Lipidomics Reveals Cisplatin-Induced Renal Lipid Alterations during Acute Kidney Injury and Their Attenuation by Cilastatin
Abstract
:1. Introduction
2. Results
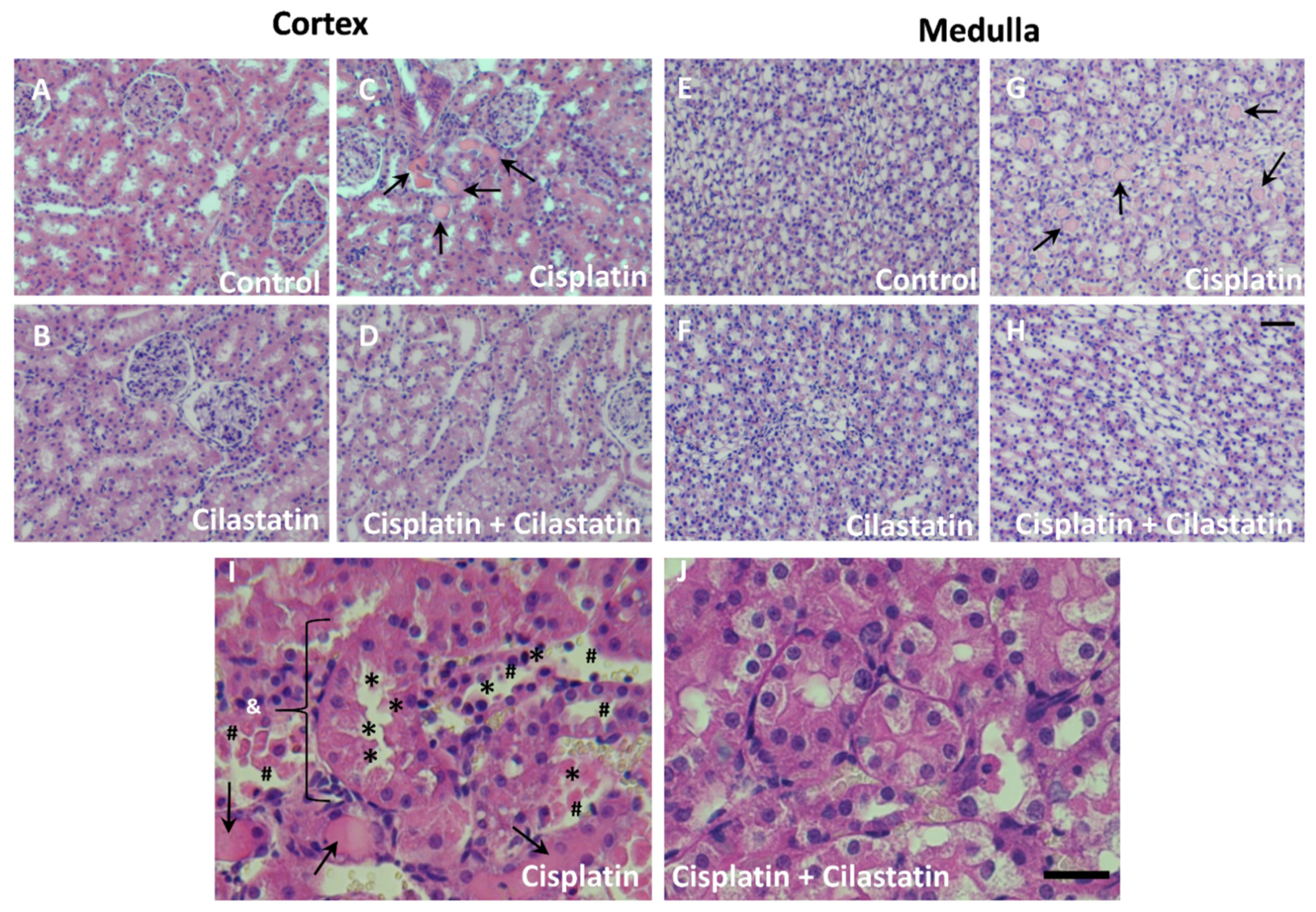
2.1. Cilastatin Protective Effect against Cisplatin-Induced Renal Damage
2.2. Cisplatin Alteration of Global Renal Lipids Classes in the Cortex and Medulla. Protective Effect of Cilastatin
2.3. Individual Renal Lipid Species Alteration by Cisplatin Treatment. Cilastatin Attenuating Effect
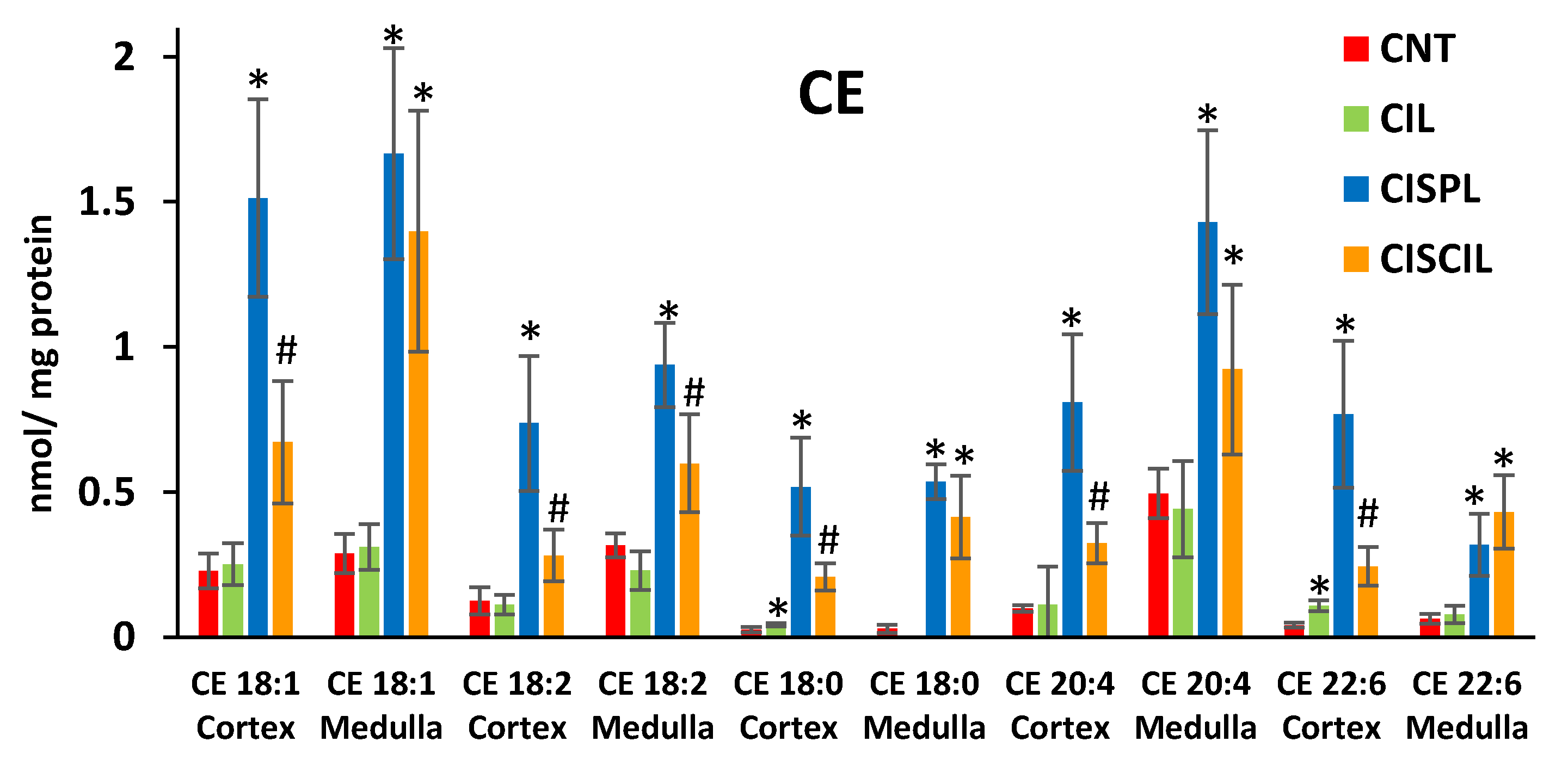
2.3.1. Cholesterol Esters
2.3.2. Sphingolipids: Cer, dhCer, HexCer, dhHexCer, SM, dhSM, and Sulf
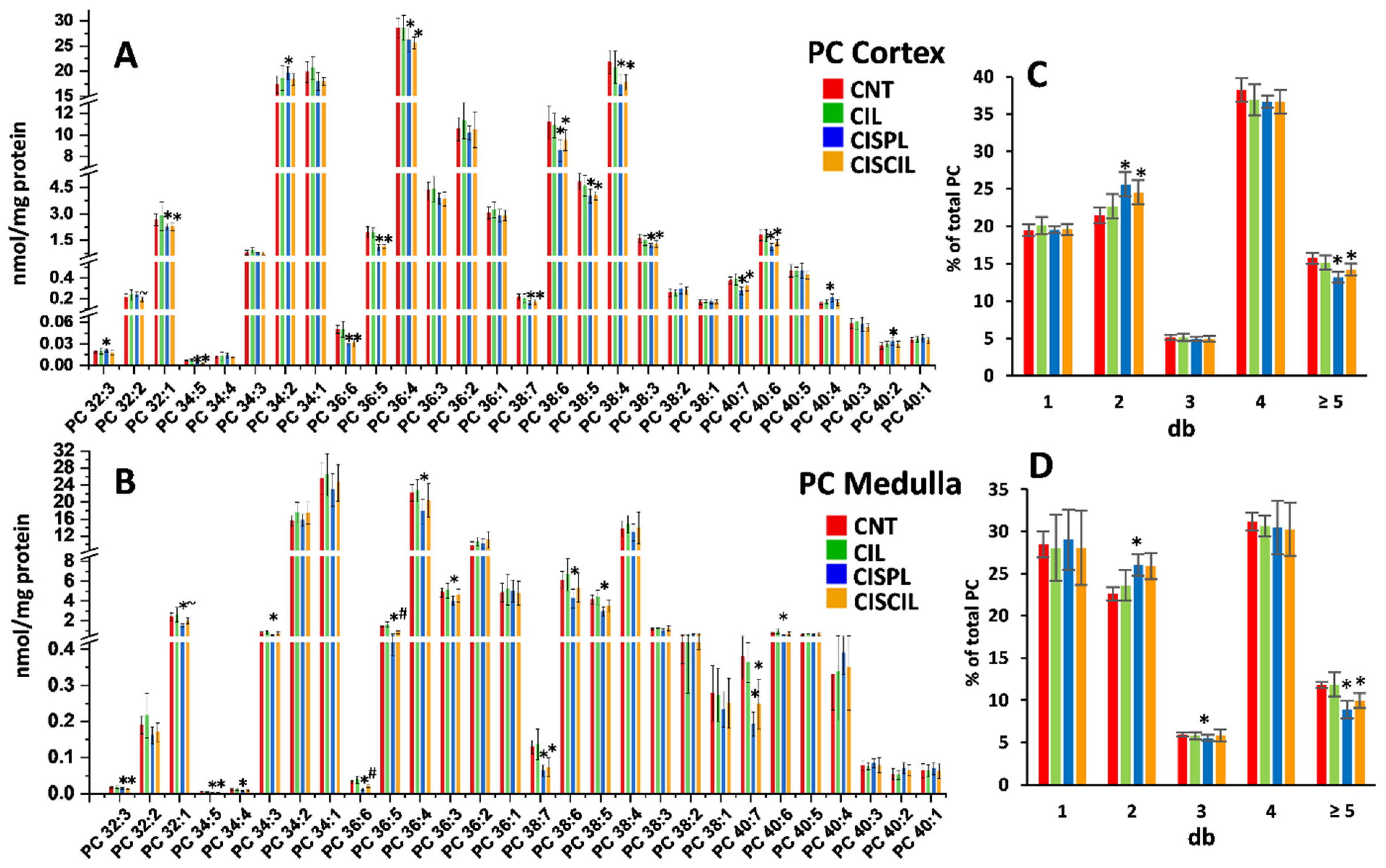
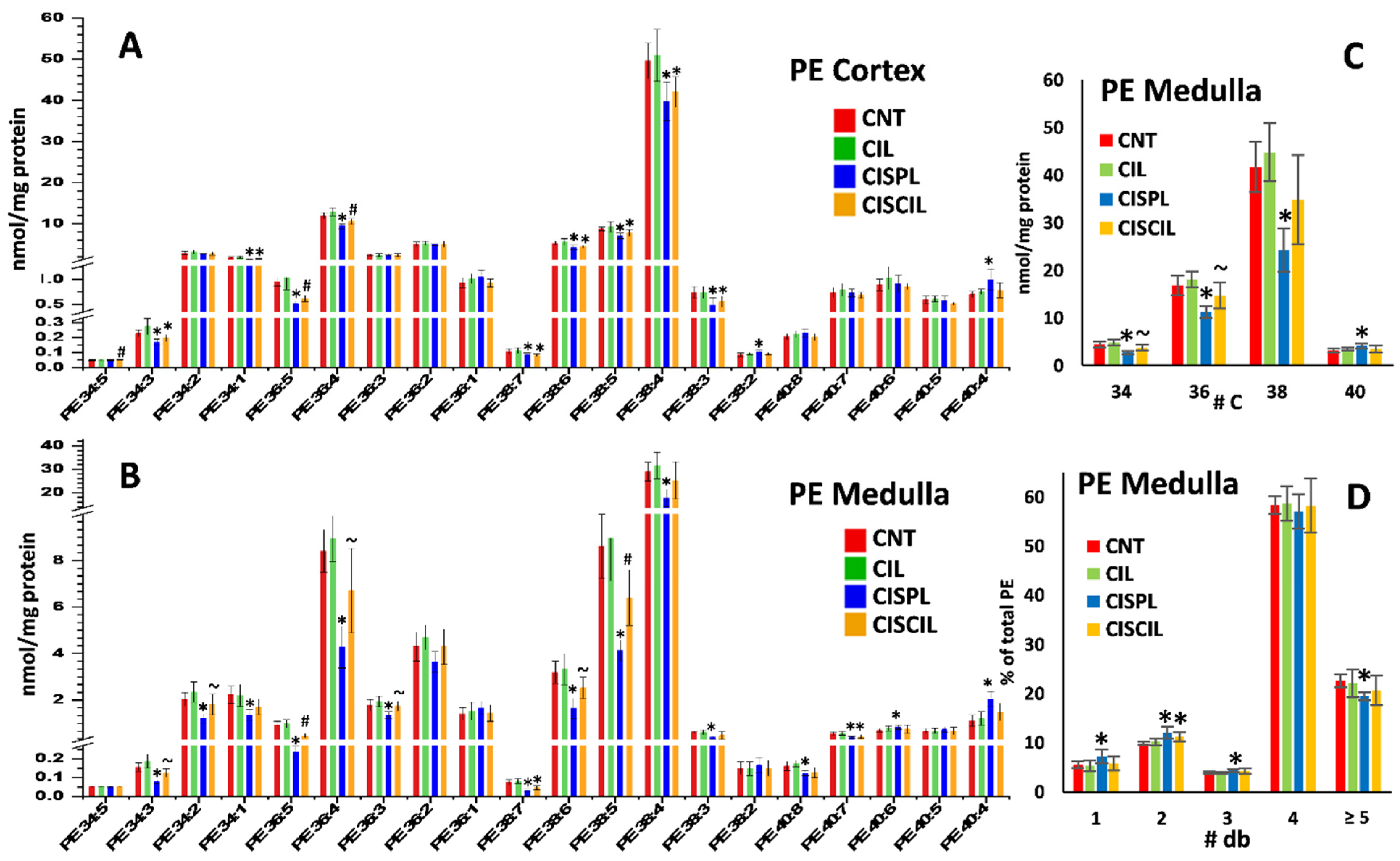
2.3.3. Phospholipids: PC, PE, and LPC
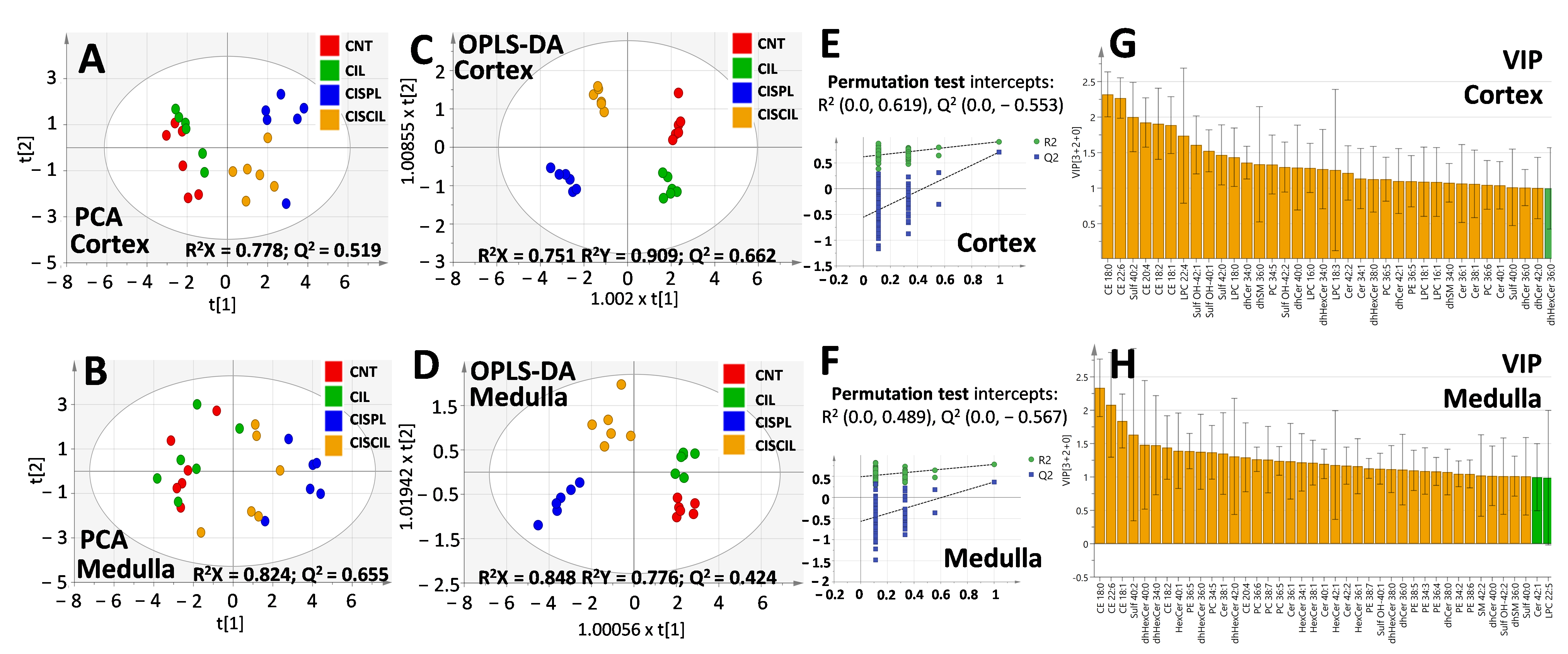
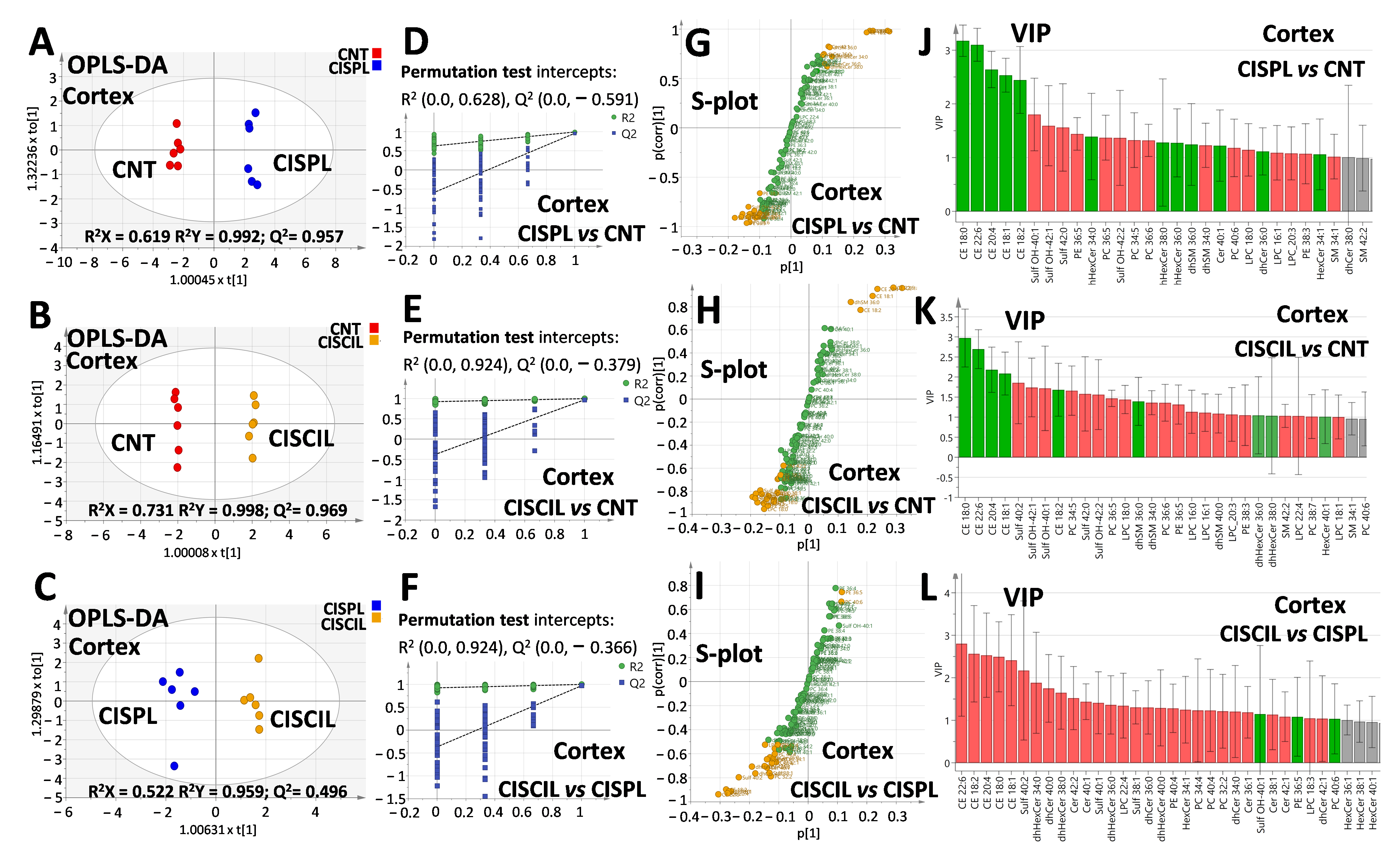
2.4. Renal Lipids as Classification Variables for Kidney Damage and Protection
2.5. Renal Lipids as Potential Biomarkers of Cisplatin-Induced Renal Damage
2.6. Renal Lipids as Potential Biomarkers of Cilastatin Nephroprotection
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Animal Model
4.3. Histological Studies
4.4. Renal Function Indicators
4.5. Tissue Homogenization
4.6. Lipidomic Analysis by LC-MS/MS
4.7. Statistical Analysis
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yao, X.; Panichpisal, K.; Kurtzman, N.; Nugent, K. Cisplatin Nephrotoxicity: A Review. Am. J. Med. Sci. 2007, 334, 115–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, R.P.; Tadagavadi, R.K.; Ramesh, G.; Reeves, W.B. Mechanisms of Cisplatin Nephrotoxicity. Toxins 2010, 2, 2490–2518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Gordaliza, E.; Giesen, C.; Lazaro, A.; Esteban-Fernandez, D.; Humanes, B.; Canas, B.; Panne, U.; Tejedor, A.; Jakubowski, N.; Gomez-Gomez, M.M. Elemental Bioimaging in Kidney by LA-ICP-MS As a Tool to Study Nephrotoxicity and Renal Protective Strategies in Cisplatin Therapies. Anal. Chem. 2011, 83, 7933–7940. [Google Scholar] [CrossRef]
- Moreno-Gordaliza, E.; Esteban-Fernández, D.; Lázaro, A.; Aboulmagd, S.; Humanes, B.; Tejedor, A.; Linscheid, M.W.; Gómez-Gómez, M.M. Lipid imaging for visualizing cilastatin amelioration of cisplatin-induced nephrotoxicity. J. Lipid Res. 2018, 59, 1561–1574. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Chen, J.-Q.; Huang, W.-Q.; Li, W.; Huang, Y.; Zhang, Z.-J.; Xu, F.-G. Renal Medulla is More Sensitive to Cisplatin than Cortex Revealed by Untargeted Mass Spectrometry-Based Metabolomics in Rats. Sci. Rep. 2017, 7, 44804. [Google Scholar] [CrossRef] [PubMed]
- Eljack, N.D.; Ma, H.Y.M.; Drucker, J.; Shen, C.; Hambley, T.W.; New, E.J.; Friedrich, T.; Clarke, R.J. Mechanisms of cell uptake and toxicity of the anticancer drug cisplatin. Metallomics 2014, 6, 2126–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McSweeney, K.R.; Gadanec, L.K.; Qaradakhi, T.; Ali, B.A.; Zulli, A.; Apostolopoulos, V. Mechanisms of Cisplatin-Induced Acute Kidney Injury: Pathological Mechanisms, Pharmacological Interventions, and Genetic Mitigations. Cancers 2021, 13, 1572. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Song, M.; Liu, Y.; Liu, H.; Sun, L.; Peng, Y.; Liu, F.; Venkatachalam, M.A.; Dong, Z. Renoprotective approaches and strategies in acute kidney injury. Pharmacol. Ther. 2016, 163, 58–73. [Google Scholar] [CrossRef] [Green Version]
- Pabla, N.; Dong, Z. Cisplatin nephrotoxicity: Mechanisms and renoprotective strategies. Kidney Int. 2008, 73, 994–1007. [Google Scholar] [CrossRef] [Green Version]
- Camano, S.; Lazaro, A.; Moreno-Gordaliza, E.; Torres, A.M.; de Lucas, C.; Humanes, B.; Lazaro, J.A.; Gomez-Gomez, M.M.; Bosca, L.; Tejedor, A. Cilastatin Attenuates Cisplatin-Induced Proximal Tubular Cell Damage. J. Pharm. Exp. 2010, 334, 419–429. [Google Scholar] [CrossRef]
- Humanes, B.; Lazaro, A.; Camano, S.; Moreno-Gordaliza, E.; Lazaro, J.A.; Blanco-Codesido, M.; Lara, J.M.; Ortiz, A.; Gomez-Gomez, M.M.; Martin-Vasallo, P.; et al. Cilastatin protects against cisplatin-induced nephrotoxicity without compromising its anticancer efficiency in rats. Kidney Int. 2012, 82, 652–663. [Google Scholar] [CrossRef] [Green Version]
- Jado, J.C.; Humanes, B.; González-Nicolás, M.Á.; Camaño, S.; Lara, J.M.; López, B.; Cercenado, E.; García-Bordas, J.; Tejedor, A.; Lázaro, A. Nephroprotective Effect of Cilastatin against Gentamicin-Induced Renal Injury In Vitro and In Vivo without Altering Its Bactericidal Efficiency. Antioxidants 2020, 9, 821. [Google Scholar] [CrossRef]
- Humanes, B.; Jado, J.C.; Camano, S.; Lopez-Parra, V.; Torres, A.M.; Alvarez-Sala, L.A.; Cercenado, E.; Tejedor, A.; Lazaro, A. Protective Effects of Cilastatin against Vancomycin-Induced Nephrotoxicity. Biomed. Res. Int. 2015, 2015, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Tejedor, A.; Torres, A.M.; Castilla, M.; Lazaro, J.A.; de Lucas, C.; Caramelo, C. Cilastatin protection against cyclosporin A-induced nephrotoxicity: Clinical evidence. Curr. Med. Res. Opin. 2007, 23, 505–513. [Google Scholar] [CrossRef]
- Humanes, B.; Camaño, S.; Lara, J.M.; Sabbisetti, V.; González-Nicolás, M.Á.; Bonventre, J.V.; Tejedor, A.; Lázaro, A. Cisplatin-induced renal inflammation is ameliorated by cilastatin nephroprotection. Nephrol. Dial. Transpl. 2017, 32, 1645–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez, M.; Castilla, M.; Torres, A.M.; Lázaro, J.A.; Sarmiento, E.; Tejedor, A. Inhibition of brush border dipeptidase with cilastatin reduces toxic accumulation of cyclosporin A in kidney proximal tubule epithelial cells. Nephrol. Dial. Transpl. 2004, 19, 2445–2455. [Google Scholar] [CrossRef] [Green Version]
- Zaballos, M.; Power, M.; Canal-Alonso, M.I.; González-Nicolás, M.; Vasquez-Jimenez, W.; Lozano-Lominchar, P.; Cabrerizo-Torrente, P.; Palencia-García, N.; Gago-Quiroga, S.; Ginel-Feito, M.D.; et al. Effect of Cilastatin on Cisplatin-Induced Nephrotoxicity in Patients Undergoing Hyperthermic Intraperitoneal Chemotherapy. Int. J. Mol. Sci. 2021, 22, 1239. [Google Scholar] [CrossRef] [PubMed]
- Obermuller, N.; Geiger, H.; Weipert, C.; Urbschat, A. Current developments in early diagnosis of acute kidney injury. Int. Urol. Nephrol. 2014, 46, 1–7. [Google Scholar] [CrossRef]
- Perez, J.D.; Colucci, J.A.; Sakata, M.M.; Cunha, T.S.; Arita, D.Y.; Casarini, D.E. Proteomic Approaches in Understanding a Detected Relationship between Chemotherapy-Induced Nephrotoxicity and Cell Respiration in HK-2 Cells. Nephron Physiol. 2011, 119, P1–P10. [Google Scholar] [CrossRef]
- Portilla, D.; Schnackenberg, L.; Beger, R.D. Metabolomics as an Extension of Proteomic Analysis: Study of Acute Kidney Injury. Semin. Nephrol. 2007, 27, 609–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.-Y.; Lin, R.-C. Metabolomics in Nephrotoxicity. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 65, pp. 69–89. [Google Scholar]
- Won, A.J.; Kim, S.; Kim, Y.G.; Kim, K.B.; Choi, W.S.; Kacew, S.; Kim, K.S.; Jung, J.H.; Lee, B.M.; Kim, H.S. Discovery of urinary metabolomic biomarkers for early detection of acute kidney injury. Mol. Biosyst. 2016, 12, 133–144. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Zhou, H.; Fan, S.; Wang, Y.; Zhang, L.; Ju, L.; Wu, X.; Wu, H.; Zhang, Y. Metabonomics study on nephrotoxicity induced by intraperitoneal and intravenous cisplatin administration using rapid resolution liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry (RRLC-Q-TOF-MS). Rsc. Adv. 2014, 4, 8260–8270. [Google Scholar] [CrossRef]
- Qu, X.; Gao, H.; Sun, J.; Tao, L.; Zhang, Y.; Zhai, J.; Song, Y.; Hu, T.; Li, Z. Identification of key metabolites during cisplatin-induced acute kidney injury using an HPLC-TOF/MS-based non-targeted urine and kidney metabolomics approach in rats. Toxicology 2020, 431, 152366. [Google Scholar] [CrossRef]
- Song, Y.; Hu, T.; Gao, H.; Zhai, J.; Gong, J.; Zhang, Y.; Tao, L.; Sun, J.; Li, Z.; Qu, X. Altered metabolic profiles and biomarkers associated with astragaloside IV-mediated protection against cisplatin-induced acute kidney injury in rats: An HPLC-TOF/MS-based untargeted metabolomics study. Biochem. Pharmacol. 2021, 183, 114299. [Google Scholar] [CrossRef]
- Xia, D.M.; Lai, X.L.; Wu, K.W.; Zhou, P.Y.; Li, L.; Guo, Z.Y.; Xu, S.G. Metabolomics study of fasudil on cisplatin-induced kidney injury. Biosci. Rep. 2019, 39, BSR20192940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pariyani, R.; Ismail, I.S.; Azam, A.; Khatib, A.; Abas, F.; Shaari, K.; Hamza, H. Urinary metabolic profiling of cisplatin nephrotoxicity and nephroprotective effects of Orthosiphon stamineus leaves elucidated by 1H NMR spectroscopy. J. Pharm. Biomed. Anal. 2017, 135, 20–30. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Vaziri, N.D.; Lin, R.-C. Lipidomics: New Insight Into Kidney Disease. In Advances in Clinical Chemistry; Gregory, S.M., Ed.; Elsevier: Amsterdam, The Netherlands, 2015; Volume 68, pp. 153–175. [Google Scholar]
- Hou, B.; He, P.; Ma, P.; Yang, X.; Xu, C.; Lam, S.M.; Shui, G.; Yang, X.; Zhang, L.; Qiang, G.; et al. Comprehensive Lipidome Profiling of the Kidney in Early-Stage Diabetic Nephropathy. Front. Endocrinol. 2020, 11, 359. [Google Scholar] [CrossRef] [PubMed]
- Bulbul, M.C.; Dagel, T.; Afsar, B.; Ulusu, N.N.; Kuwabara, M.; Covic, A.; Kanbay, M. Disorders of Lipid Metabolism in Chronic Kidney Disease. Blood Purif. 2018, 46, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Portilla, D.; Li, S.; Nagothu, K.K.; Megyesi, J.; Kaissling, B.; Schnackenberg, L.; Safirstein, R.L.; Beger, R.D. Metabolomic study of cisplatin-induced nephrotoxicity. Kidney Int. 2006, 69, 2194–2204. [Google Scholar] [CrossRef] [Green Version]
- Nagothu, K.K.; Bhatt, R.; Kaushal, G.P.; Portilla, D. Fibrate prevents cisplatin-induced proximal tubule cell death. Kidney Int. 2005, 68, 2680–2693. [Google Scholar] [CrossRef] [Green Version]
- Dupre, T.V.; Doll, M.A.; Shah, P.P.; Sharp, C.N.; Siow, D.; Megyesi, J.; Shayman, J.; Bielawska, A.; Bielawski, J.; Beverly, L.J.; et al. Inhibiting glucosylceramide synthase exacerbates cisplatin-induced acute kidney injury. J. Lipid Res. 2017, 58, 1439–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Gordaliza, E.; Esteban-Fernández, D.; Lázaro, A.; Humanes, B.; Aboulmagd, S.; Tejedor, A.; Linscheid, M.W.; Gómez-Gómez, M.M. MALDI-LTQ-Orbitrap mass spectrometry imaging for lipidomic analysis in kidney under cisplatin chemotherapy. Talanta 2017, 164, 16–26. [Google Scholar] [CrossRef]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Zager, R.A.; Johnson, A.; Anderson, K.; Wright, S. Cholesterol ester accumulation: An immediate consequence of acute in vivo ischemic renal injury. Kidney Int. 2001, 59, 1750–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [Green Version]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef]
- Li, Y.; Lin, M.; He, S.; Jin, J. Sphingolipid metabolism affects the anticancer effect of cisplatin. World J. Transl. Med. 2016, 5, 37–45. [Google Scholar] [CrossRef]
- Dupre, T.V.; Siskind, L.J. The role of sphingolipids in acute kidney injury. Adv. Biol. Regul. 2018, 70, 31–39. [Google Scholar] [CrossRef]
- Slotte, J.P. Biological functions of sphingomyelins. Prog. Lipid Res. 2013, 52, 424–437. [Google Scholar] [CrossRef]
- Takahashi, T.; Suzuki, T. Role of sulfatide in normal and pathological cells and tissues. J. Lipid Res. 2012, 53, 1437–1450. [Google Scholar] [CrossRef] [Green Version]
- Stillwell, W. Chapter 5—Membrane Polar Lipids. In An Introduction to Biological Membranes, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 63–87. [Google Scholar]
- Rao, S.; Walters, K.B.; Wilson, L.; Chen, B.; Bolisetty, S.; Graves, D.; Barnes, S.; Agarwal, A.; Kabarowski, J.H. Early lipid changes in acute kidney injury using SWATH lipidomics coupled with MALDI tissue imaging. Am. J. Physiol. Ren. Physiol. 2016, 310, F1136–F1147. [Google Scholar] [CrossRef] [Green Version]
- Ichimura, T.; Hung, C.C.; Yang, S.A.; Stevens, J.L.; Bonventre, J.V. Kidney injury molecule-1: A tissue and urinary biomarker for nephrotoxicant-induced renal injury. Am. J. Physiol.-Ren. Physiol. 2004, 286, F552. [Google Scholar] [CrossRef]
- Mishra, J.; Mori, K.; Ma, Q.; Kelly, C.; Barasch, J.; Devarajan, P. Neutrophil Gelatinase-Associated Lipocalin: A Novel Early Urinary Biomarker for Cisplatin Nephrotoxicity. Am. J. Nephrol. 2004, 24, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Delgado, J.; Lazaro, A.; Gomez-Garre, D.; Osende, J.I.; Gonzalez-Rubio, M.L.; Herraiz, M.; Manzarbeitia, F.; Fortes, J.; Fernandez-Cruz, A.; Egido, J. Long-term organ protection by doxazosin and/or quinapril as antihypertensive therapy. J. Nephrol. 2006, 19, 588–598. [Google Scholar] [PubMed]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Babiy, B.; Busto, R.; Pastor, Ó. A normalized signal calibration with a long-term reference improves the robustness of RPLC-MRM/MS lipidomics in plasma. Anal. Bioanal. Chem. 2021, 413, 4077–4090. [Google Scholar] [CrossRef]
- Adams, K.J.; Pratt, B.; Bose, N.; Dubois, L.G.; St. John-Williams, L.; Perrott, K.M.; Ky, K.; Kapahi, P.; Sharma, V.; MacCoss, M.J.; et al. Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J. Proteome Res. 2020, 19, 1447–1458. [Google Scholar] [CrossRef]
- Liebisch, G.; Vizcaíno, J.A.; Köfeler, H.; Trötzmüller, M.; Griffiths, W.J.; Schmitz, G.; Spener, F.; Wakelam, M.J.O. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]




| Group | S Cr (mg/dL) | BUN (mg/dL) | GFR (mL/min/100 g) | Prot urine (mg/dL) | FE Na+ (%) | FE K+ (%) | Vol Urine 24 h (mL) |
|---|---|---|---|---|---|---|---|
| Control | 0.31 ± 0.06 | 33 ± 7 | 2.3 ± 0.5 | 30 ± 6 | 0.5 ± 0.1 | 20 ± 5 | 14 ± 2 |
| Cilastatin | 0.33 ± 0.05 | 30 ± 6 | 2.0 ± 0.5 | 28 ± 7 | 0.53 ± 0.09 | 21 ± 3 | 14 ± 6 |
| Cisplatin | 1.4 ± 0.8 a | 110 ± 70 a | 0.4 ± 0.2 a | 40 ± 8 a | 2 ± 1 a | 70 ± 40 a | 26 ± 4 a |
| Cisplatin + cilastatin | 0.6 ± 0.2 | 50 ± 10 | 1.0 ± 0.3 b | 30 ± 5 | 0.8 ± 0.2 | 31 ± 9 | 20 ± 10 |
| Lipid (Cortex) | FC | Trend | p-Value | p(corr) | VIP Score | AUC ROC |
|---|---|---|---|---|---|---|
| CE 18:0 | 20.16 | ↑ | 0.00079 | 0.974 | 3.18 | 1 |
| CE 18:1 | 6.62 | ↑ | 0.00020 | 0.983 | 2.53 | 1 |
| CE 18:2 | 5.86 | ↑ | 2.28 × 10−6 | 0.964 | 2.44 | 1 |
| CE 20:4 | 8.12 | ↑ | 0.00216 | 0.985 | 2.64 | 1 |
| CE 22:6 | 18.35 | ↑ | 0.00088 | 0.985 | 3.10 | 1 |
| Cer 40:1 | 1.70 | ↑ | 0.00074 | 0.823 | 1.22 | 0.944 |
| dhCer 36:0 | 1.59 | ↑ | 0.01193 | 0.748 | 1.12 | 0.917 |
| dhHexCer 34:0 | 2.00 | ↑ | 0.00384 | 0.721 | 1.39 | 0.972 |
| dhHexCer 36:0 | 1.79 | ↑ | 0.01704 | 0.651 | 1.27 | 0.861 |
| dhHexCer 38:0 | 1.85 | ↑ | 0.01409 | 0.620 | 1.28 | 0.861 |
| dhSM 34:0 | 0.61 | ↓ | 0.00866 | −0.866 | 1.23 | 0.944 |
| dhSM 36:0 | 1.77 | ↑ | 0.00158 | 0.817 | 1.24 | 1 |
| HexCer 34:1 | 1.50 | ↑ | 0.00491 | 0.737 | 1.06 | 0.944 |
| LPC 16:1 | 0.70 | ↓ | 0.00216 | −0.903 | 1.09 | 1 |
| LPC 18:0 | 0.67 | ↓ | 0.00001 | −0.909 | 1.14 | 1 |
| LPC 20:3 | 0.69 | ↓ | 0.00018 | −0.900 | 1.08 | 1 |
| PC 34:5 | 0.57 | ↓ | 0.00010 | −0.873 | 1.33 | 1 |
| PC 36:5 | 0.55 | ↓ | 0.00016 | −0.902 | 1.37 | 1 |
| PC 36:6 | 0.58 | ↓ | 0.00001 | −0.955 | 1.32 | 1 |
| PC 40:6 | 0.62 | ↓ | 0.00433 | −0.840 | 1.18 | 0.972 |
| PE 36:5 | 0.54 | ↓ | 6.33 × 10−7 | −0.965 | 1.44 | 1 |
| PE 38:3 | 0.66 | ↓ | 0.00782 | −0.662 | 1.07 | 0.944 |
| SM 34:1 | 0.71 | ↓ | 0.00050 | −0.868 | 1.02 | 1 |
| Sulf 42:0 | 0.43 | ↓ | 0.00799 | −0.870 | 1.56 | 1 |
| Sulf OH-40:1 | 0.36 | ↓ | 0.00196 | −0.912 | 1.80 | 1 |
| Sulf OH-42:1 | 0.43 | ↓ | 0.00219 | −0.903 | 1.59 | 1 |
| Sulf OH-42:2 | 0.51 | ↓ | 0.00896 | −0.799 | 1.37 | 0.944 |
| Lipid (Medulla) | FC | Trend | p-Value | p(corr) | VIP Score | AUC ROC |
|---|---|---|---|---|---|---|
| CE 18:0 | 18.44 | ↑ | 1.98 × 10−9 | 0.969 | 2.60 | 1 |
| CE 18:1 | 5.77 | ↑ | 0.0002 | 0.977 | 2.01 | 1 |
| CE 18:2 | 2.95 | ↑ | 0.0001 | 0.989 | 1.60 | 1 |
| CE 20:4 | 2.88 | ↑ | 0.0005 | 0.942 | 1.54 | 1 |
| CE 22:6 | 5.05 | ↑ | 0.0002 | 0.914 | 1.82 | 1 |
| Cer 36:1 | 1.74 | ↑ | 0.0019 | 0.804 | 1.13 | 0.917 |
| Cer 38:1 | 2.02 | ↑ | 0.0003 | 0.861 | 1.26 | 0.972 |
| Cer 40:1 | 2.52 | ↑ | 0.0022 | 0.928 | 1.47 | 1 |
| Cer 42:1 | 2.26 | ↑ | 8.63 × 10−6 | 0.965 | 1.36 | 1 |
| Cer 42:2 | 2.77 | ↑ | 0.0022 | 0.951 | 1.52 | 1 |
| dhCer 34:0 | 1.83 | ↑ | 7.71 × 10−6 | 0.948 | 1.19 | 1 |
| dhCer 36:0 | 2.19 | ↑ | 0.0001 | 0.916 | 1.35 | 1 |
| dhCer 38:0 | 2.06 | ↑ | 0.0006 | 0.840 | 1.27 | 0.944 |
| dhCer 40:0 | 1.71 | ↑ | 0.0152 | 0.797 | 1.09 | 0.917 |
| dhCer 42:1 | 1.86 | ↑ | 0.0003 | 0.894 | 1.17 | 1 |
| dhHexCer 36:0 | 2.27 | ↑ | 0.0022 | 0.877 | 1.35 | 1 |
| dhHexCer 38:0 | 1.77 | ↑ | 0.0130 | 0.650 | 1.16 | 0.917 |
| dhSM 36:0 | 1.84 | ↑ | 1.23 × 10−5 | 0.933 | 1.18 | 1 |
| dhSM 40:0 | 1.56 | ↑ | 0.0071 | 0.743 | 1.00 | 0.944 |
| HexCer 34:1 | 2.46 | ↑ | 1.25 × 10−6 | 0.903 | 1.42 | 1 |
| HexCer 36:1 | 2.47 | ↑ | 2.59 × 10−6 | 0.907 | 1.41 | 1 |
| HexCer 38:1 | 1.86 | ↑ | 0.0024 | 0.741 | 1.19 | 0.972 |
| PC 34:5 | 0.39 | ↓ | 2.83 × 10−6 | −0.933 | 1.46 | 1 |
| PC 36:5 | 0.37 | ↓ | 0.0022 | −0.961 | 1.55 | 1 |
| PC 36:6 | 0.37 | ↓ | 0.0001 | −0.954 | 1.55 | 1 |
| PC 38:7 | 0.49 | ↓ | 0.0001 | −0.933 | 1.28 | 1 |
| PC 40:7 | 0.51 | ↓ | 0.0022 | −0.953 | 1.26 | 1 |
| PE 34:1 | 0.61 | ↓ | 0.0022 | −0.881 | 1.08 | 1 |
| PE 34:2 | 0.59 | ↓ | 0.0001 | −0.933 | 1.10 | 1 |
| PE 34:3 | 0.49 | ↓ | 1.28 × 10−5 | −0.973 | 1.29 | 1 |
| PE 36:4 | 0.51 | ↓ | 0.0022 | −0.951 | 1.27 | 1 |
| PE 36:5 | 0.26 | ↓ | 0.0022 | −0.989 | 1.77 | 1 |
| PE 38:3 | 0.64 | ↓ | 0.0002 | −0.922 | 1.02 | 1 |
| PE 38:4 | 0.61 | ↓ | 0.0005 | −0.889 | 1.05 | 0.972 |
| PE 38:5 | 0.48 | ↓ | 0.0022 | −0.969 | 1.31 | 1 |
| PE 38:6 | 0.51 | ↓ | 0.0022 | −0.920 | 1.26 | 1 |
| PE 38:7 | 0.45 | ↓ | 0.0001 | −0.955 | 1.37 | 1 |
| PE 40:4 | 1.83 | ↑ | 0.0043 | 0.841 | 1.16 | 0.972 |
| SM 34:1 | 0.61 | ↓ | 0.0022 | −0.872 | 1.07 | 1 |
| Sulf 40:2 | 0.40 | ↓ | 0.0022 | −0.777 | 1.48 | 1 |
| Lipid (Cortex) | FC CISCIL vs. CISPL | FC CISPL vs. CNT | FC CISCIL vs. CNT | p Value CISCIL vs. CISPL | p(corr) | VIP Score | AUC ROC |
|---|---|---|---|---|---|---|---|
| CE 18:0 | 0.40 a | 20.16 a | 8.08 a | 0.0051 | −0.912 | 2.49 | 1 |
| CE 18:1 | 0.44 a | 6.62 a | 2.94 a | 0.0004 | −0.912 | 2.41 | 1 |
| CE 18:2 | 0.38 a | 5.86 a | 2.24 a | 0.0004 | −0.896 | 2.57 | 1 |
| CE 20:4 | 0.40 a | 8.12 a | 3.25 a | 0.0007 | −0.931 | 2.53 | 1 |
| CE 22:6 | 0.32 a | 18.35 a | 5.83 a | 0.0031 | −0.935 | 2.80 | 1 |
| Cer 40:1 | 0.71 a | 1.70 a | 1.20 a | 0.0060 | −0.714 | 1.44 | 0.889 |
| dhHexCer 34:0 | 0.57 a | 2.00 a | 1.14 | 0.0117 | −0.709 | 1.88 | 0.917 |
| PC 32:2 | 0.80 a | 1.12 | 0.89 | 0.0053 | −0.787 | 1.21 | 0.944 |
| PE 36:5 | 1.21 a | 0.54 a | 0.65 a | 0.0118 | 0.746 | 1.08 | 0.889 |
| Sulf 38:1 | 0.78 a | 1.16 | 0.90 | 0.0101 | −0.758 | 1.31 | 0.861 |
| Sulf 40:2 | 0.40 a | 1.00 | 0.41 a | 0.0028 | −0.797 | 2.17 | 1 |
| Lipid (Medulla) | FC CISCIL vs. CISPL | FC CISPL vs. CNT | FC CISCIL vs. CNT | p-Value CISCIL vs. CISPL | p(corr) | VIP Score | AUC ROC |
|---|---|---|---|---|---|---|---|
| CE 18:2 | 0.64 a | 2.95 a | 1.89 a | 0.0039 | −0.864 | 1.53 | 0.917 |
| Cer 42:2 | 0.69 a | 2.77 a | 1.92 a | 0.0087 | −0.720 | 1.36 | 0.944 |
| dhHexCer 36:0 | 0.62 a | 2.27 a | 1.40 | 0.0022 | −0.637 | 1.45 | 1 |
| HexCer 34:1 | 0.72 a | 2.46 a | 1.78 a | 0.0123 | −0.659 | 1.26 | 0.972 |
| PC 32:1 | 1.32 a | 0.63 a | 0.84 | 0.0047 | 0.741 | 1.04 | 0.944 |
| PC 36:5 | 1.58 a | 0.37 a | 0.58 a | 0.0087 | 0.817 | 1.44 | 0.944 |
| PC 36:6 | 1.71 a | 0.37 a | 0.63 a | 0.0030 | 0.875 | 1.63 | 0.944 |
| PE 34:2 | 1.51 a | 0.59 a | 0.89 | 0.0082 | 0.689 | 1.24 | 0.944 |
| PE 34:3 | 1.63 a | 0.49 a | 0.80 | 0.0022 | 0.935 | 1.54 | 1 |
| PE 36:3 | 1.29 a | 0.76 a | 0.98 | 0.0040 | 0.794 | 1.04 | 0.944 |
| PE 36:4 | 1.57 a | 0.51 a | 0.80 | 0.0087 | 0.654 | 1.29 | 0.944 |
| PE 36:5 | 1.94 a | 0.26 a | 0.51 a | 0.0022 | 0.960 | 1.78 | 1 |
| PE 38:5 | 1.55 a | 0.48 a | 0.74 a | 0.0022 | 0.854 | 1.39 | 1 |
| PE 38:6 | 1.54 a | 0.51 a | 0.79 | 0.0130 | 0.811 | 1.44 | 0.917 |
| SM 42:1 | 1.49 a | 0.71 a | 1.07 | 0.0115 | 0.756 | 1.33 | 0.889 |
| Sulf 40:2 | 2.47 a | 0.40 a | 0.98 | 0.0049 | 0.720 | 2.01 | 0.917 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Gordaliza, E.; Marazuela, M.D.; Pastor, Ó.; Lázaro, A.; Gómez-Gómez, M.M. Lipidomics Reveals Cisplatin-Induced Renal Lipid Alterations during Acute Kidney Injury and Their Attenuation by Cilastatin. Int. J. Mol. Sci. 2021, 22, 12521. https://doi.org/10.3390/ijms222212521
Moreno-Gordaliza E, Marazuela MD, Pastor Ó, Lázaro A, Gómez-Gómez MM. Lipidomics Reveals Cisplatin-Induced Renal Lipid Alterations during Acute Kidney Injury and Their Attenuation by Cilastatin. International Journal of Molecular Sciences. 2021; 22(22):12521. https://doi.org/10.3390/ijms222212521
Chicago/Turabian StyleMoreno-Gordaliza, Estefanía, Maria Dolores Marazuela, Óscar Pastor, Alberto Lázaro, and María Milagros Gómez-Gómez. 2021. "Lipidomics Reveals Cisplatin-Induced Renal Lipid Alterations during Acute Kidney Injury and Their Attenuation by Cilastatin" International Journal of Molecular Sciences 22, no. 22: 12521. https://doi.org/10.3390/ijms222212521






