OsABF1 Represses Gibberellin Biosynthesis to Regulate Plant Height and Seed Germination in Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Results
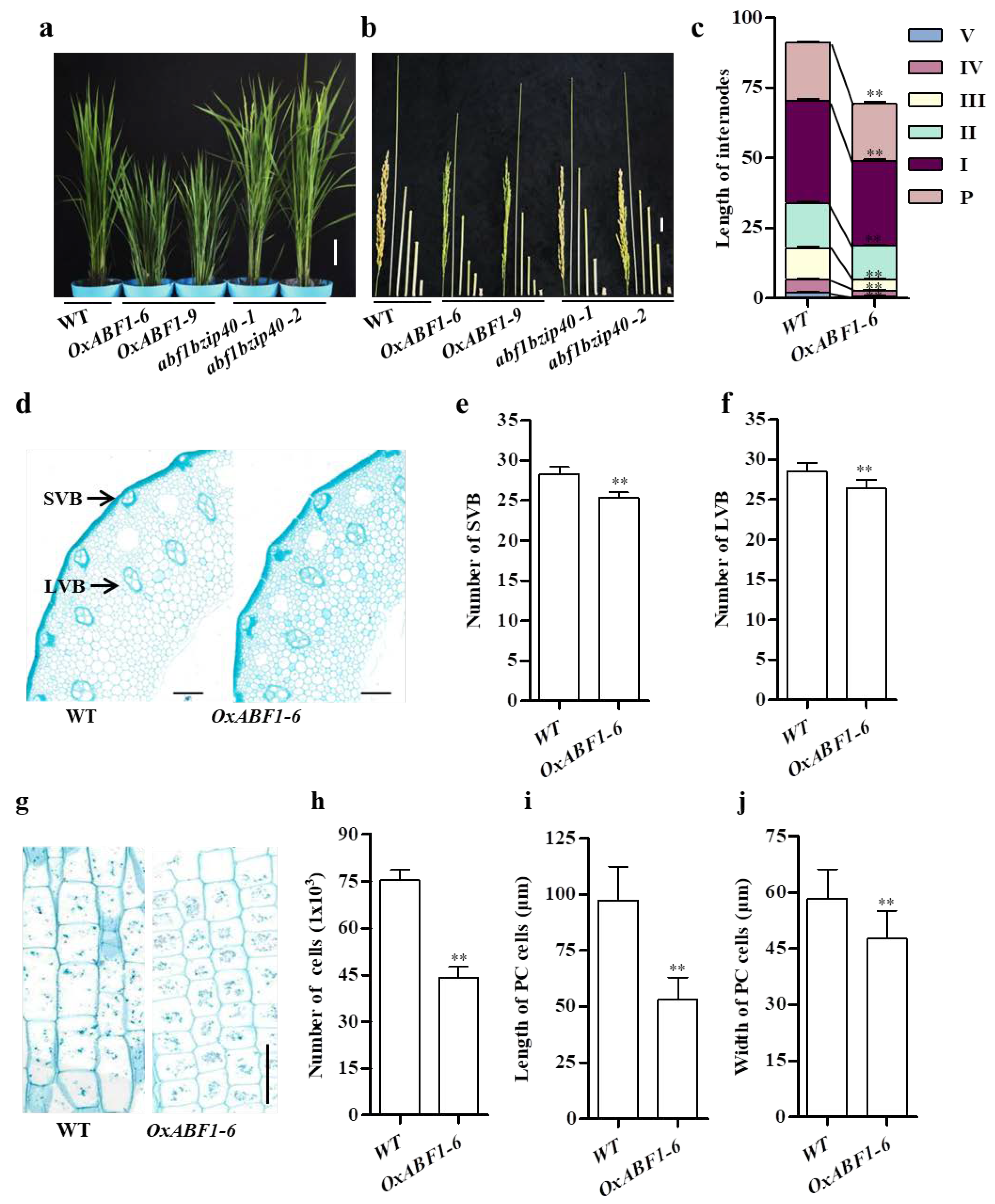
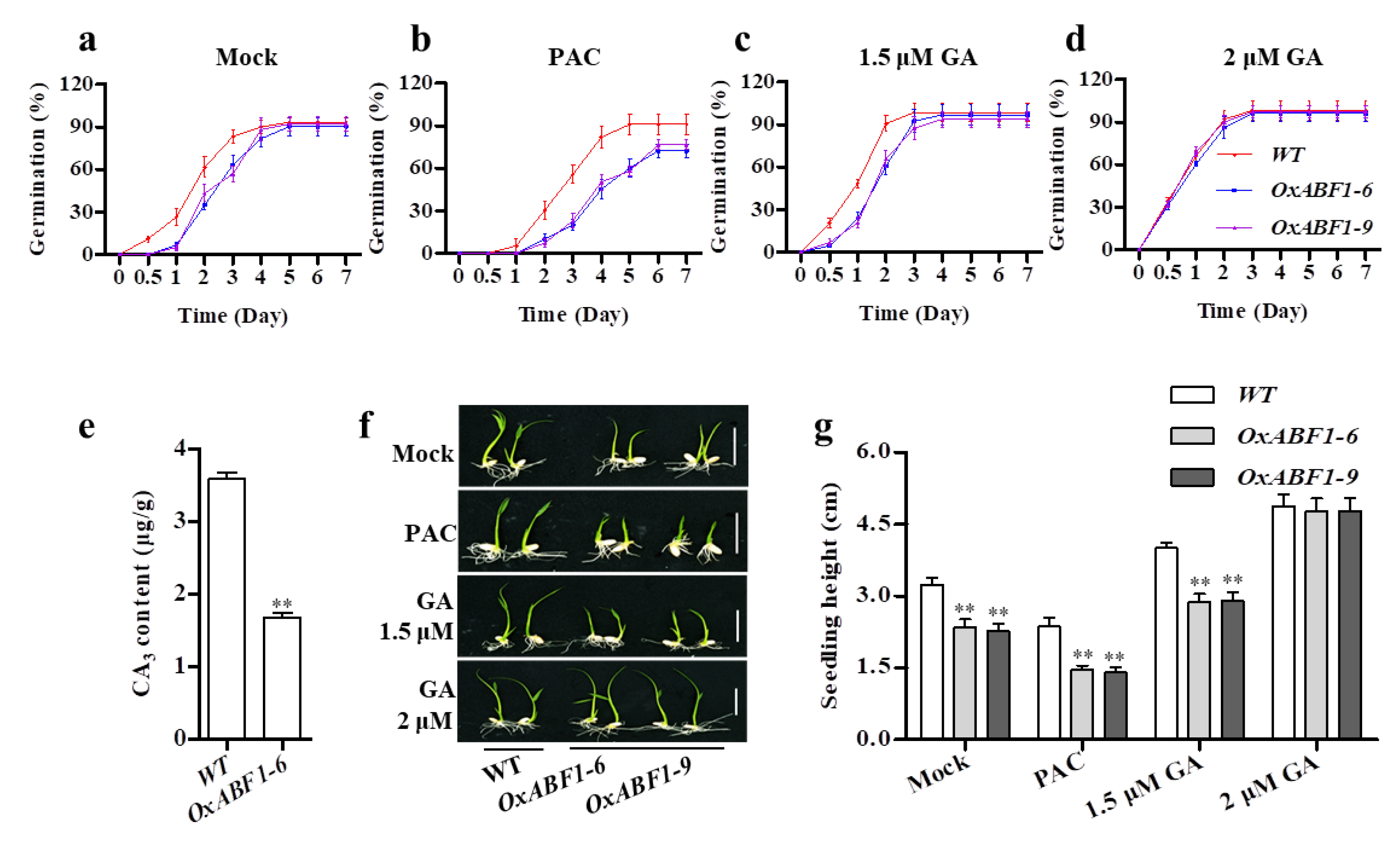
2.1. OsABF1 Overexpression Lines Exhibited GA-Deficient Phenotypes
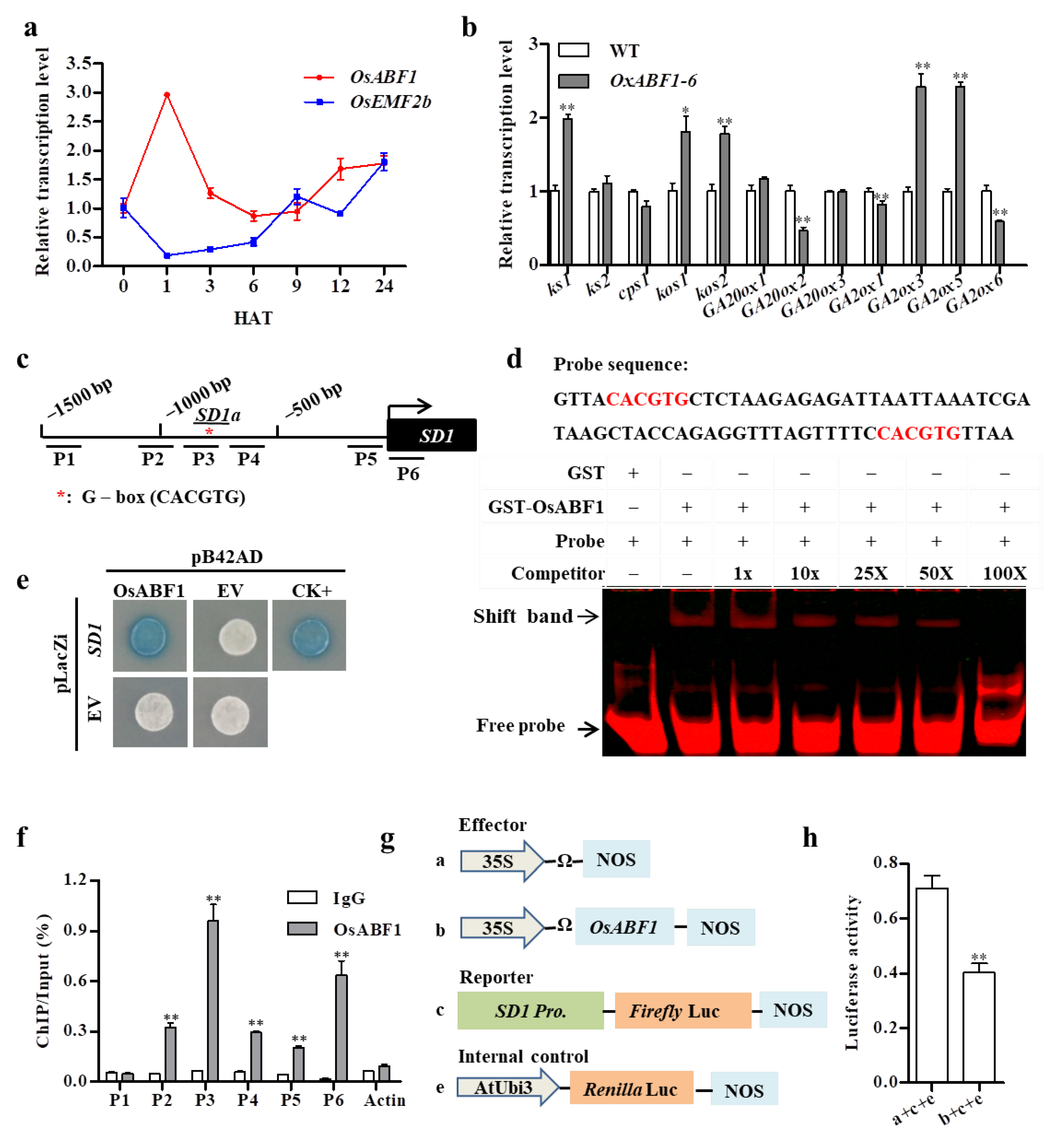
2.2. OsABF1 Directly Repressed the Transcription of SD1
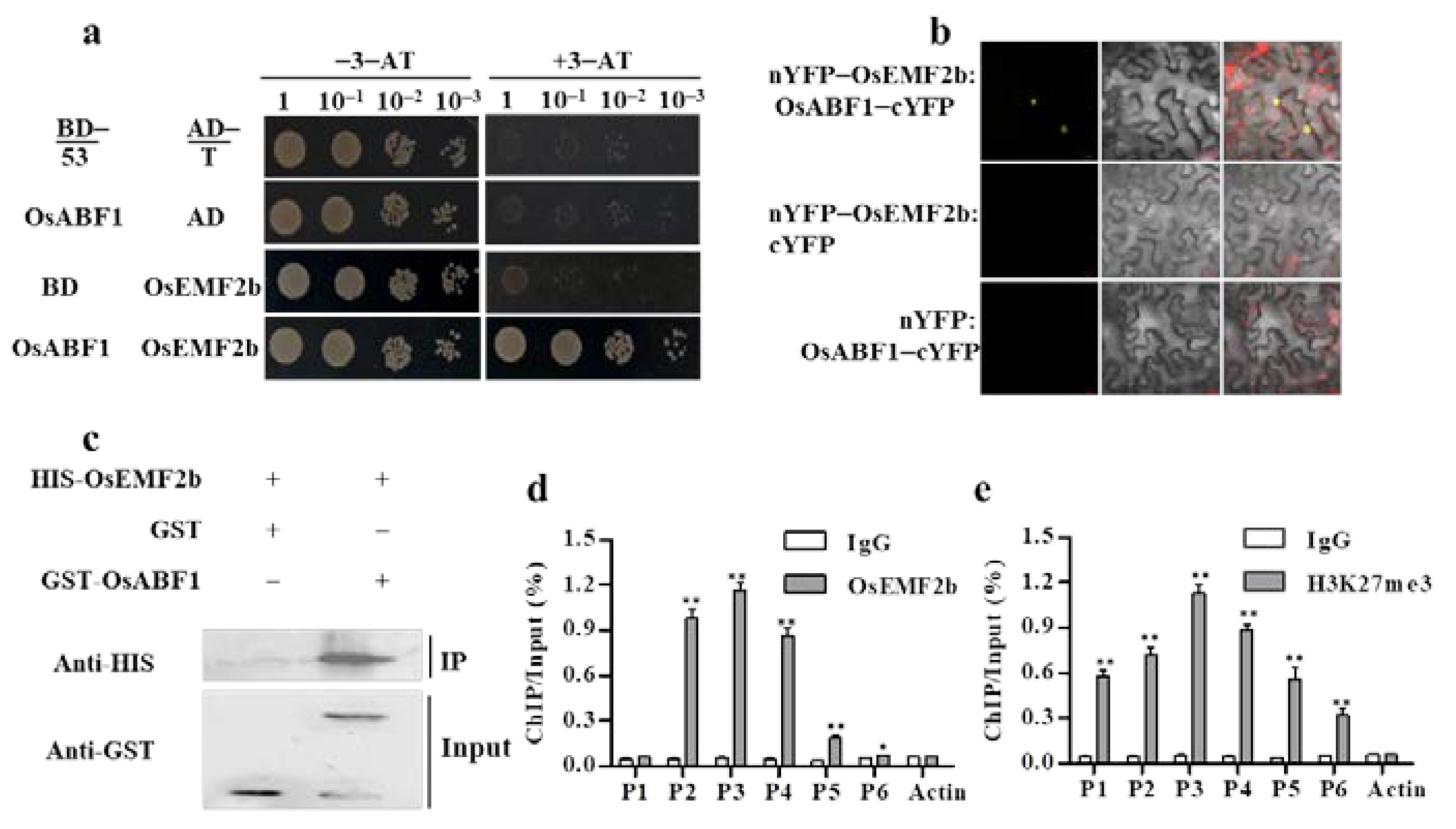
2.3. OsABF1 Bound to OsEMF2b, Causing the Deposition of H3K27me3 on the SD1 Promoter
2.4. OsEMF2b and OsABF1 Had Antagonistic Roles in GA Biosynthesis Regulation
3. Discussion
3.1. OsABF1 Targets SD1 to Regulate GA Biosynthesis
3.2. OsABF1 May Mediate the Antagonistic Relationship of ABA and GA
3.3. OsABF1 Interacts with OsEMF2b-PRC2 Complex to Suppress SD1 Transcription through H3K27me3
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Seed Germination and Seedling Growth Assays
4.3. Hormone Quantification
4.4. Paraffin Sections
4.5. Quantitative Reverse Transcription (qRT)-PCR
4.6. Yeast One-Hybrid Assay
4.7. Electrophoresis Mobility Shift Assay (EMSA)
4.8. Luciferase Transient Transcriptional Activity Assay
4.9. ChIP-qPCR
4.10. Yeast Two-Hybrid Analysis and Bimolecular Fluorescence Complementation
4.11. In Vitro Pull-Down Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gao, S.; Chu, C. Gibberellin Metabolism and Signaling: Targets for Improving Agronomic Performance of Crops. Plant Cell Physiol. 2020, 61, 1902–1911. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [Green Version]
- Achard, P.; Genschik, P. Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 2009, 60, 1085–1092. [Google Scholar] [CrossRef] [Green Version]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [CrossRef] [Green Version]
- Silverstone, A.L.; Sun, T. Gibberellins and the Green Revolution. Trends Plant Sci. 2000, 5, 1–2. [Google Scholar] [CrossRef]
- Sponsel, V.M. The localization, metabolism and biological activity of gibberellins in maturing and germinating seeds of Pisum sativum cv. Progress No. 9. Planta 1983, 159, 454–468. [Google Scholar] [CrossRef]
- Graebe, J.E. Gibberellin Biosynthesis from Gibberellin A12-Aldehyde; Springer: Berlin/Heidelberg, Germany, 1986. [Google Scholar]
- Ye, H.; Feng, J.; Zhang, L.; Zhang, J.; Mispan, M.S.; Cao, Z.; Beighley, D.H.; Yang, J.; Gu, X.Y. Map-Based Cloning of Seed Dormancy1–2 Identified a Gibberellin Synthesis Gene Regulating the Development of Endosperm-Imposed Dormancy in Rice. Plant Physiol. 2015, 169, 2152–2165. [Google Scholar]
- Annunziata, M.G. The Long and the Short of It: GA 2-oxidaseA9 Regulates Plant Height in Wheat. Plant Physiol. 2018, 177, 3–4. [Google Scholar] [CrossRef] [Green Version]
- Ge, W.; Steber, C.M. Positive and negative regulation of seed germination by the Arabidopsis GA hormone receptors, GID1a, b, and c. Plant Direct. 2018, 2, e00083. [Google Scholar] [CrossRef]
- Chen, H.; Ruan, J.; Chu, P.; Fu, W.; Liang, Z.; Li, Y.; Tong, J.; Xiao, L.; Liu, J.; Li, C.; et al. AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J. 2020, 101, 310–323. [Google Scholar] [CrossRef]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Withanage, S.P.; Hossain, M.A.; Kumar, M.S.; Roslan, H.A.; Abdullah, M.P.; Napis, S.B.; Shukor, N.A. Overexpression of Arabidopsis thaliana gibberellic acid 20 oxidase (AtGA20ox) gene enhance the vegetative growth and fiber quality in kenaf (Hibiscus cannabinus L.) plants. Breed. Sci. 2015, 65, 177–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plackett, A.R.; Powers, S.J.; Fernandez-Garcia, N.; Urbanova, T.; Takebayashi, Y.; Seo, M.; Jikumaru, Y.; Benlloch, R.; Nilsson, O.; Ruiz-Rivero, O.; et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 2012, 24, 941–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G.; et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008, 53, 488–504. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Yu, L.; Tang, M.; Tian, D.; Yang, S.; Zhang, X.; Traw, M.B. Pleiotropic changes revealed by in situ recovery of the semi-dwarf gene sd1 in rice. Plant Physiol. 2020, 248, 153141. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Liu, J.H.; Zhao, W.S.; Chen, X.J.; Guo, Z.J.; Peng, Y.L. Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Plant Microbe Interact. 2013, 26, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Toyomasu, T.; Kawaide, H.; Sekimoto, H.; Von Numers, C.; Phillips, A.L.; Hedden, P.; Kamiya, Y. Cloning and characterization of a cDNA encoding gibberellin 20-oxidase from rice (Oryza sativa) seedlings. Physiol. Plantarum. 2010, 99, 111–118. [Google Scholar] [CrossRef]
- Oikawa, T.; Koshioka, M.; Kojima, K.; Yoshida, H.; Kawata, M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 2004, 55, 687–700. [Google Scholar] [CrossRef]
- Ferrero, V.; Viola, I.L.; Ariel, F.D.; Gonzalez, D.H. Class I TCP Transcription Factors Target the Gibberellin Biosynthesis Gene GA20ox1 and the Growth-Promoting Genes HBI1 and PRE6 during Thermomorphogenic Growth in Arabidopsis. Plant Cell Physiol. 2019, 60, 1633–1645. [Google Scholar] [CrossRef]
- Andrés, F.; Porri, A.; Torti, S.; Mateos, J.; Romera-Branchat, M.; García-Martínez, J.L.; Fornara, F.; Gregis, V.; Kater, M.M.; Coupland, G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 2014, 111, E2760–E2769. [Google Scholar] [CrossRef] [Green Version]
- Fukazawa, J.; Nakata, M.; Ito, T.; Yamaguchi, S.; Takahashi, Y. The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J. 2010, 62, 1035–1045. [Google Scholar]
- Fukazawa, J.; Sakai, T.; Ishida, S.; Yamaguchi, I.; Kamiya, Y.; Takahashi, Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 2000, 12, 901–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E, Z.G.; Zhang, Y.P.; Zhou, J.H.; Wang, L. Mini review roles of the bZIP gene family in rice. Genet. Mol. Res. 2014, 13, 3025. [Google Scholar] [CrossRef] [PubMed]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Chu, J.; Ma, C.; Jiang, Y.; Ma, Y.; Xiong, J.; Cheng, Z.M. Overexpression of an ABA-dependent grapevine bZIP transcription factor, VvABF2, enhances osmotic stress in Arabidopsis. Plant Cell Rep. 2019, 38, 587–596. [Google Scholar] [CrossRef]
- Yang, S.; Xu, K.; Chen, S.; Li, T.; Xia, H.; Chen, L.; Liu, H.; Luo, L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019, 19, 260. [Google Scholar] [CrossRef] [Green Version]
- Zinsmeister, J.; Lalanne, D.; Terrasson, E.; Chatelain, E.; Vandecasteele, C.; Vu, B.L.; Dubois-Laurent, C.; Geoffriau, E.; Signor, C.L.; Dalmais, M.; et al. ABI5 Is a Regulator of Seed Maturation and Longevity in Legumes. Plant Cell 2016, 28, 2735–2754. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336–1353. [Google Scholar] [CrossRef]
- Giuliano, G.; Pichersky, E.; Malik, V.S.; Timko, M.P.; Scolnik, P.A.; Cashmore, A.R. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 1988, 85, 7089–7093. [Google Scholar] [CrossRef] [Green Version]
- Donald, R.G.; Schindler, U.; Batschauer, A.; Cashmore, A.R. The plant G box promoter sequence activates transcription in Saccharomyces cerevisiae and is bound in vitro by a yeast activity similar to GBF, the plant G box binding factor. EMBO J. 1990, 9, 1727–1735. [Google Scholar] [CrossRef]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burman, N.; Bhatnagar, A.; Khurana, J.P. OsbZIP48, a HY5 Transcription Factor Ortholog, Exerts Pleiotropic Effects in Light-Regulated Development. Plant Physiol. 2018, 176, 1262–1285. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, S.; Kim, S.H.; Bahk, S.; Ali, A.; Nguyen, X.C.; Yun, D.J.; Chung, W.S. The Auxin Signaling Repressor IAA8 Promotes Seed Germination Through Down-Regulation of ABI3 Transcription in Arabidopsis. Front. Plant Sci. 2020, 11, 111. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.H.; Yu, L.H.; Xiang, C.B. ARABIDOPSIS NITRATE REGULATED 1 acts as a negative modulator of seed germination by activating ABI3 expression. New Phytol. 2020, 225, 835–847. [Google Scholar] [CrossRef]
- Utsugi, S.; Ashikawa, I.; Nakamura, S.; Shibasaka, M. TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J. Plant Res. 2020, 133, 245–256. [Google Scholar] [CrossRef]
- Hu, Y.; Han, X.; Yang, M.; Zhang, M.; Pan, J.; Yu, D. The Transcription Factor INDUCER OF CBF EXPRESSION1 Interacts with ABSCISIC ACID INSENSITIVE5 and DELLA Proteins to Fine-Tune Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhao, T.; Gomez, A.; Li, C.; Yu, C.; Li, H.; Lin, J.; Yang, Y.; Liu, B.; et al. A Drought-Inducible Transcription Factor Delays Reproductive Timing in Rice. Plant Physiol. 2016, 171, 334–343. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Li, C.; Liu, J.; Lv, Y.; Yu, C.; Li, H.; Zhao, T.; Liu, B. The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J. Exp. Bot. 2017, 68, 4695–4707. [Google Scholar] [CrossRef] [Green Version]
- Amir Hossain, M.; Lee, Y.; Cho, J.I.; Ahn, C.H.; Lee, S.K.; Jeon, J.S.; Kang, H.; Lee, C.H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 2010, 72, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zhou, C.; Zhao, Y.; Zhou, S.; Wang, W.; Zhou, D.X. The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front. Plant Sci. 2014, 5, 591. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Lee, S.; Hang, R.; Kim, S.R.; Lee, Y.S.; Cao, X.; Amasino, R.; An, G. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 2013, 73, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xie, S.; Ouyang, Y.; Yao, J. Rice PcG gene OsEMF2b controls seed dormancy and seedling growth by regulating the expression of OsVP1. Plant Sci. 2017, 260, 80–89. [Google Scholar] [CrossRef]
- Zhong, J.; Peng, Z.; Peng, Q.; Cai, Q.; Peng, W.; Chen, M.; Yao, J. Regulation of plant height in rice by the Polycomb group genes OsEMF2b, OsFIE2 and OsCLF. Plant Sci. 2018, 267, 157–167. [Google Scholar] [CrossRef]
- Qiao, F.; Zhao, K.-J. The Influence of RNAi Targeting of OsGA20ox2 Gene on Plant Height in Rice. Plant Mol. Biol. Rep. 2011, 29, 952–960. [Google Scholar] [CrossRef]
- Hu, X.; Cui, Y.; Dong, G.; Feng, A.; Wang, D.; Zhao, C.; Zhang, Y.; Hu, J.; Zeng, D.; Guo, L.; et al. Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces. Sci. Rep. 2019, 9, 19096. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef] [Green Version]
- Barreto, L.C.; Herken, D.M.D.; Silva, B.M.R.; Munné-Bosch, S.; Garcia, Q.S. ABA and GA(4) dynamic modulates secondary dormancy and germination in Syngonanthus verticillatus seeds. Planta 2020, 251, 86. [Google Scholar] [CrossRef]
- Shuai, H.; Meng, Y.; Luo, X.; Chen, F.; Zhou, W.; Dai, Y.; Qi, Y.; Du, J.; Yang, F.; Liu, J.; et al. Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Sci. Rep. 2017, 7, 12620. [Google Scholar] [CrossRef] [Green Version]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [Green Version]
- Vishal, B.; Kumar, P.P. Regulation of Seed Germination and Abiotic Stresses by Gibberellins and Abscisic Acid. Front. Plant Sci. 2018, 9, 838. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhou, W.; Yang, W. APETALA 2-domain-containing transcription factors: Focusing on abscisic acid and gibberellins antagonism. New Phytol 2018, 217, 977–983. [Google Scholar] [CrossRef]
- Muñiz García, M.N.; Stritzler, M.; Capiati, D.A. Heterologous expression of Arabidopsis ABF4 gene in potato enhances tuberization through ABA-GA crosstalk regulation. Planta 2014, 239, 615–631. [Google Scholar] [CrossRef]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H.; et al. The SnRK2-APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Hou, Y.; Wang, S.; Tong, X.; Wang, Y. WRKY72 Negatively Regulates Seed Germination Through Interfering Gibberellin Pathway in Rice. Rice Sci. 2021, 28, 1–5. [Google Scholar]
- Hou, Y.; Wang, Y.; Tang, L.; Tong, X.; Wang, L.; Liu, L.; Huang, S.; Zhang, J. SAPK10-Mediated Phosphorylation on WRKY72 Releases Its Suppression on Jasmonic Acid Biosynthesis and Bacterial Blight Resistance. iScience 2019, 16, 499–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omidvar, V.; Abdullah, S.; Chai, L.H.; Mahmood, M.; Al-Shanfari, A.B. Isolation and characterization of two ABRE-binding proteins: EABF and EABF1 from the oil palm. Mol. Biol. Rep. 2012, 39, 8907–8918. [Google Scholar] [CrossRef]
- Bondos, S.E.; Hsiao, H.C. Roles for Intrinsic Disorder and Fuzziness in Generating Context-specific Function in Ultrabithorax, a Hox Transcription Factor. Adcances Exp. Med. Biol. 2012, 725, 86–105. [Google Scholar]
- Yan, B.; Lv, Y.; Zhao, C.; Wang, X. Knowing When to Silence: Roles of Polycomb-Group Proteins in SAM Maintenance, Root Development, and Developmental Phase Transition. Int. J. Mol. Sci. 2020, 21, 5871. [Google Scholar] [CrossRef]
- Xiao, J.; Jin, R.; Yu, X.; Shen, M.; Wagner, J.D.; Pai, A.; Song, C.; Zhuang, M.; Klasfeld, S.; He, C.; et al. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 2017, 49, 1546–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qüesta, J.I.; Song, J.; Geraldo, N.; An, H.; Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 2016, 353, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Luo, X.; Li, Z.; Yang, W.; Wang, Y.; Liu, R.; Du, J.; He, Y. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 2016, 48, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, J.; Qian, Q.; Xue, H.W. LC2 and OsVIL2 promote rice flowering by photoperoid-induced epigenetic silencing of OsLF. Mol. Plant 2013, 6, 514–527. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, H.; Guo, S.; Wang, B.; Li, Z.; Chong, K.; Xu, Y. OsmiR396d Affects Gibberellin and Brassinosteroid Signaling to Regulate Plant Architecture in Rice. Plant Physiol. 2018, 176, 946–959. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Dan, Z.; Gao, F.; Chen, P.; Fan, F.; Li, S. Rice GROWTH-REGULATING FACTOR7 Modulates Plant Architecture through Regulating GA and Indole-3-Acetic Acid Metabolism. Plant Physiol. 2020, 184, 393–406. [Google Scholar] [CrossRef]
- Zhao, J.; Long, T.; Wang, Y.; Tong, X.; Tang, J.; Li, J.; Wang, H.; Tang, L.; Li, Z.; Shu, Y.; et al. RMS2 Encoding a GDSL Lipase Mediates Lipid Homeostasis in Anthers to Determine Rice Male Fertility. Plant Physiol. 2020, 182, 2047–2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Shi, Y.; Zhang, X.; Xu, X.; Wu, J.L. The OsABCI7 transporter interacts with OsHCF222 to stabilize the thylakoid membrane in rice. Plant Physiol. 2020, 184, 283–299. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; Xu, H.; Wang, Y.; Wang, H.; Li, Z.; Liu, X.; Shu, Y.; Li, G.; Liu, W.; Ying, J.; et al. OsABF1 Represses Gibberellin Biosynthesis to Regulate Plant Height and Seed Germination in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2021, 22, 12220. https://doi.org/10.3390/ijms222212220
Tang L, Xu H, Wang Y, Wang H, Li Z, Liu X, Shu Y, Li G, Liu W, Ying J, et al. OsABF1 Represses Gibberellin Biosynthesis to Regulate Plant Height and Seed Germination in Rice (Oryza sativa L.). International Journal of Molecular Sciences. 2021; 22(22):12220. https://doi.org/10.3390/ijms222212220
Chicago/Turabian StyleTang, Liqun, Huayu Xu, Yifeng Wang, Huimei Wang, Zhiyong Li, Xixi Liu, Yazhou Shu, Guan Li, Wanning Liu, Jiezheng Ying, and et al. 2021. "OsABF1 Represses Gibberellin Biosynthesis to Regulate Plant Height and Seed Germination in Rice (Oryza sativa L.)" International Journal of Molecular Sciences 22, no. 22: 12220. https://doi.org/10.3390/ijms222212220
APA StyleTang, L., Xu, H., Wang, Y., Wang, H., Li, Z., Liu, X., Shu, Y., Li, G., Liu, W., Ying, J., Tong, X., Yao, J., Xiao, W., Tang, S., Ni, S., & Zhang, J. (2021). OsABF1 Represses Gibberellin Biosynthesis to Regulate Plant Height and Seed Germination in Rice (Oryza sativa L.). International Journal of Molecular Sciences, 22(22), 12220. https://doi.org/10.3390/ijms222212220









