different manners. Biol. Cell 2021, 113, 235–249. [Google Scholar] [CrossRef]
- Kollárová, E.; Forero, A.B.; Stillerová, L.; Perostová, S.; Cvrčková, F. Arabidopsis class II formins AtFH13 and AtFH14 can form heterodimers but exhibit distinct patterns of cellular localization. Int. J. Mol. Sci 2020, 21, 348. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Ren, S.; Zhang, X.; Gao, M.; Ye, S.; Qi, Y.; Zheng, Y.; Wang, J.; Zeng, L.; Li, Q.; et al. BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 2011, 23, 661–680. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Liang, W.; Zhang, X.; Ren, H.; Zhang, D. Rice actin-binding protein RMD is a key link in the auxin-actin regulatory loop that controls cell growth. Proc. Natl. Acad. Sci. USA 2014, 111, 10377–10382. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Liang, W.; Sturrock, C.J.; Pandey, B.K.; Giri, J.; Mairhofer, S.; Wang, D.; Muller, L.; Tan, H.; York, L.M.; et al. Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun. 2018, 9, 2346. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Li, G.; Nowak, J.; Zhang, X.; Xu, D.; Yang, X.; Huang, G.; Liang, W.; Yang, L.; Wang, C.; et al. The rice actin-binding protein RMD regulates light-dependent shoot gravitropism. Plant Physiol. 2019, 181, 630–644. [Google Scholar] [CrossRef] [Green Version]
- Kollárová, E.; Baquero Forero, A.; Cvrčková, F. The arabidopsis thaliana class II formin FH13 modulates pollen tube growth. Front. Plant Sci. 2021, 12, 599961. [Google Scholar] [CrossRef]
- Diao, M.; Ren, S.L.; Wang, Q.N.; Qian, L.C.; Shen, J.F.; Liu, Y.L.; Huang, S.J. Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata. Elife 2018, 7, e36316. [Google Scholar] [CrossRef] [PubMed]
- Deeks, M.J.; Fendrych, M.; Smertenko, A.; Bell, K.S.; Oparka, K.; Cvrčková, F.; Žárský, V.; Hussey, P. The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. J. Cell Sci. 2010, 123, 1209–1215. [Google Scholar] [CrossRef] [Green Version]
- Hohmann, T.; Dehghani, F. The cytoskeleton-a complex interacting meshwork. Cells 2019, 8, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, Y.; Wu, J.; Meng, L.; Ren, H. AtFH16, an Arabidopsis type II formin, binds and bundles both microfilaments and microtubules, and preferentially binds to microtubules. J. Integr. Plant Biol. 2013, 55, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Cvrčková, F. Formins and membranes: Anchoring cortical actin to the cell wall and beyond. Front. Plant Sci. 2013, 4, 436. [Google Scholar] [CrossRef] [Green Version]
- Van Gisbergen, P.A.C.; Li, M.; Wu, S.-Z.; Bezanilla, M. Class II formin targeting to the cell cortex by binding PI(3,5)P2 is essential for polarized growth. J. Cell Biol. 2012, 198, 235–250. [Google Scholar] [CrossRef] [Green Version]
- Cvrčková, F.; Oulehlová, D.; Žárský, V. Formins: Linking cytoskeleton and endomembranes in plant cells. J. Mol. Sci 2014, 16, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raftopoulou, M.; Etienne-Manneville, S.; Self, A.; Nicholls, S.; Hall, A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science 2004, 303, 1179–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosero, A.; Žárský, V.; Cvrčková, F. AtFH1 formin mutation affects actin filament and microtubule dynamics in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef] [Green Version]
- Olyslaegers, G.; Verbelen, J.P. Improved staining of F-actin and co-localization of mitochondria in plant cells. J. Microsc. 2002, 192, 73–77. [Google Scholar] [CrossRef]
- Lee, S.T.; Huang, W.L. Cytokinin, auxin, and abscisic acid affects sucrose metabolism conduce to de novo shoot organogenesis in rice (Oryza sativa L.) callus. Bot. Stud. 2013, 54, 5. [Google Scholar] [CrossRef] [Green Version]
- Pardee, J.D. Purification of muscle actin. Enzymology 1982, 85, 164–181. [Google Scholar]
- Pollard, T.D. Measurement of rate constants for actin filament elongation in solution. Anal. Biochem. 1983, 134, 406–412. [Google Scholar] [CrossRef]
- Doolittle, L.K.; Rosen, M.K.; Padrick, S.B. Measurement and analysis of in vitro actin polymerization. Methods Mol. Biol. 2013, 1046, 273–293. [Google Scholar] [PubMed] [Green Version]
- Kuhn, J.R.; Pollard, T.D. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 2005, 88, 1387–1402. [Google Scholar] [CrossRef] [Green Version]
- Okabe, S. Incorporation and turnover of biotin-labeled actin microinjected into fibroblastic cells: An immunoelectron microscopic study. J. Cell Biol 1989, 109, 1581–1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higgs, H.N.; Blanchoin, L.; Pollard, T.D. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 1999, 38, 15212–15222. [Google Scholar] [CrossRef] [PubMed]
- Kovar, D.R.; Pollard, T.D. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA 2004, 101, 14725–14730. [Google Scholar] [CrossRef] [Green Version]
- Amann, K.J.; Pollard, T.D. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. USA 2001, 98, 15009–15013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Blanchoin, L.; Kovar, D.R.; Staiger, C.J. Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 2003, 278, 44832–44842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khurana, P.; Henty, J.L.; Huang, S.; Staiger, A.M.; Blanchoin, L.; Staiger, C.J. Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 2010, 22, 2727–2748. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Robinson, R.C.; Gao, L.Y.; Matsumoto, T.; Brunet, A.; Blanchoin, L.; Staiger, C.J. Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 2005, 17, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, K.; Williamson, R.E.; Wasteneys, G.O. New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 2000, 124, 1493–1506. [Google Scholar] [CrossRef] [Green Version]
- Montes-Rodriguez, A.; Kost, B. Direct comparison of the performance of commonly employed in vivo F-actin markers (Lifeact-YFP, YFP-mTn and YFP-FABD2) in tobacco pollen tubes. Front. Plant Sci. 2017, 8, 1349. [Google Scholar] [CrossRef] [Green Version]
- McCullock, T.W.; MacLean, D.M.; Kammermeier, P.J. Comparing the performance of mScarlet-I, mRuby3, and mCherry as FRET acceptors for mNeonGreen. PLoS ONE 2020, 15, e0219886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Li, Y.; Li, L.; Lin, J.; Zheng, C.; Zhang, L. Overexpression of PwTUA1, a pollen-specific tubulin gene, increases pollen tube elongation by altering the distribution of alpha-tubulin and promoting vesicle transport. J. Exp. Bot. 2009, 60, 2737–2749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Cao, J.Y.; Xu, Y.P.; Cai, X.Z. Artificial Agrobacterium tumefaciens strains exhibit diverse mechanisms to repress Xanthomonas oryzae pv. oryzae-induced hypersensitive response and non-host resistance in Nicotiana benthamiana. Mol. Plant Pathol. 2017, 18, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, S.; Ren, Z.; Liu, C.; Du, P.; Li, J.; Liu, Z.; Zhang, F.; Hou, H.; Shi, J.; Liang, W.; et al. OsFH3 Encodes a Type II Formin Required for Rice Morphogenesis. Int. J. Mol. Sci. 2021, 22, 13250. https://doi.org/10.3390/ijms222413250
Chang S, Ren Z, Liu C, Du P, Li J, Liu Z, Zhang F, Hou H, Shi J, Liang W, et al. OsFH3 Encodes a Type II Formin Required for Rice Morphogenesis. International Journal of Molecular Sciences. 2021; 22(24):13250. https://doi.org/10.3390/ijms222413250
Chicago/Turabian StyleChang, Shuwei, Zhanhong Ren, Chang Liu, Pingzhou Du, Jingbin Li, Zengyu Liu, Fengli Zhang, Haili Hou, Jianxin Shi, Wanqi Liang, and et al. 2021. "OsFH3 Encodes a Type II Formin Required for Rice Morphogenesis" International Journal of Molecular Sciences 22, no. 24: 13250. https://doi.org/10.3390/ijms222413250
APA StyleChang, S., Ren, Z., Liu, C., Du, P., Li, J., Liu, Z., Zhang, F., Hou, H., Shi, J., Liang, W., Yang, L., Ren, H., & Zhang, D. (2021). OsFH3 Encodes a Type II Formin Required for Rice Morphogenesis. International Journal of Molecular Sciences, 22(24), 13250. https://doi.org/10.3390/ijms222413250







