Putative RNA Ligase RtcB Affects the Switch between T6SS and T3SS in Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Results
2.1. RtcB Controls the Expression of Multiple Phenotypes and Virulence Factors
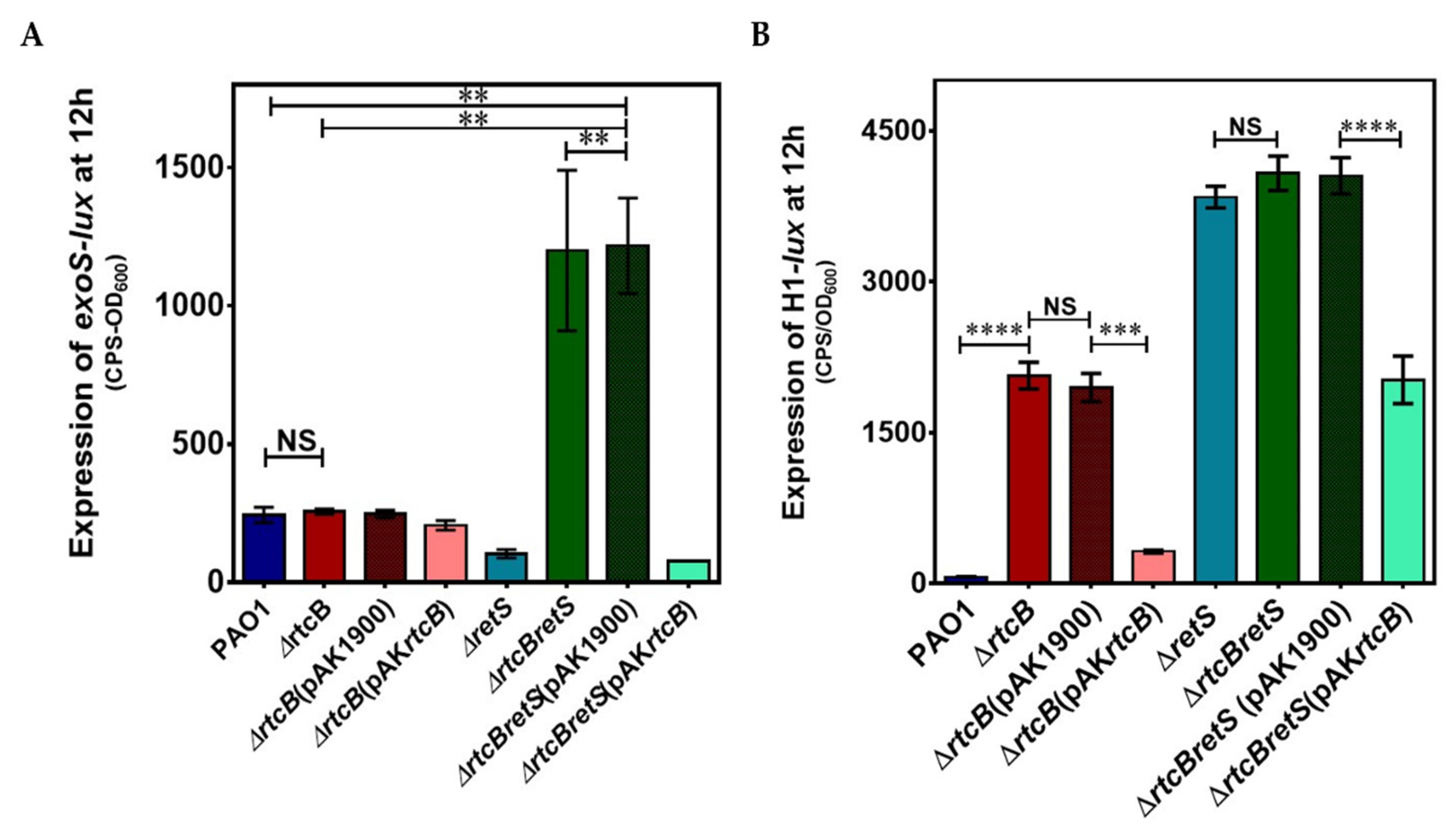
2.2. Putative RNA Ligase RtcB Affects T6SS and T3SS Transcription and Its Deletion Inactivates the Gac-Rsm Controlled Switch between the Two Secretion Systems
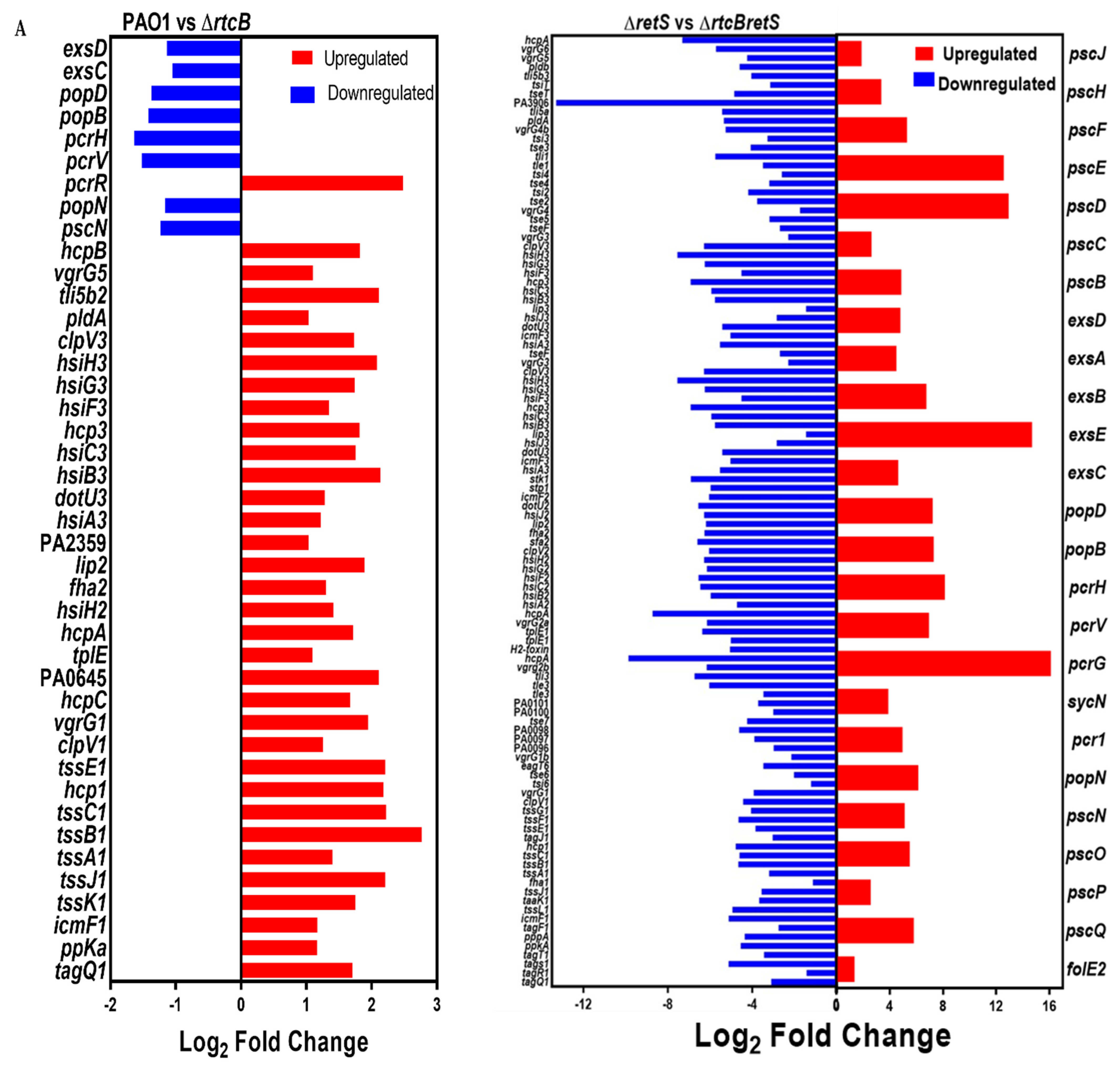
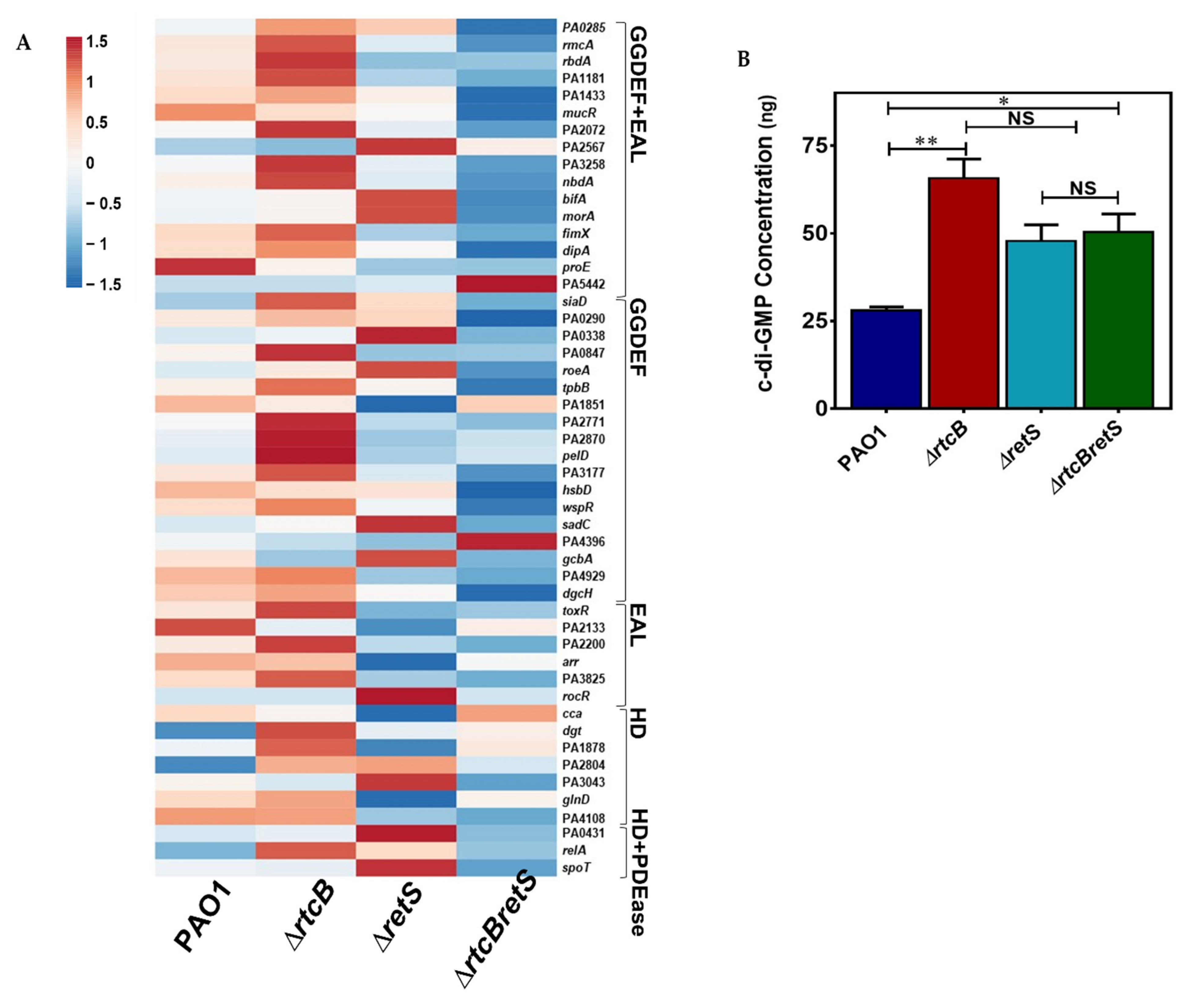
2.3. Transcriptional Profiling of the rtcB Knockout Mutant
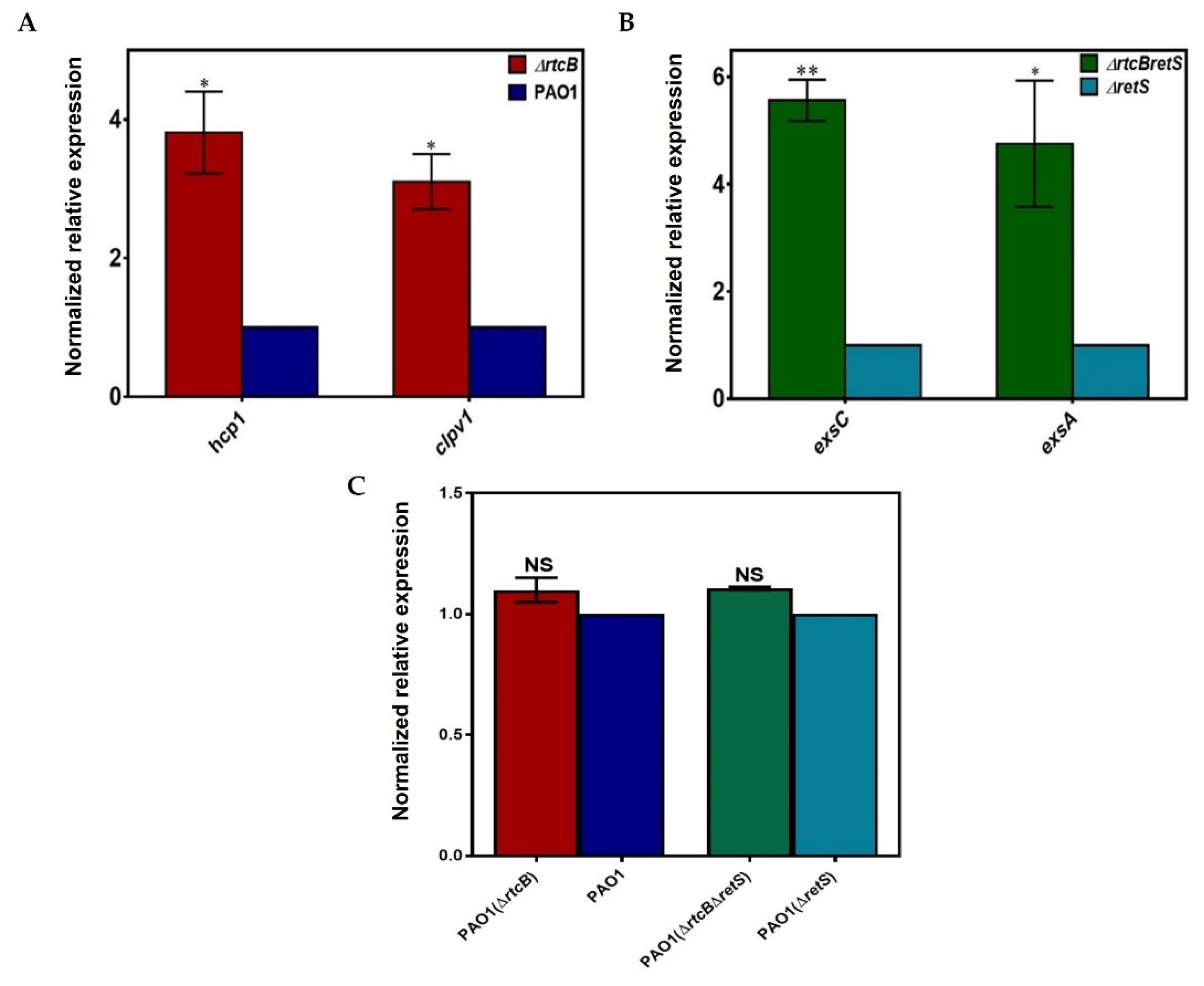
2.4. Deletion of rtcB Does Not Affect Transcription of rsmA but Upregulates the Expression of clpV1, hcp1, exsC, and exsA
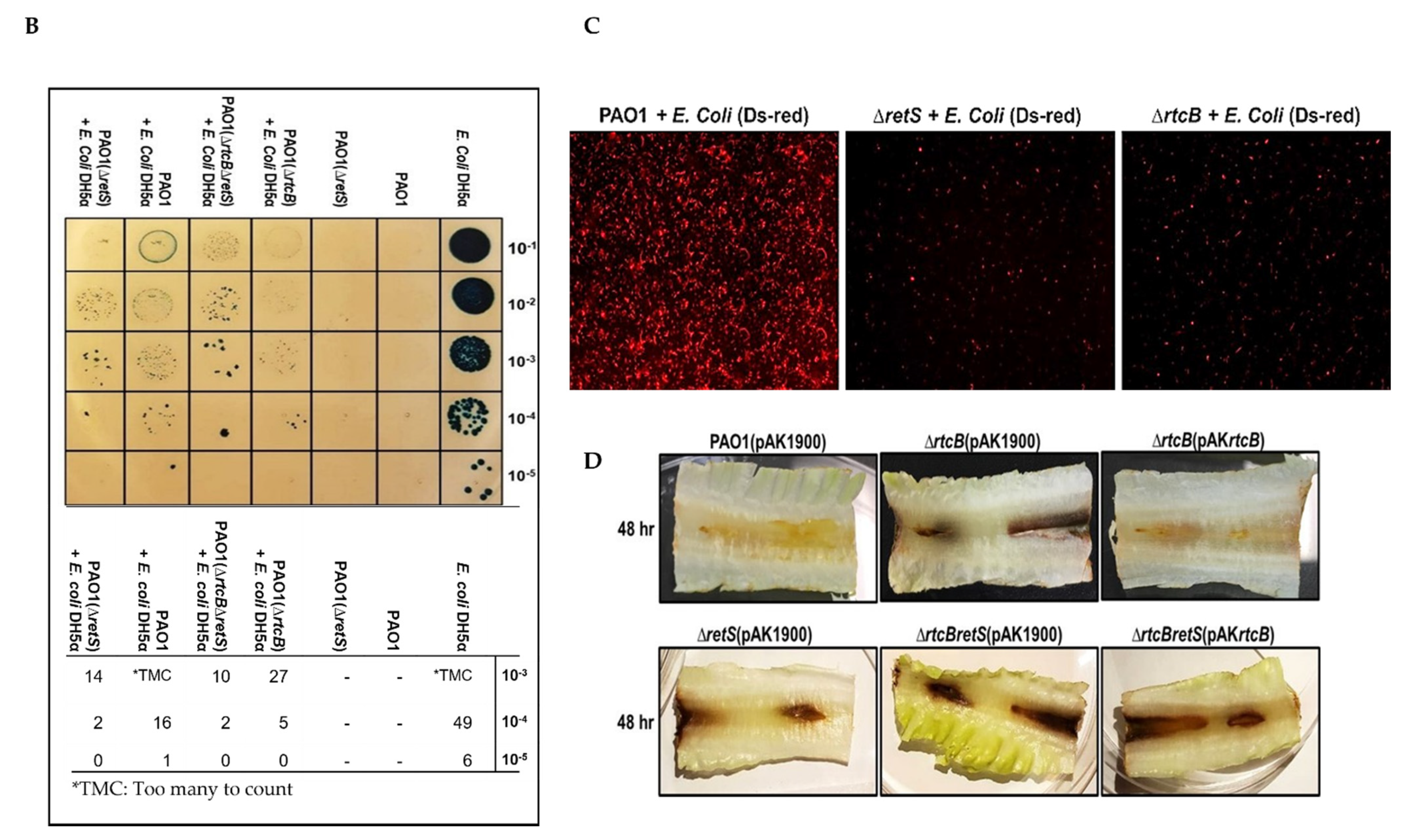
2.5. The Three T6SSs Are Activated upon Deletion of rtcB and Promote the Pathogenicity of P. aeruginosa
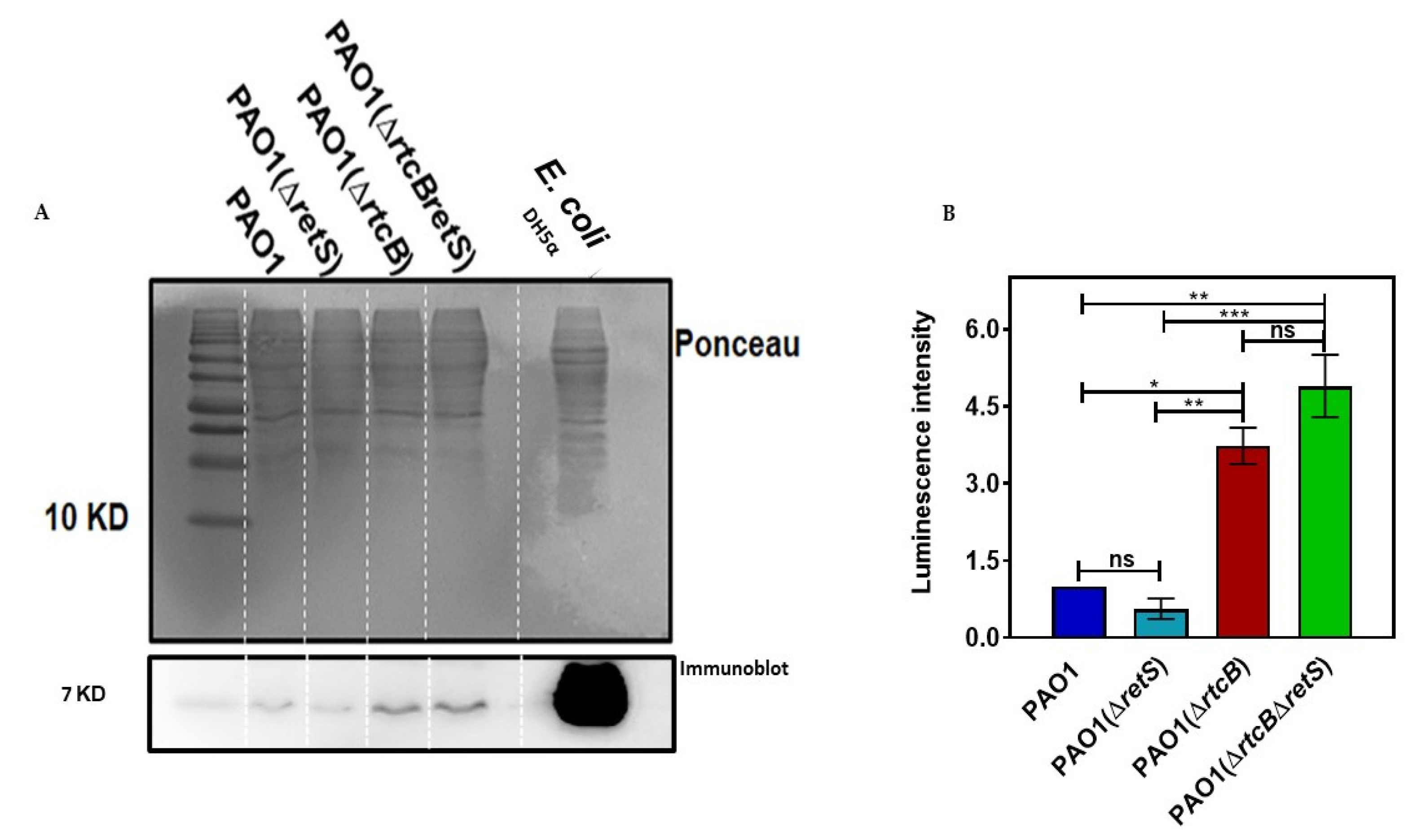
2.6. RsmA Protein Concentration Is Increased by Deletion of rtcB
2.7. Complementation of PAO1(∆rtcB∆retS) with pAKrsmY and pAKrsmY Diminishes the Expression of T6SS and Represses T3SS
2.8. Intracellular Levels of c-di-GMP Are Significantly Increased in rtcB Deletion Mutant
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Growth Conditions, and Antibiotic Reagents
4.2. Construction of Gene Expression Reporters
4.3. Construction of Mutant Strains
4.4. Pyocyanin Assay
4.5. Biofilm Formation and Bacterial Motility Assay
4.6. RNA Isolation and RNA-Sequencing and Analysis of Differentially Expressed Genes
4.7. Quantitative Real-Time PCR Analysis
4.8. Bacterial Competition Assay
4.9. Plant Virulence Assay
4.10. Immunoblotting Assay
4.11. Construction of Overexpression Vectors
4.12. C-di-GMP Quantification
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fuentefria, D.B.; Ferreira, A.E.; Corção, G. Antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater and superficial water: Are they genetically related? J. Environ. Manag. 2011, 92, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Klockgether, J.; Cramer, N.; Wiehlmann, L.; Davenport, C.F.; Tümmler, B. Pseudomonas aeruginosa Genomic Structure and Diversity. Front. Microbiol. 2011, 2, 150. Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2011.00150/full (accessed on 21 June 2021). [CrossRef] [PubMed] [Green Version]
- Kipnis, E.; Sawa, T.; Wiener-Kronish, J. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Mal. Infect. 2006, 36, 78–91. [Google Scholar] [CrossRef]
- Sharma, A.K.; Dhasmana, N.; Dubey, N.; Kumar, N.; Gangwal, A.; Gupta, M.; Singh, Y. Bacterial Virulence Factors: Secreted for Survival. Indian J. Microbiol. 2017, 57, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol. Spectr. 2016, 4, 12. [Google Scholar] [CrossRef] [Green Version]
- Kepseu, W.D.; Van Gijsegem, F.; Sepulchre, J.-A. Modelling the Onset of Virulence in Pathogenic Bacteria. Methods Mol. Biol. 2011, 804, 501–517. [Google Scholar] [CrossRef]
- Valentini, M.; Gonzalez, D.; Mavridou, D.; Filloux, A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 2018, 41, 15–20. [Google Scholar] [CrossRef]
- Berube, B.J.; Rangel, S.M.; Hauser, A.R. Pseudomonas aeruginosa: Breaking down barriers. Curr. Genet. 2015, 62, 109–113. [Google Scholar] [CrossRef]
- Lam, J.; Chan, R.; Lam, K.; Costerton, J.W. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 1980, 28, 546–556. [Google Scholar] [CrossRef]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A Signaling Network Reciprocally Regulates Genes Associated with Acute Infection and Chronic Persistence in Pseudomonas aeruginosa. Dev. Cell 2004, 7, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Goodman, A.L.; Merighi, M.; Hyodo, M.; Ventre, I.; Filloux, A.; Lory, S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009, 23, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Brencic, A.; McFarland, K.A.; McManus, H.R.; Castang, S.; Mogno, I.; Dove, S.L.; Lory, S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009, 73, 434–445. [Google Scholar]
- Moscoso, J.A.; Mikkelsen, H.; Heeb, S.; Williams, P.; Filloux, A. The Pseudomonas aeruginosa sensor RetS switches Type III and Type VI secretion via c-di-GMP signalling. Environ. Microbiol. 2011, 13, 3128–3138. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, L.; Wood, T.; Howard, S.A.; Maggiorelli, F.; Nolan, L.; Wettstadt, S.; Filloux, A. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 7707–7712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merritt, J.H.; Brothers, K.M.; Kuchma, S.L.; O’Toole, G.A. SadC Reciprocally Influences Biofilm Formation and Swarming Motility via Modulation of Exopolysaccharide Production and Flagellar Function. J. Bacteriol. 2007, 189, 8154–8164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [Green Version]
- Chua, S.L.; Sivakumar, K.; Rybtke, M.; Yuan, M.; Andersen, J.B.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T.; Cao, B.; Kjelleberg, S.; et al. C-di-GMP regulates Pseudomonas aeruginosa stress response to tellurite during both planktonic and biofilm modes of growth. Sci. Rep. 2015, 5, 10052. [Google Scholar] [CrossRef] [Green Version]
- Manwar, M.R.; Shao, C.; Shi, X.; Wang, J.; Lin, Q.; Tong, Y.; Kang, Y.; Yu, J. The bacterial RNA ligase RtcB accelerates the repair process of fragmented rRNA upon releasing the antibiotic stress. Sci. China Life Sci. 2019, 63, 251–258. [Google Scholar] [CrossRef]
- Englert, M.; Sheppard, K.; Aslanian, A.; Yates, J.R.; Söll, D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. USA 2011, 108, 1290–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Temmel, H.; Müller, C.; Sauert, M.; Vesper, O.; Reiss, A.; Popow, J.; Martinez, J.; Moll, I. The RNA ligase RtcB reverses MazF-induced ribosome heterogeneity in Escherichia coli. Nucleic Acids Res. 2017, 45, 4708–4721. [Google Scholar] [PubMed] [Green Version]
- Genschik, P.; Drabikowski, K.; Filipowicz, W. Characterization of the Escherichia coli RNA 3′-Terminal Phosphate Cyclase and Its ς54-Regulated Operon. J. Biol. Chem. 1998, 273, 25516–25526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2015, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazlauskiene, M.; Kostiuk, G.; Venclovas, Č.; Tamulaitis, G.; Siksnys, V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 2017, 357, 605–609. [Google Scholar] [CrossRef] [Green Version]
- Niewoehner, O.; Doval, C.G.; Rostøl, J.T.; Berk, C.; Schwede, F.; Bigler, L.; Hall, J.; Marraffini, L.A.; Jinek, M. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 2017, 548, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Anantharaman, V.; Grishin, N.V.; Koonin, E.V.; Aravind, L. CARF and WYL domains: Ligand-binding regulators of prokaryotic defense systems. Front. Genet. 2014, 5, 102. [Google Scholar] [CrossRef]
- Hughes, K.J.; Chen, X.; Burroughs, A.M.; Aravind, L.; Wolin, S.L. An RNA Repair Operon Regulated by Damaged tRNAs. Cell Rep. 2020, 33, 108527. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Chakravarty, A.K.; Maughan, B.; Shuman, S. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 2011, 286, 43134–43143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, N.; Shuman, S. RtcB Is the RNA Ligase Component of an Escherichia coli RNA Repair Operon. J. Biol. Chem. 2011, 286, 7727–7731. [Google Scholar] [CrossRef] [Green Version]
- Yahr, T.L.; Greenberg, E. The Genetic Basis for the Commitment to Chronic versus Acute Infection in Pseudomonas aeruginosa. Mol. Cell 2004, 16, 497–498. [Google Scholar] [CrossRef]
- Zolfaghar, I.; Angus, A.A.; Kang, P.J.; To, A.; Evans, D.J.; Fleiszig, S.M. Mutation of retS, encoding a putative hybrid two-component regulatory protein in Pseudomonas aeruginosa, attenuates multiple virulence mechanisms. Microbes Infect. 2005, 7, 1305–1316. [Google Scholar] [CrossRef]
- Laskowski, M.A.; Osborn, E.; Kazmierczak, B.I. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol. Microbiol. 2004, 54, 1090–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [Green Version]
- Tamar, E.; Koler, M.; Vaknin, A. The role of motility and chemotaxis in the bacterial colonization of protected surfaces. Sci. Rep. 2016, 6, 19616. [Google Scholar] [CrossRef]
- Kilmury, S.L.N.; Burrows, L.L. The Pseudomonas aeruginosa PilSR Two-Component System Regulates Both Twitching and Swimming Motilities. mBio 2018, 9, e01310-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joge, S.; Kollaran, A.M.; Kotian, H.S.; Badal, D.; Prakash, D.; Mishra, A.; Varma, M.; Singh, V. Two-component systems regulate swarming in Pseudomonas aeruginosa PA14. bioRxiv 2018, 16, 445015. [Google Scholar]
- Miller, R.M.; Tomaras, A.P.; Barker, A.P.; Voelker, D.R.; Chan, E.D.; Vasil, A.I.; Vasil, M.L. Pseudomonas aeruginosa Twitching Motility-Mediated Chemotaxis towards Phospholipids and Fatty Acids: Specificity and Metabolic Requirements. J. Bacteriol. 2008, 190, 4038–4049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pletzer, D.; Sun, E.; Ritchie, C.; Wilkinson, L.; Liu, L.T.; Trimble, M.J.; Wolfmeier, H.; Blimkie, T.M.; Hancock, R.E.W. Surfing motility is a complex adaptation dependent on the stringent stress response in Pseudomonas aeruginosa LESB58. PLOS Pathog. 2020, 16, e1008444. [Google Scholar] [CrossRef] [Green Version]
- Diaz, M.R.; King, J.M.; Yahr, T.L. Intrinsic and extrinsic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Front. Microbiol. 2011, 2, 89. [Google Scholar] [CrossRef] [Green Version]
- Hachani, A.; Lossi, N.S.; Hamilton, A.; Jones, C.; Bleves, S.; Albesa-Jové, D.; Filloux, A. Type VI Secretion System in Pseudomonas aeruginosa. J. Biol. Chem. 2011, 286, 12317–12327. [Google Scholar] [CrossRef] [Green Version]
- Hsu, F.; Schwarz, S.; Mougous, J.D. TagR Promotes PpkA-Catalyzed Type VI Secretion Activation in Pseudomonas aeruginosa. Mol. Microbiol. 2009, 72, 1111–1125. [Google Scholar] [CrossRef] [Green Version]
- Hood, R.D.; Singh, P.; Hsu, F.; Güvener, T.; Carl, M.A.; Trinidad, R.R.; Silverman, J.M.; Ohlson, B.B.; Hicks, K.G.; Plemel, R.L.; et al. A Type VI Secretion System of Pseudomonas aeruginosa Targets a Toxin to Bacteria. Cell Host Microbe 2010, 7, 25–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, T.; Howard, S.A.; Förster, A.; Nolan, L.; Manoli, E.; Bullen, N.P.; Yau, H.; Hachani, A.; Hayward, R.D.; Whitney, J.C.; et al. The Pseudomonas aeruginosa T6SS Delivers a Periplasmic Toxin that Disrupts Bacterial Cell Morphology. Cell Rep. 2019, 29, 187–201.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sana, T.; Hachani, A.; Bucior, I.; Soscia, C.; Garvis, S.; Termine, E.; Engel, J.; Filloux, A.; Bleves, S. The Second Type VI Secretion System of Pseudomonas aeruginosa Strain PAO1 Is Regulated by Quorum Sensing and Fur and Modulates Internalization in Epithelial Cells. J. Biol. Chem. 2012, 287, 27095–27105. [Google Scholar] [CrossRef] [Green Version]
- Sana, T.; Soscia, C.; Tonglet, C.M.; Garvis, S.; Bleves, S. Divergent Control of Two Type VI Secretion Systems by RpoN in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e76030. [Google Scholar] [CrossRef]
- Janssen, K.H.; Diaz, M.R.; Gode, C.J.; Wolfgang, M.C.; Yahr, T.L. RsmV a small non-coding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. bioRxiv 2018, 200, e00277-18. [Google Scholar] [CrossRef] [Green Version]
- Janssen, K.; Diaz, M.R.; Golden, M.; Graham, J.W.; Sanders, W.; Wolfgang, M.C.; Yahr, T.L. Functional Analyses of the RsmY and RsmZ Small Noncoding Regulatory RNAs in Pseudomonas aeruginosa. J. Bacteriol. 2018, 200, e00736-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sibley, C.D.; Parkins, M.D.; Rabin, H.R.; Duan, K.; Norgaard, J.C.; Surette, M.G. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 2008, 105, 15070–15075. [Google Scholar] [CrossRef] [Green Version]
- Filkins, L.M.; O’Toole, G.A. Cystic Fibrosis Lung Infections: Polymicrobial, Complex, and Hard to Treat. PLoS Pathog. 2015, 11, e1005258. [Google Scholar] [CrossRef] [Green Version]
- Bernard, C.S.; Brunet, Y.R.; Gueguen, E.; Cascales, E. Nooks and Crannies in Type VI Secretion Regulation. J. Bacteriol. 2010, 192, 3850–3860. [Google Scholar] [CrossRef] [Green Version]
- Cianciotto, N.P. Type II secretion: A protein secretion system for all seasons. Trends Microbiol. 2005, 13, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Sijbrandi, R.; Urbanus, M.L.; Hagen-Jongman, C.M.T.; Bernstein, H.D.; Oudega, B.; Otto, B.R.; Luirink, J. Signal Recognition Particle (SRP)-mediated Targeting and Sec-dependent Translocation of an Extracellular Escherichia coli Protein. J. Biol. Chem. 2003, 278, 4654–4659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostroff, R.M.; Wretlind, B.; Vasil, M.L. Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect. Immun. 1989, 57, 1369–1373. [Google Scholar] [CrossRef] [Green Version]
- Elsheikh, L.E.; Kronevi, T.; Wretlind, B.; Abaas, S.; Iglewski, B.H. Assessment of elastase as a Pseudomonas aeruginosa virulence factor in experimental lung infection in mink. Vet. Microbiol. 1987, 13, 281–289. [Google Scholar] [CrossRef]
- Filloux, A. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta BBA—Mol. Cell Res. 2004, 1694, 163–179. [Google Scholar] [CrossRef] [Green Version]
- Ball, G.; Durand, E.; Lazdunski, A.; Filloux, A. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 2002, 43, 475–485. [Google Scholar] [CrossRef]
- Martinez, A.; Ostrovsky, P.; Nunn, D.N. Identification of an additional member of the secretin superfamily of proteins in Pseudomonas aeruginosa that is able to function in type II protein secretion. Mol. Microbiol. 1998, 28, 1235–1246. [Google Scholar] [CrossRef] [Green Version]
- Hartl, F.U.; Lecker, S.; Schiebel, E.; Hendrick, J.P.; Wickner, W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 1990, 63, 269–279. [Google Scholar] [CrossRef]
- Engel, J.; Balachandran, P. Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 2009, 12, 61–66. [Google Scholar] [CrossRef]
- Klauser, T.; Pohlner, J.; Meyer, T.F. The secretion pathway of IgA protease-type proteins in gram-negative bacteria. BioEssays 1993, 15, 799–805. [Google Scholar] [CrossRef]
- Grijpstra, J.; Arenas, J.; Rutten, L.; Tommassen, J. Autotransporter secretion: Varying on a theme. Res. Microbiol. 2013, 164, 562–582. [Google Scholar] [CrossRef] [PubMed]
- Reichhardt, C.; Wong, C.; da Silva, D.P.; Wozniak, D.J.; Parsek, M.R. CdrA Interactions within the Pseudomonas aeruginosa Biofilm Matrix Safeguard It from Proteolysis and Promote Cellular Packing. mBio 2018, 9, e01376-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forster, A.; Planamente, S.; Manoli, E.; Lossi, N.S.; Freemont, P.S.; Filloux, A. Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes. J. Biol. Chem. 2014, 289, 33032–33043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lennings, J.; Mayer, C.; Makhlouf, M.; Brötz-Oesterhelt, H.; Schwarz, S. Polar localization of the ATPase ClpV-5 occurs independent of type VI secretion system apparatus proteins in Burkholderia thailandensis. BMC Res. Notes 2019, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Genet. 2009, 7, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Lykken, G.L.; Chen, G.; Brutinel, E.D.; Chen, L.; Yahr, T.L. Characterization of ExsC and ExsD Self-Association and Heterocomplex Formation. J. Bacteriol. 2006, 188, 6832–6840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, L.H.; DeMoss, R.D. On the biosynthesis of pyocyanine. J. Bacteriol. 1959, 77, 776–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colley, B.; Dederer, V.; Carnell, M.; Kjelleberg, S.; Rice, S.A.; Klebensberger, J. SiaA/D Interconnects c-di-GMP and RsmA Signaling to Coordinate Cellular Aggregation of Pseudomonas aeruginosa in Response to Environmental Conditions. Front. Microbiol. 2016, 7, 179. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.B.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Duan, K.; Dammel, C.; Stein, J.; Rabin, H.; Surette, M.G. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 2003, 50, 1477–1491. [Google Scholar] [CrossRef] [Green Version]
- Ditta, G.; Stanfield, S.; Corbin, D.; Helinski, D.R. Broad host range DNA cloning system for gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 1980, 77, 7347–7351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
- Sharp, R.; Jansons, I.S.; Gertman, E.; Kropinski, A.M. Genetic and sequence analysis of the cos region of the temperate Pseudomonas aeruginosa bacteriophage, D3. Gene 1996, 177, 47–53. [Google Scholar] [CrossRef]
- Becher, A.; Schweizer, H.P. Integration-Proficient Pseudomonas aeruginosa Vectors for Isolation of Single-Copy Chromosomal lacZ and lux Gene Fusions. BioTechniques 2000, 29, 948–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, T.T.; Kutchma, A.J.; Becher, A.; Schweizer, H.P. Integration-Proficient Plasmids for Pseudomonas aeruginosa: Site-Specific Integration and Use for Engineering of Reporter and Expression Strains. Plasmid 2000, 43, 59–72. [Google Scholar] [CrossRef]
- Liang, H.; Li, L.; Dong, Z.; Surette, M.G.; Duan, K. The YebC Family Protein PA0964 Negatively Regulates the Pseudomonas aeruginosa Quinolone Signal System and Pyocyanin Production. J. Bacteriol. 2008, 190, 6217–6227. [Google Scholar] [CrossRef] [Green Version]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Rashid, M.H.; Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 4885–4890. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hachani, A.; Lossi, N.S.; Filloux, A. A Visual Assay to Monitor T6SS-mediated Bacterial Competition. J. Vis. Exp. 2013, 5, e50103. [Google Scholar] [CrossRef] [PubMed]
- Rahme, L.G.; Tan, M.-W.; Le, L.; Wong, S.M.; Tompkins, R.G.; Calderwood, S.B.; Ausubel, F.M. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 1997, 94, 13245–13250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harlow, E.; Lane, D. Bradford assay. CSH Protoc. 2006, 2006, 4644. [Google Scholar] [CrossRef] [PubMed]
- Poole, K.; Neshat, S.; Krebes, K.; Heinrichs, D.E. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J. Bacteriol. 1993, 175, 4597–4604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irie, Y.; Parsek, M.R. LC/MS/MS-Based Quantitative Assay for the Secondary Messenger Molecule, c-di-GMP. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.-L., Eds.; Springer: New York, NY, USA, 2014; pp. 271–279. [Google Scholar]



| Gene Locus | Secretion System | Secreted Protein | p Value | FDR | Log 2-Fold Change |
|---|---|---|---|---|---|
| PAO1(∆rtcB) | |||||
| PA2939 | T2SS | PaAP | 6.97 × 10−15 | 3.55 × 10−13 | 1 |
| PA4625 | T5SS | CdrA | 5.24 × 10−43 | 1.53 × 10−40 | 2.1 |
| PAO1(∆rtcB∆retS) | |||||
| PA0572 | T2SS | Protease | 2.25 × 10−15 | 4.64 × 10−14 | −1.5 |
| PA0852 | T2SS | CbpD | 4.32 × 10−21 | 1.18 × 10−19 | −1.6 |
| PA1249 | T2SS | AprA | 5.56 × 10−6 | 4.76 × 10−5 | −1.2 |
| PA1948 | T2SS | ToxA | 5.76 × 10−6 | 4.91 × 10−5 | 1.1 |
| PA2862 | T2SS | LipA | 1.08 × 10−7 | 1.17 × 10−6 | 4.8 |
| PA2939 | T2SS | PaAP | 3.42 × 10−17 | 7.92 × 10−16 | −1.5 |
| PA2676 | T2SS | HplS | 0.010726 | 0.041385 | −1.8 |
| PA2677 | T2SS | HplR | 5.62 × 10−8 | 6.40 × 10−7 | −3 |
| PA2678 | T2SS | Unknown protein | 1.53 × 10−5 | 0.000119 | −3.3 |
| PA3105 | T2SS | XcpQ | 1.43 × 10−23 | 4.40 × 10−22 | −2.1 |
| PA3724 | T2SS | LasB | 1.48 × 10−11 | 2.35 × 10−10 | 1.2 |
| PA3822 | Sec-SRP | Unknown protein | 1.14 × 10−5 | 9.19 × 10−5 | 1 |
| PA4082 | T5SS | CupB5 | 2.84 × 10−11 | 4.39 × 10−10 | −2.3 |
| PA4624 | T5SS | CdrB | 1.47 × 10−8 | 1.82 × 10−7 | −1.3 |
| PA5210 | T2SS | Probable ATPase | 3.32 × 10−9 | 4.30 × 10−8 | −1.1 |
| Bacterial Strains or Plasmid | Relevant Characteristics/Sequence | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80lacZ∆M15 ∆(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk−, mk+) phoA supE44 λ− thi−1 gyrA96 relA1 | Invitrogen |
| SM10-λ pir | Mobilizing strain, RP4 integrated into the chromosome; Knr | [69] |
| Mach1™-T1R | F- Φ80lacZΔM15 ΔlacΧ74 hsdR(rK- mK+) ΔrecA1398 endA1 tonA | Invitrogen |
| P. aeruginosa strains | ||
| PAO1 | Wild type, lab strain | In-house |
| PAO1 (∆retS) | retS replacement mutant of PAO1 | In-house |
| PAO1 (∆rtcB) | rtcB replacement mutant of PAO1 | This study |
| PAO1 (∆rtcB∆retS) | rtcB retS replacement mutant of PAO1 | This study |
| Plasmids | ||
| pMS402 | Expression reporter plasmid carrying the promoterless luxCDABE; Knr Tmpr | [70] |
| CTX-6.1 | Integration plasmid origins of plasmid mini-CTX-lux; Tcr | In-house |
| pRK2013 | Broad-host-range helper vector; Tra+, Knr | [71] |
| pEX18Tc | oriT+sacB+ gene replacement vector with multiple-cloning site from pUC18; Tcr | [72] |
| pAK1900 | E. coli-P. aeruginosa shuttle cloning vector, Ampr | [73] |
| pAK-rtcB | pAK1900 with a 1260 bp fragment of PA4583 between BamHI and HindIII; Ampr, Cbr | This study |
| pEX18Tc-rtcB | pEX18Tc carrying the upstream and downstream fragment of rtcB | This study |
| pKD-H1T6SS | pMS402 containing H1-T6SS promoter region; Knr, Tmpr | In-house |
| CTX-H1T6SS | Integration plasmid, CTX6.1 with a fragment of pKD-H1-T6SS containing H1 promoter region and luxCDABE gene; Knr, Tmpr, Tcr | This study |
| CTX-ExoS | Integration plasmid, CTX6.1 with a fragment of pKD-ExoS containing exoS promoter region and luxCDABE gene; Knr, Tmpr, Tcr | In-house |
| pAK-rsmY | pAK1900 with a 124 bp fragment of rsmY between BamHI and HindIII; Ampr, Cbr | This study |
| pAK-rsmZ | pAK1900 with a 116 bp fragment of rsmZ between BamHI and HindIII; Ampr, Cbr | This study |
| Primer | Sequence (5′→3′) a | Restriction Site |
|---|---|---|
| rtcB -UP-S | acagcgGAATTCCTGGTGGAGCGCTACTTCACC | EcoRI |
| rtcB -UP-AS | acagcgGGATCCGTCCAGAGCTTGATCGGCTT | BamHI |
| rtcB -DW-S | acagcgGGATCCCGATGGCCTACAAGGACATCG | BamHI |
| rtcB -DW-AS | acagcgAAGCTTGATGTTGCGGATGCGCAGC | HindIII |
| pAK-rtcB-S | acagcgAAGCTTGGAGCACAAGGAAAGCACGATG | HindIII |
| pAK-rtcB-AS | acagcgGGATCCCGTCATCCTTTCACGCACACC | BamHI |
| pKD-rtcB-S | acagcgCTCGAGGCTCGAACGTTGCCTTAACGGC | XhoI |
| pKD-rtcB-AS | acagcgGGATCCGGAATACAGCAGGCCGCGTTCG | BamHI |
| pAK-rsmY-S | acagcgAAGCTTCATGCTGGGAAGGCTCGCGATG | XhoI |
| pAK-rsmY-AS | acagcgGAATTCCTGAAGGTCGGCCTGGTCTACC | BamHI |
| pAK-rsmZ-S | acagcgGGATCCGTGACGCGCTGTTCCAGTGACG | XhoI |
| pAK-rsmZ-AS | acagcgGGAATTCATCGAGCTGAACAGC | BamHI |
| rpoD-S | GATCTCCATGGAAACCCCGATC | |
| rpoD-AS | GAGGACTTCGCGGGTGGATTC | |
| exsA-S | TTCTGCTCGAGGGCGAACTGAC | |
| exsA-AS | CGGCTGTCCTTTCCCTTGGTAC | |
| clpv1-S | CTGAACAGCCTGGCCTACAAGG | |
| clpv1-AS | GAGTCCGGCAACTGGAGGATC | |
| hcp1-S | GACGTCAAGGGTGAGTCCAAGG | |
| hcp1-AS | CAGGTTGGGCGTGGACTTGTC | |
| exsC-S | CAAGGTCAACCGACTGCTTGC | |
| exsC-AS | CATCGGCCTCCAGCAACAGAC | |
| rsmA-S | AGACCCTGATGGTAGGTGACGA | |
| rsmA-AS | TGGATGCGCTGGTAAATTTCCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dadashi, M.; Chen, L.; Nasimian, A.; Ghavami, S.; Duan, K. Putative RNA Ligase RtcB Affects the Switch between T6SS and T3SS in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 12561. https://doi.org/10.3390/ijms222212561
Dadashi M, Chen L, Nasimian A, Ghavami S, Duan K. Putative RNA Ligase RtcB Affects the Switch between T6SS and T3SS in Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2021; 22(22):12561. https://doi.org/10.3390/ijms222212561
Chicago/Turabian StyleDadashi, Maryam, Lin Chen, Ahmad Nasimian, Saeid Ghavami, and Kangmin Duan. 2021. "Putative RNA Ligase RtcB Affects the Switch between T6SS and T3SS in Pseudomonas aeruginosa" International Journal of Molecular Sciences 22, no. 22: 12561. https://doi.org/10.3390/ijms222212561
APA StyleDadashi, M., Chen, L., Nasimian, A., Ghavami, S., & Duan, K. (2021). Putative RNA Ligase RtcB Affects the Switch between T6SS and T3SS in Pseudomonas aeruginosa. International Journal of Molecular Sciences, 22(22), 12561. https://doi.org/10.3390/ijms222212561







