AMBRA1 Negatively Regulates the Function of ALDH1B1, a Cancer Stem Cell Marker, by Controlling Its Ubiquitination
Abstract
:1. Introduction
2. Results
2.1. AMBRA1 Interacts with ALDH1B1 and Suppresses the Self-Association of ALDH1B1
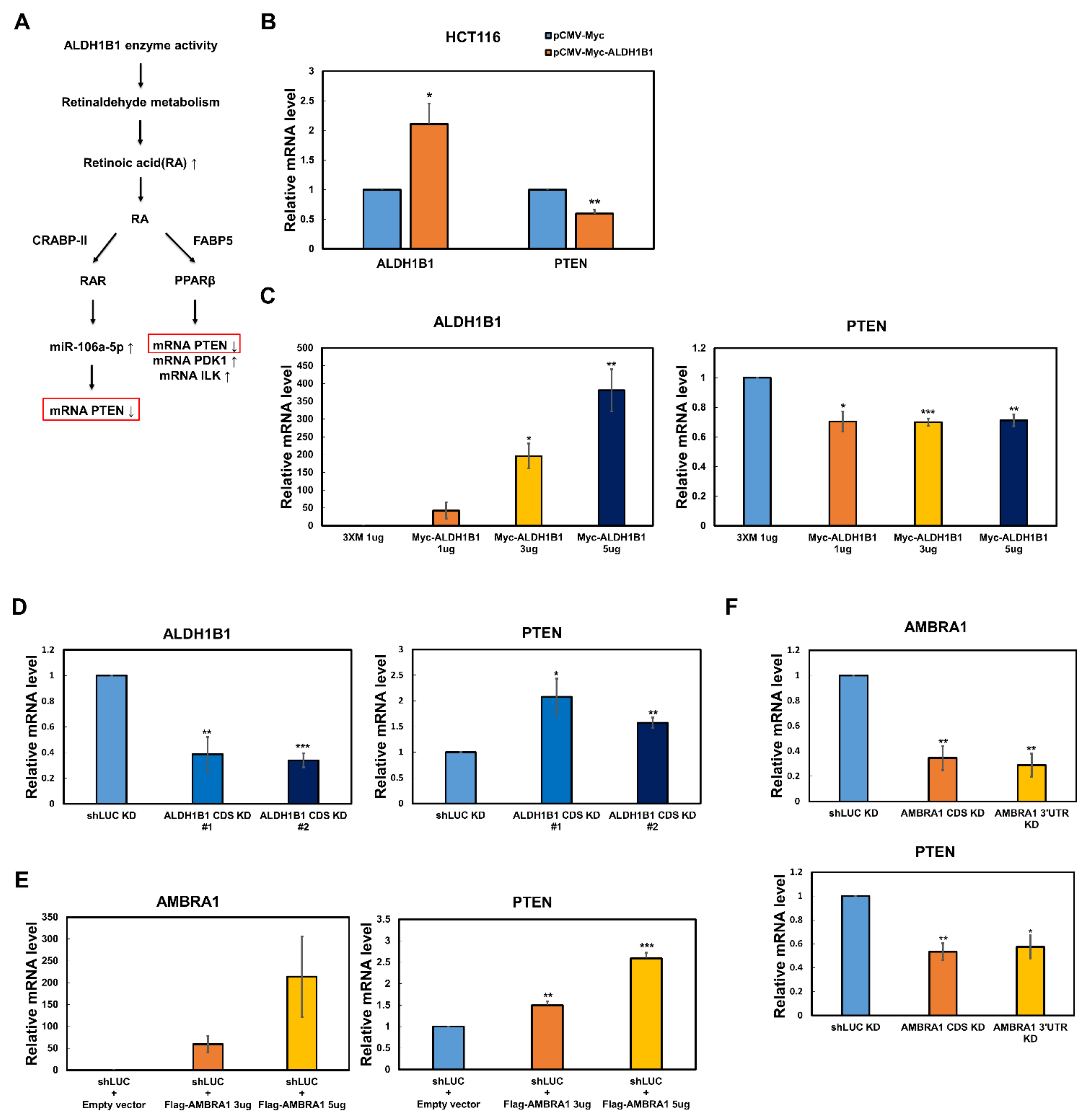
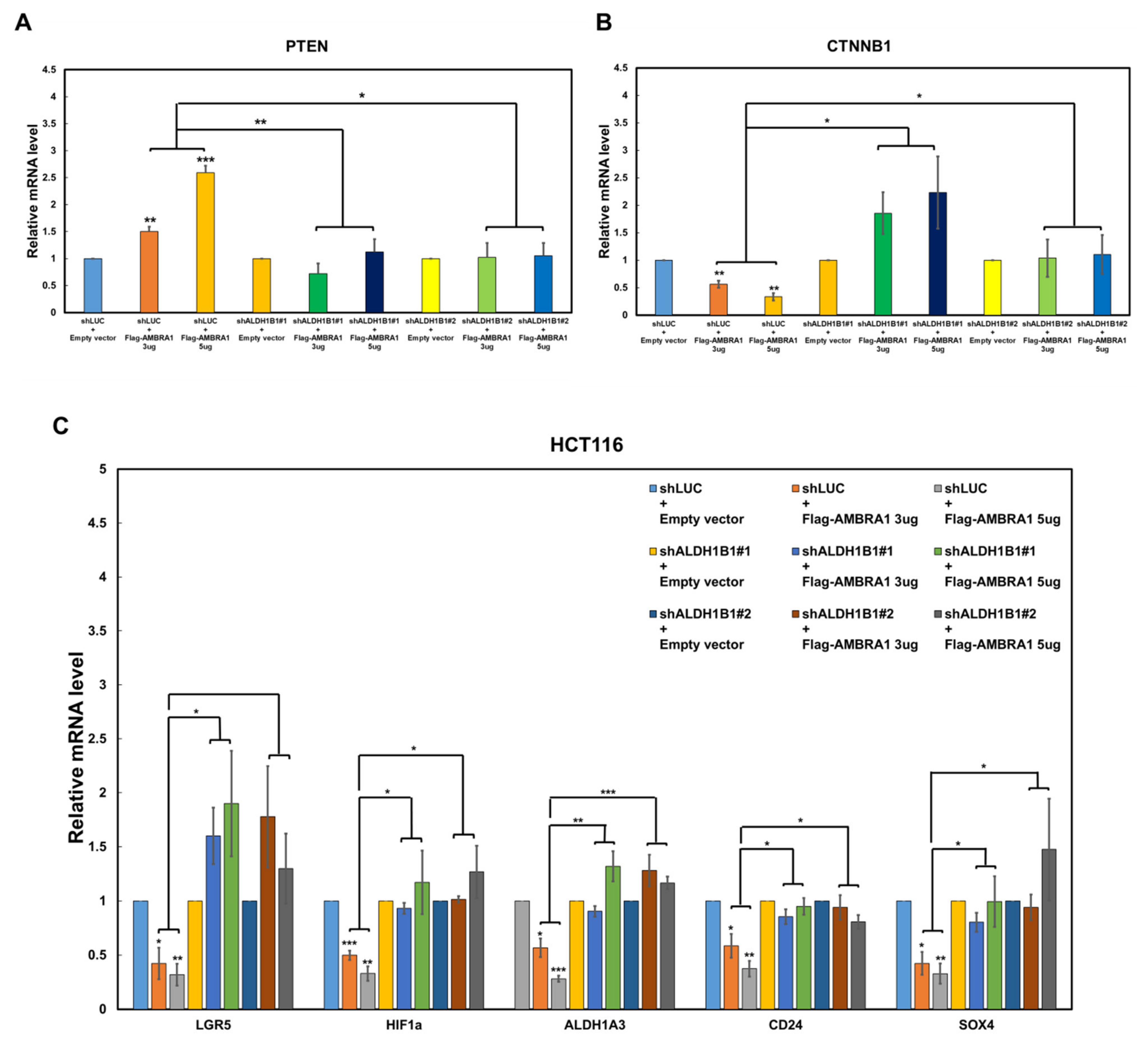
2.2. The PTEN mRNA Level Is Reversely Regulated by ALDH1B1 and AMBRA1
2.3. AMBRA1 Regulates the Transcription of PTEN, CTNNB1, and CSC-Related β-Catenin Target Genes via ALDH1B1
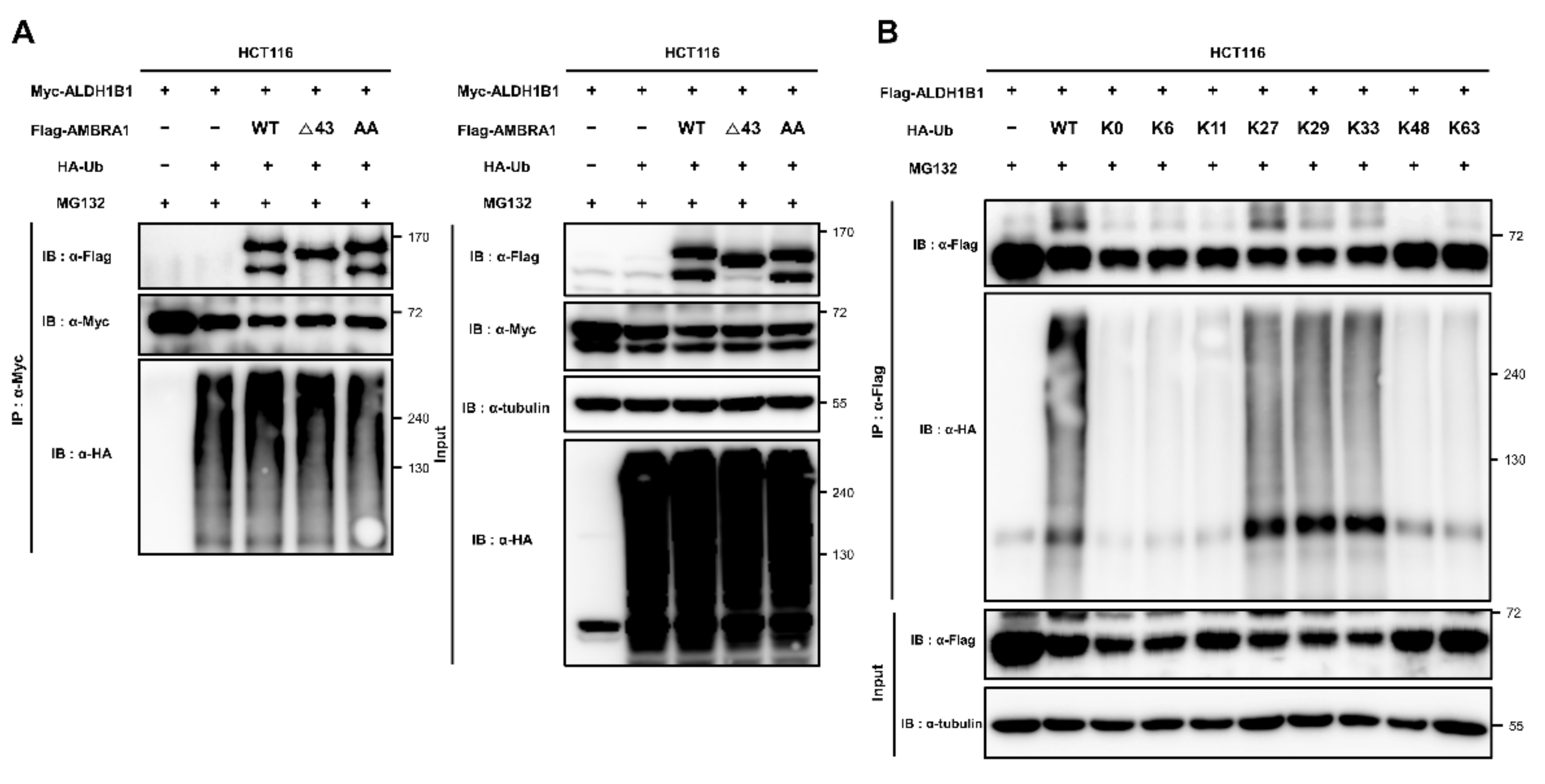
2.4. ALDH1B1 Is Ubiquitinated by AMBRA1, DDB1, and TRAF6
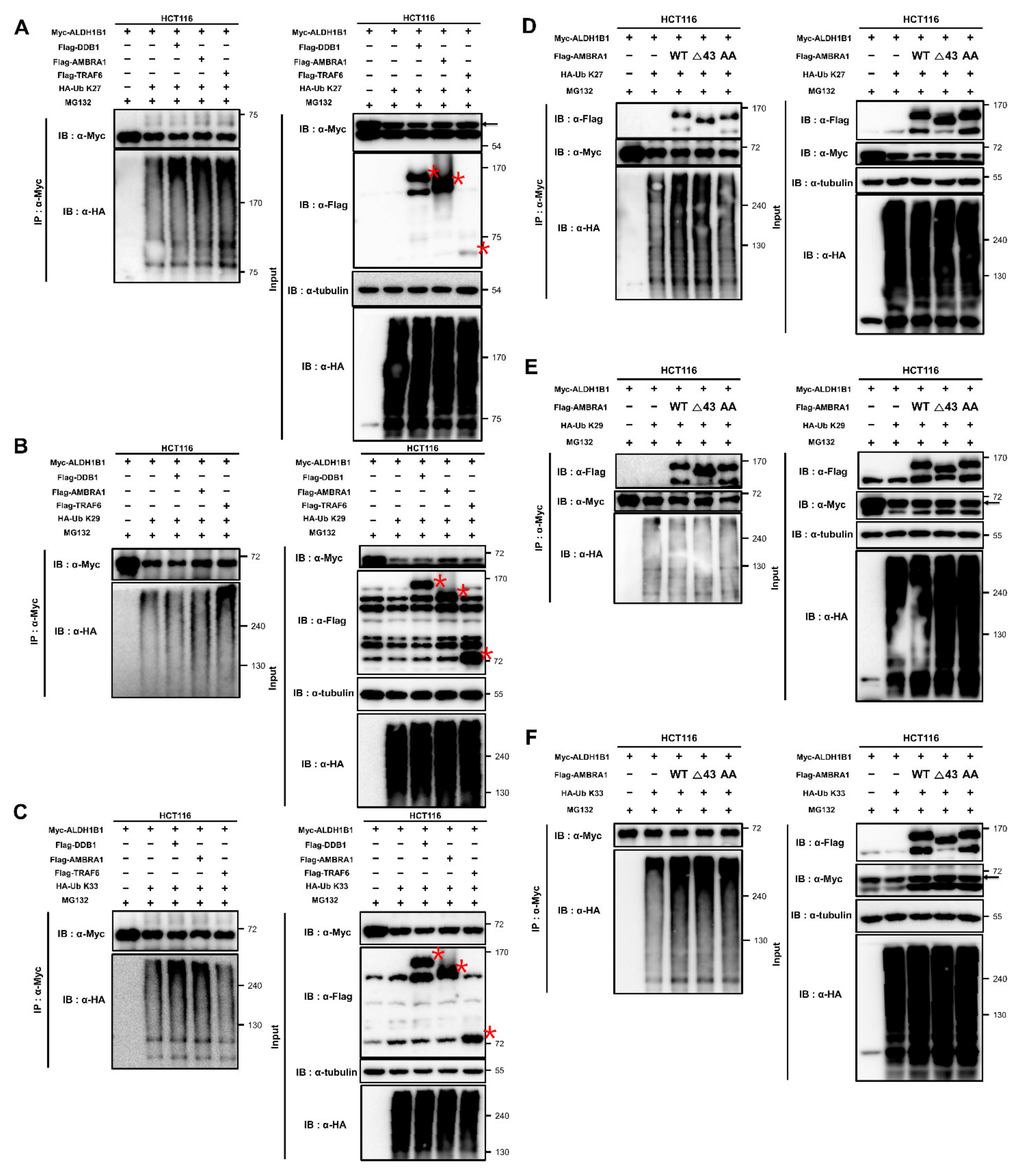
2.5. ALDH1B1 Is Ubiquitinated via Cooperation of AMBRA1 with TRAF6 and through K27, K29, and K33 Linkages
2.6. AMBRA1 Ubiquitinates ALDH1B1 through K27 and K33 Linkages by Cooperating with Different Partners
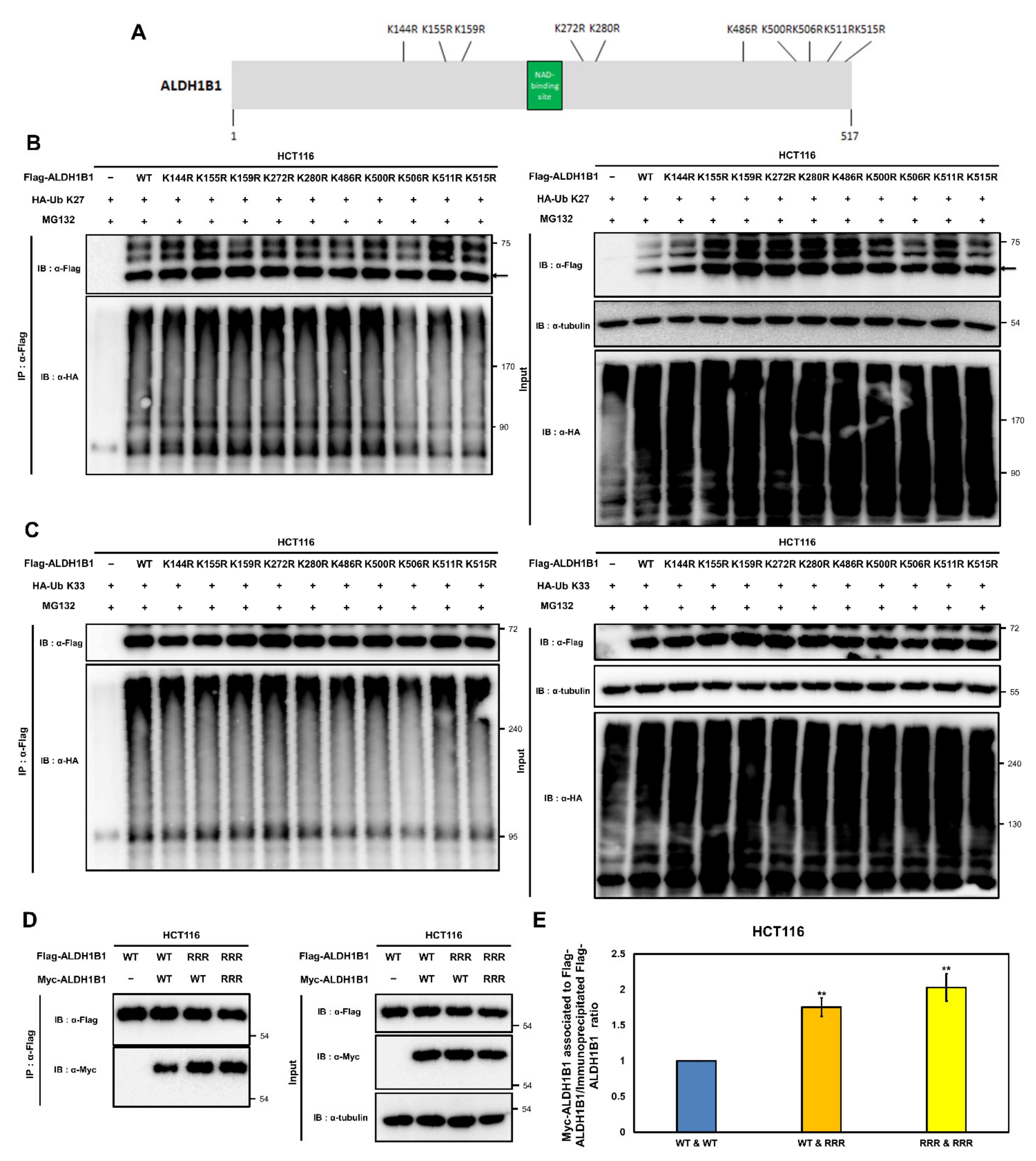
2.7. K506, K511, and K515 Are Potential K27- and K33-Mediated Ubiquitination Acceptor Sites of ALDH1B1 Involved in the Self-Association of ALDH1B1
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Cloning and Site-Directed/Deletion Mutagenesis
4.3. Transfection of Plasmid Constructs and Infection of Viral Particles
4.4. Construction of AMBRA1- and ALDH1B1-Knockdown Cell Lines
4.5. Generation of ALDH1B1 Stable Overexpression Cell Lines
4.6. Immunoblot Analysis
4.7. Co-Immunoprecipitation Assay
4.8. Cell-Based Ubiquitination Assay and MG132 Treatment
4.9. RT-qPCR
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta—Proteins Proteom. 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [Green Version]
- Cianfanelli, V.; De Zio, D.; Di Bartolomeo, S.; Nazio, F.; Strappazzon, F.; Cecconi, F. Ambra1 at a glance. J. Cell Sci. 2015, 128, 2003–2008. [Google Scholar] [CrossRef] [Green Version]
- Nazio, F.; Strappazzon, F.; Antonioli, M.; Bielli, P.; Cianfanelli, V.; Bordi, M.; Gretzmeier, C.; Dengjel, J.; Piacentini, M.; Fimia, G.M.; et al. MTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013, 15, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Antonioli, M.; Albiero, F.; Nazio, F.; Vescovo, T.; Perdomo, A.B.; Corazzari, M.; Marsella, C.; Piselli, P.; Gretzmeier, C.; Dengjel, J.; et al. AMBRA1 interplay with cullin E3 Ubiquitin ligases regulates autophagy dynamics. Dev. Cell 2014, 31, 734–746. [Google Scholar] [CrossRef] [PubMed]
- van Humbeeck, C.; Cornelissen, T.; Hofkens, H.; Mandemakers, W.; Gevaert, K.; de Strooper, B.; Vandenberghe, W. Parkin interacts with ambra1 to induce mitophagy. J. Neurosci. 2011, 31, 10249–10261. [Google Scholar] [CrossRef] [Green Version]
- Strappazzon, F.; Nazio, F.; Corrado, M.; Cianfanelli, V.; Romagnoli, A.; Fimia, G.M.; Campello, S.; Nardacci, R.; Piacentini, M.; Campanella, M.; et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015, 22, 419–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maria Fimia, G.; Stoykova, A.; Romagnoli, A.; Giunta, L.; Di Bartolomeo, S.; Nardacci, R.; Corazzari, M.; Fuoco, C.; Ucar, A.; Schwartz, P.; et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007, 447, 1121–1125. [Google Scholar] [CrossRef] [Green Version]
- Skobo, T.; Benato, F.; Grumati, P.; Meneghetti, G.; Cianfanelli, V.; Castagnaro, S.; Chrisam, M.; Di Bartolomeo, S.; Bonaldo, P.; Cecconi, F.; et al. Zebrafish ambra1a and ambra1b knockdown impairs skeletal muscle development. PLoS ONE 2014, 9, e109480. [Google Scholar] [CrossRef] [Green Version]
- Cianfanelli, V.; Fuoco, C.; Lorente, M.; Salazar, M.; Quondamatteo, F.; Gherardini, P.F.; De Zio, D.; Nazio, F.; Antonioli, M.; D’Orazio, M.; et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat. Cell Biol. 2015, 17, 20–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simoneschi, D.; Rona, G.; Zhou, N.; Jeong, Y.T.; Jiang, S.; Milletti, G.; Arbini, A.A.; O’Sullivan, A.; Wang, A.A.; Nithikasem, S.; et al. CRL4AMBRA1 is a master regulator of D-type cyclins. Nature 2021, 592, 789–793. [Google Scholar] [CrossRef]
- Chaikovsky, A.C.; Li, C.; Jeng, E.E.; Loebell, S.; Lee, M.C.; Murray, C.W.; Cheng, R.; Demeter, J.; Swaney, D.L.; Chen, S.H.; et al. The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. Nature 2021, 592, 794–798. [Google Scholar] [CrossRef]
- Maiani, E.; Milletti, G.; Nazio, F.; Holdgaard, S.G.; Bartkova, J.; Rizza, S.; Cianfanelli, V.; Lorente, M.; Simoneschi, D.; Di Marco, M.; et al. AMBRA1 regulates cyclin D to guard S-phase entry and genomic integrity. Nature 2021, 592, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Orlicky, D.J.; Matsumoto, A.; Singh, S.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem. Biophys. Res. Commun. 2011, 405, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, E.; Farghali, O.; El Badawy, Z.; Ahmed, S.; Ahmed, A. Immunohistochemical Expression of Cancer Stem Cell marker Aldehyde dehydrogenase 1B1 (ALDH1B1) in Colorectal Carcinoma. Sohag Med. J. 2018, 22, 109–115. [Google Scholar] [CrossRef]
- Singh, S.; Arcaroli, J.; Chen, Y.; Thompson, D.C.; Messersmith, W.; Jimeno, A.; Vasiliou, V. ALDH1B1 is crucial for colon tumorigenesis by modulating Wnt/β-catenin, Notch and PI3K/Akt signaling pathways. PLoS ONE 2015, 10, e0121648. [Google Scholar] [CrossRef]
- Wang, X.; Li, C.; Yao, W.; Tian, Z.; Liu, Z.; Ge, H. MicroRNA-761 suppresses tumor progression in osteosarcoma via negatively regulating ALDH1B1. Life Sci. 2020, 262, 118544. [Google Scholar] [CrossRef]
- Singh, S.; Brocker, C.; Koppaka, V.; Ying, C.; Jackson, B.; Thompson, D.C.; Vasiliou, V. Electrophilic Stress. Free Radic. Biol. Med. 2014, 1, 89–101. [Google Scholar] [CrossRef]
- Chen, S.; Jang, G.M.; Hüttenhain, R.; Gordon, D.E.; Du, D.; Newton, B.W.; Johnson, J.R.; Hiatt, J.; Hultquist, J.F.; Johnson, T.L.; et al. CRL4-AMBRA1 targets Elongin C for ubiquitination and degradation to modulate CRL5 signaling. EMBO J. 2018, 37, e97508. [Google Scholar] [CrossRef]
- Lutfullah, G.; Qaisar Khan, N.; Amin, F.; Kakakhel, L.; Azhar, N. Structural Modeling Studies of Aldehyde Dehydrogenase X: Insights into Key Interactions in the Tetrameric Assembly of the Isoenzyme. Protein Pept. Lett. 2011, 18, 41–57. [Google Scholar] [CrossRef]
- Shi, L.; Liang, Y.; Yang, L.; Li, B.; Zhang, B.; Zhen, C.; Fan, J.; Tang, S.; Road, N.D.; Road, N.D.; et al. All-trans Retinoic Acid Regulates Inhibition of Autophagic in Developing Cleft Palates. bioRxiv 2020. [Google Scholar] [CrossRef]
- Applegate, C.C.; Lane, M.A. Role of retinoids in the prevention and treatment of colorectal cancer. World J. Gastrointest. Oncol. 2015, 7, 184–203. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Arcaroli, J.; Chen, Y.; Gasparetto, M.; Neumeister, V.; Thompson, D.C.; Singh, S.; Smith, C.; Messersmith, W.; Vasiliou, V. Aldehyde dehydrogenase 1B1: A novel immunohistological marker for colorectal cancer. Br. J. Cancer 2017, 117, 1537–1543. [Google Scholar] [CrossRef] [PubMed]
- Ciccone, V.; Morbidelli, L.; Ziche, M.; Donnini, S. How to conjugate the stemness marker ALDH1A1 with tumor angiogenesis, progression, and drug resistance. Cancer Drug Resist. 2020, 3, 26–37. [Google Scholar] [CrossRef] [Green Version]
- de Sousa e Melo, F.; Vermeulen, L. Wnt signaling in cancer stem cell biology. Cancers 2016, 8, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Rita, A.; Peschiaroli, A.; D′Acunzo, P.; Strobbe, D.; Hu, Z.; Gruber, J.; Nygaard, M.; Lambrughi, M.; Melino, G.; Papaleo, E.; et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 2018, 9, 3755. [Google Scholar] [CrossRef]
- Liu, L.; Yin, S.; Brobbey, C.; Gan, W. Ubiquitination in cancer stem cell: Roles and targeted cancer therapy. STEMedicine 2020, 1, e37. [Google Scholar] [CrossRef]
- Crabb, D.W.; Stewart, M.J.; Xiao, Q. Hormonal and Chemical Influences on the Expression of Class 2 Aldehyde Dehydrogenases in Rat H4IIEC3 and Human HuH7 Hepatoma Cells. Alcohol. Clin. Exp. Res. 1995, 19, 1414–1419. [Google Scholar] [CrossRef]
- Zucchelli, S.; Codrich, M.; Marcuzzi, F.; Pinto, M.; Vilotti, S.; Biagioli, M.; Ferrer, I.; Gustincich, S. TRAF6 promotes atypical ubiquitination of mutant DJ-1 and alpha-synuclein and is localized to Lewy bodies in sporadic Parkinson’s disease brains. Hum. Mol. Genet. 2010, 19, 3759–3770. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef]
- Wang, X.; Yu, Y.; He, Y.; Cai, Q.; Gao, S.; Yao, W.; Liu, Z.; Tian, Z.; Han, Q.; Wang, W.; et al. Upregulation of ALDH1B1 promotes tumor progression in osteosarcoma. Oncotarget 2018, 9, 2502–2514. [Google Scholar] [CrossRef] [Green Version]
- Merlos-Suárez, A.; Barriga, F.M.; Jung, P.; Iglesias, M.; Céspedes, M.V.; Rossell, D.; Sevillano, M.; Hernando-Momblona, X.; Da Silva-Diz, V.; Muñoz, P.; et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011, 8, 511–524. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Zhang, J. K27-linked noncanonic ubiquitination in immune regulation. J. Leukoc. Biol. 2021. online ahead of print. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, S.-H.; Jang, Y.-K. AMBRA1 Negatively Regulates the Function of ALDH1B1, a Cancer Stem Cell Marker, by Controlling Its Ubiquitination. Int. J. Mol. Sci. 2021, 22, 12079. https://doi.org/10.3390/ijms222112079
Baek S-H, Jang Y-K. AMBRA1 Negatively Regulates the Function of ALDH1B1, a Cancer Stem Cell Marker, by Controlling Its Ubiquitination. International Journal of Molecular Sciences. 2021; 22(21):12079. https://doi.org/10.3390/ijms222112079
Chicago/Turabian StyleBaek, Seung-Heon, and Yeun-Kyu Jang. 2021. "AMBRA1 Negatively Regulates the Function of ALDH1B1, a Cancer Stem Cell Marker, by Controlling Its Ubiquitination" International Journal of Molecular Sciences 22, no. 21: 12079. https://doi.org/10.3390/ijms222112079
APA StyleBaek, S.-H., & Jang, Y.-K. (2021). AMBRA1 Negatively Regulates the Function of ALDH1B1, a Cancer Stem Cell Marker, by Controlling Its Ubiquitination. International Journal of Molecular Sciences, 22(21), 12079. https://doi.org/10.3390/ijms222112079





