The Papain-like Cysteine Protease HpXBCP3 from Haematococcus pluvialis Involved in the Regulation of Growth, Salt Stress Tolerance and Chlorophyll Synthesis in Microalgae
Abstract
:1. Introduction
2. Results
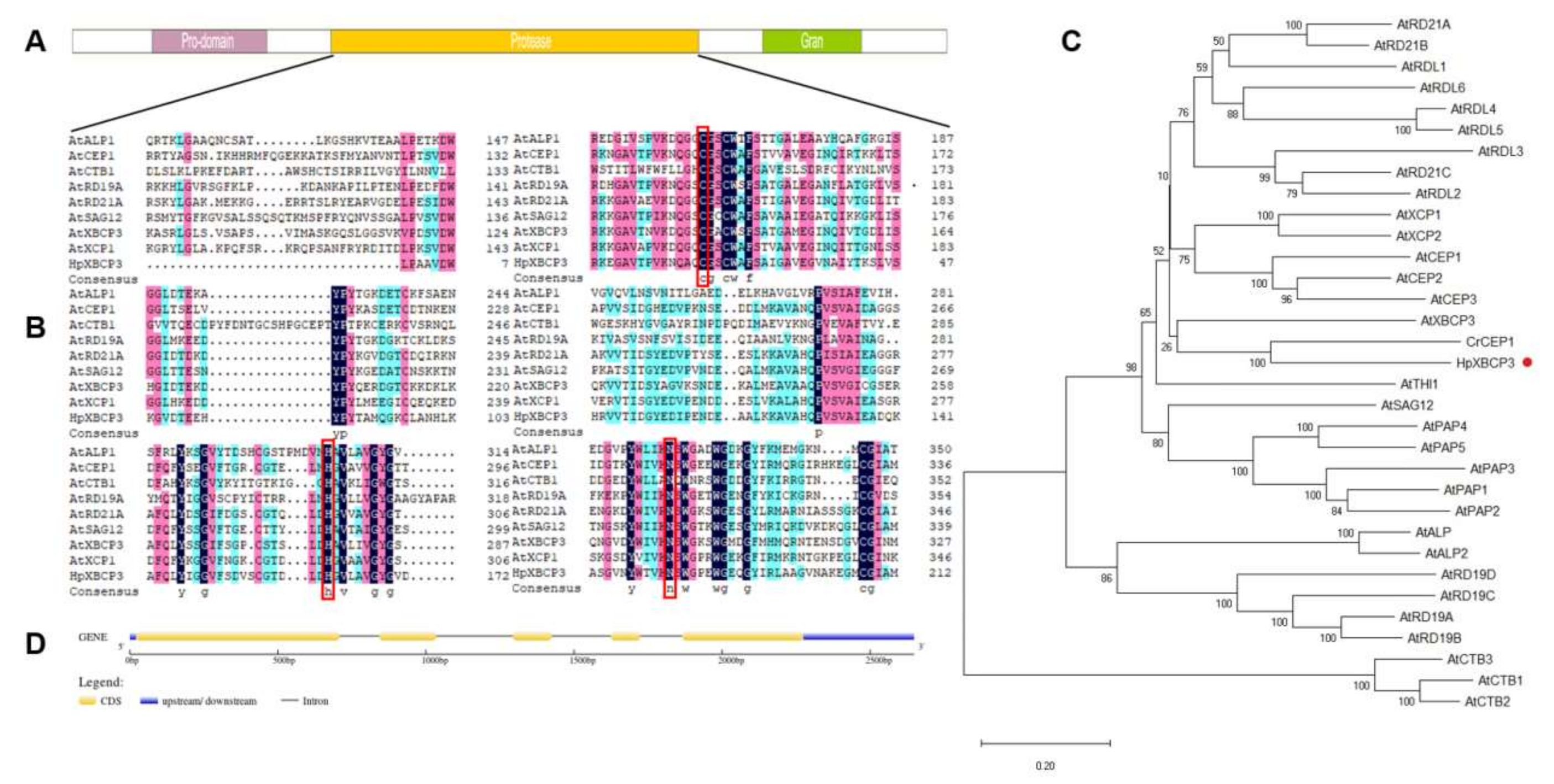
2.1. Cloning and Sequence Analysis of HpXBCP3 from H. pluvialis
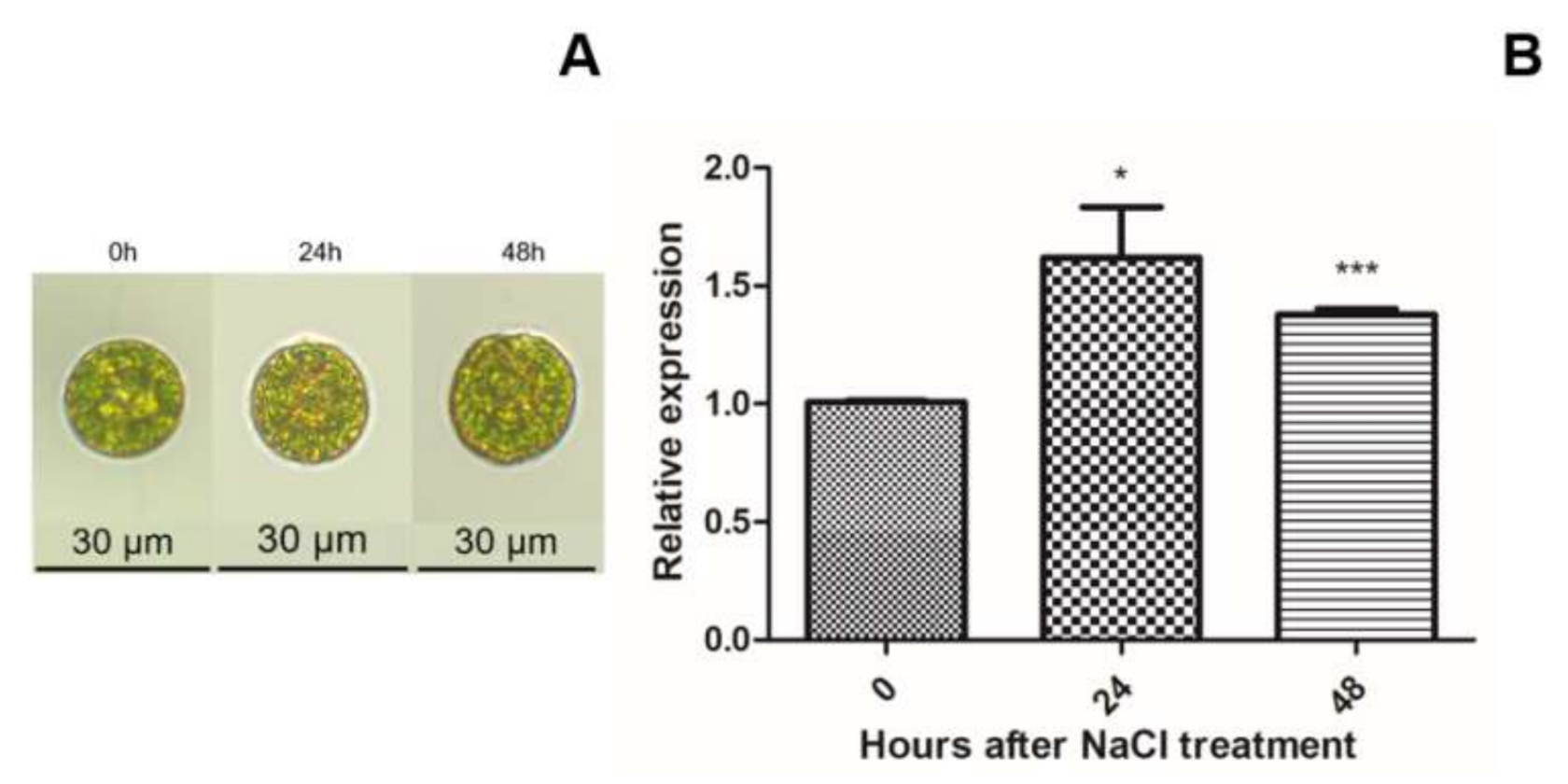
2.2. HpXBCP3 Responded to NaCl Stress
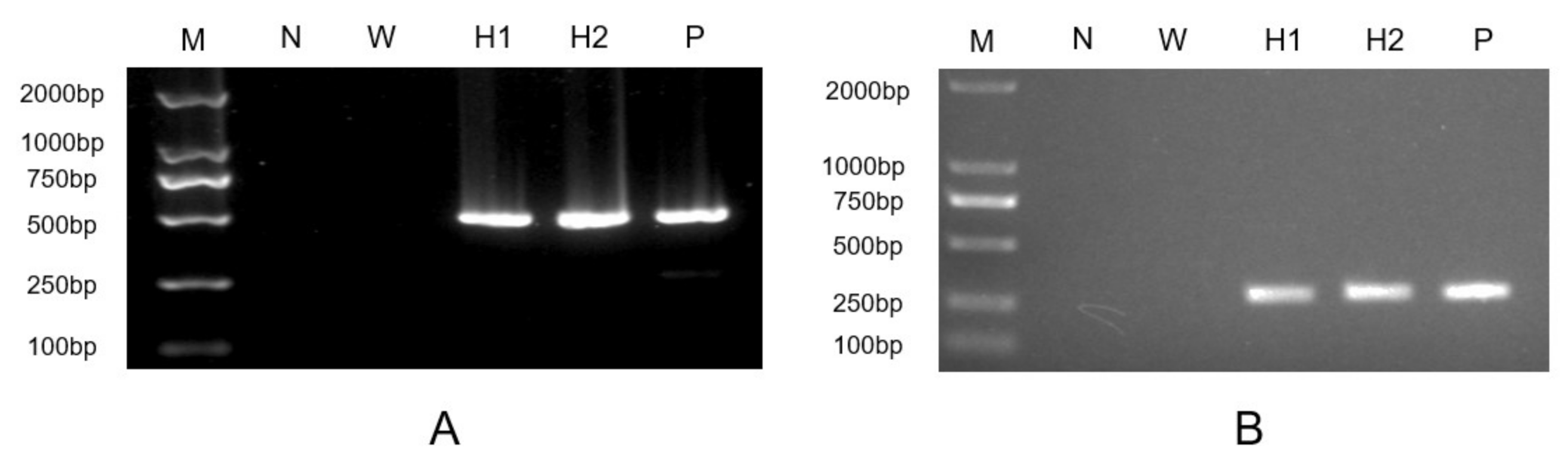
2.3. Expressing HpXBCP3 in C. reinhardtii
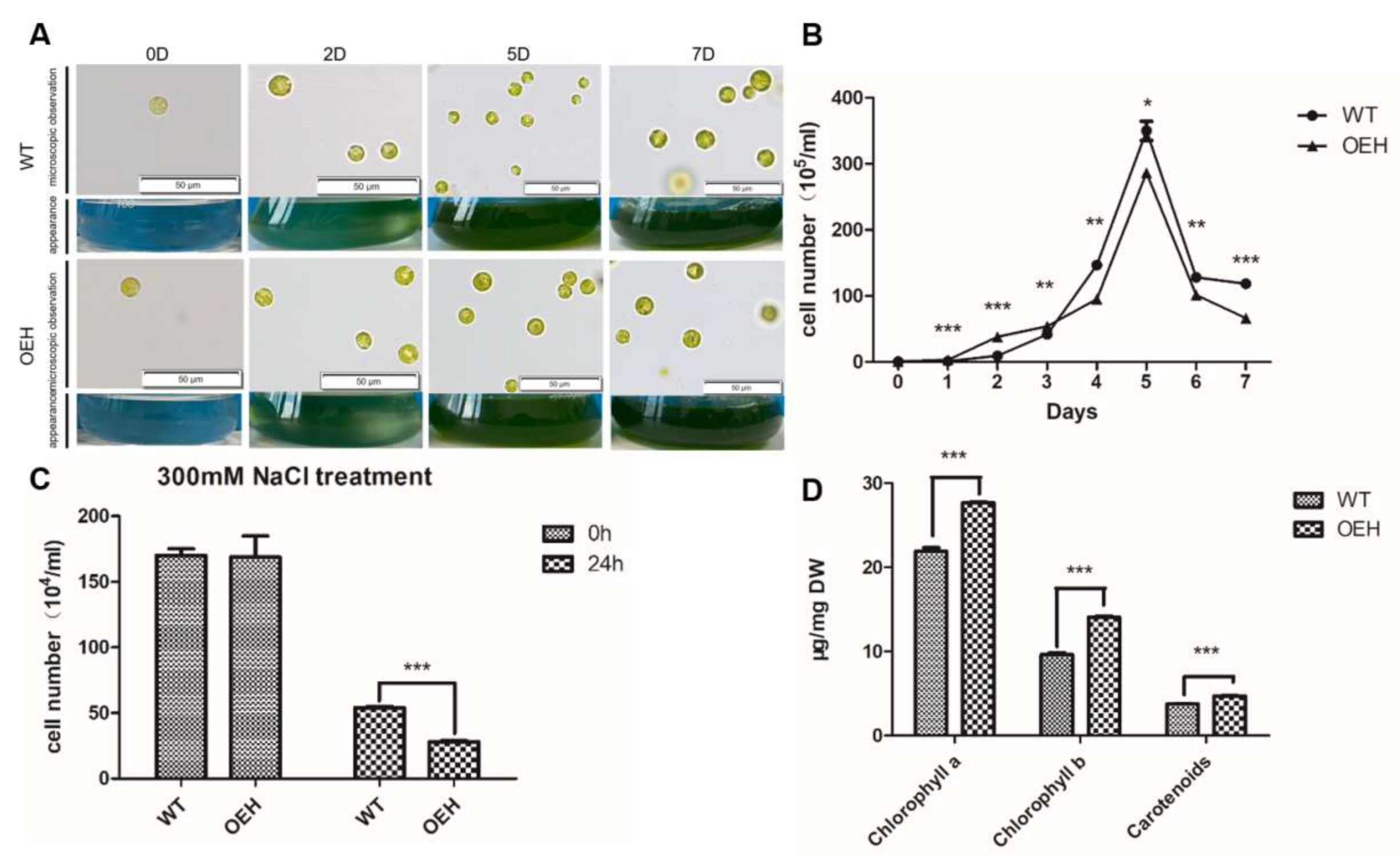
2.4. Physiological Changes of Transgenic C. reinhardtii Expressing HpXBCP3
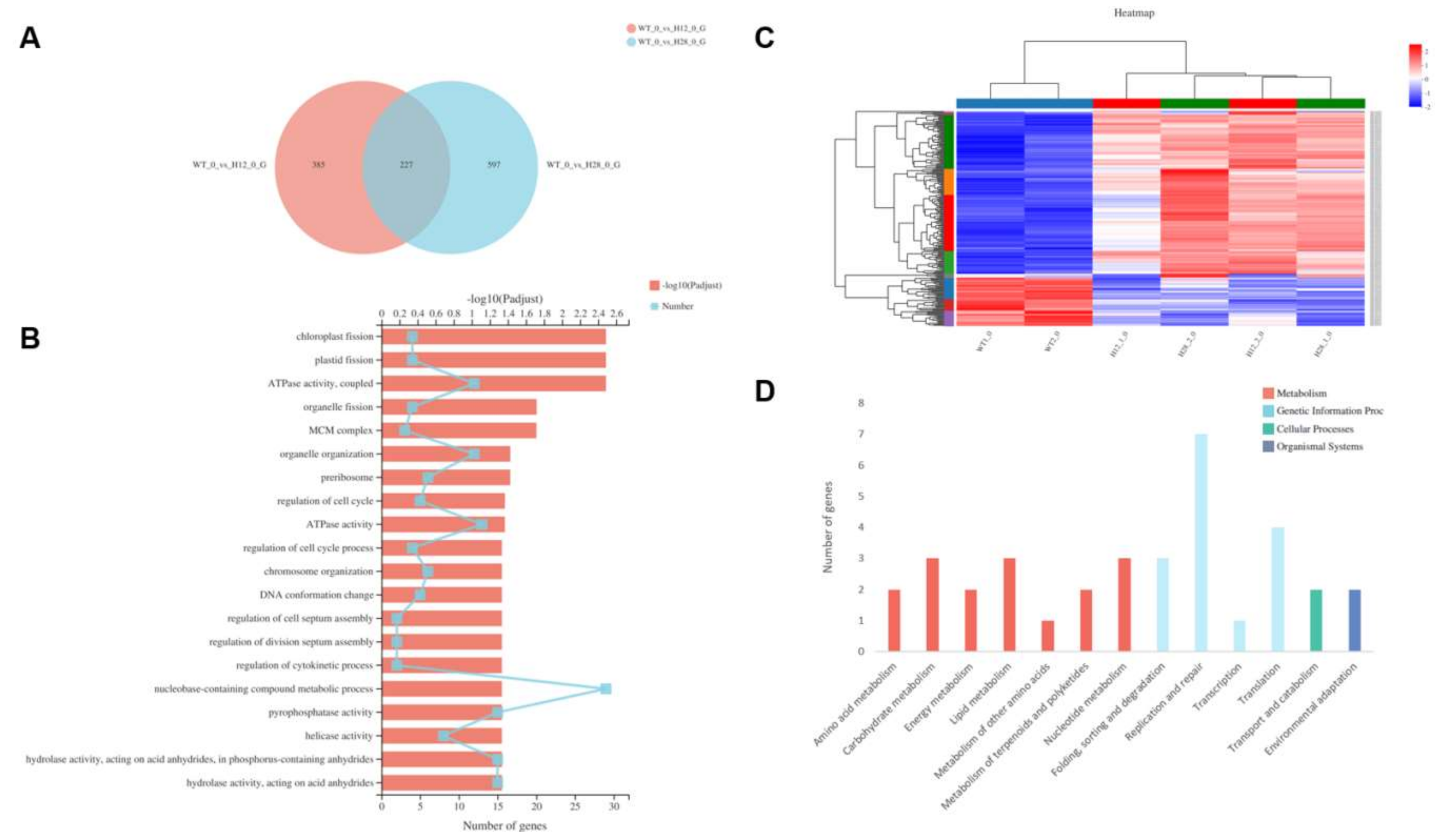
2.5. Transcriptomic Analysis of OEH
2.5.1. GO Significant Enrichment Analysis of DEGs
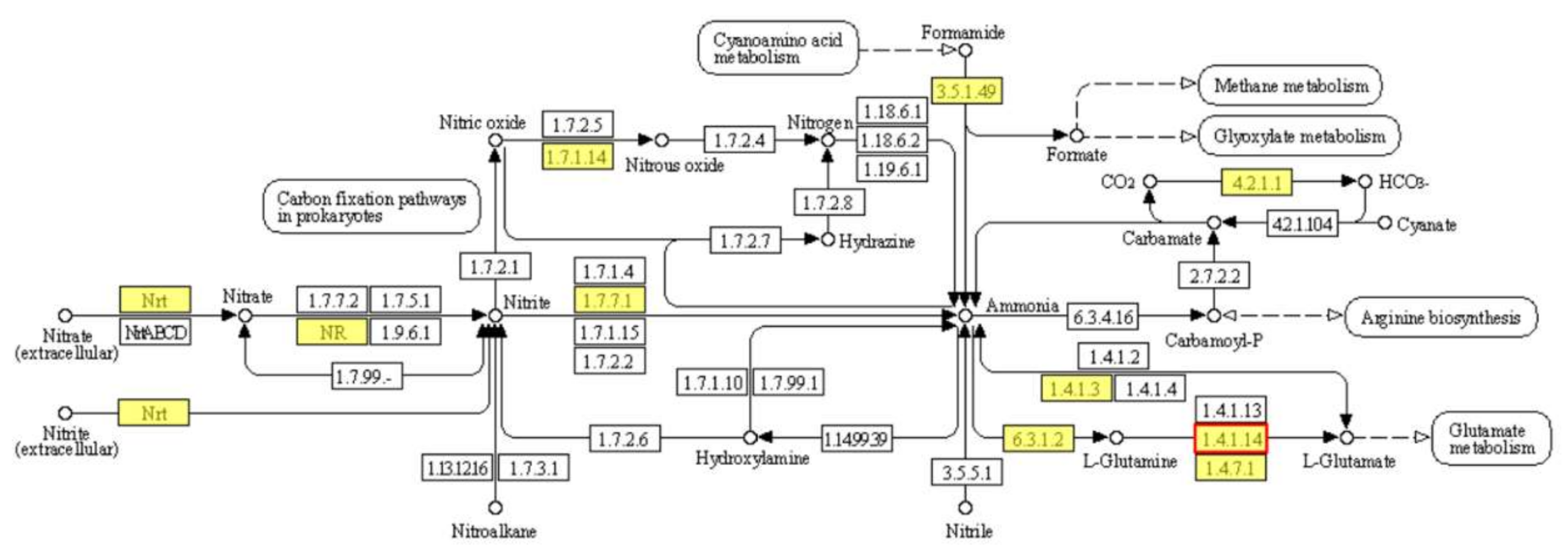
2.5.2. KEGG Pathway Significant Enrichment Analysis of DEGs
3. Discussion
3.1. Expression of HpXBCP3 in C. reinhardtii Caused Abnormal Growth
3.2. HpXBCP3 Responded to NaCl Stress in Microalgae
3.3. Expression of HpXBCP3 Leaded to Increased Chlorophyll Content
4. Materials and Methods
4.1. Algal Strains and Culture Conditions
4.2. Microscope Observation and Growth Curve Determination
4.3. Cloning of HpXBCP3 and Bioinformatic Analysis
4.4. Detecting the Gene Expression Pattern Responding to Salt Stress by qRT-PCR
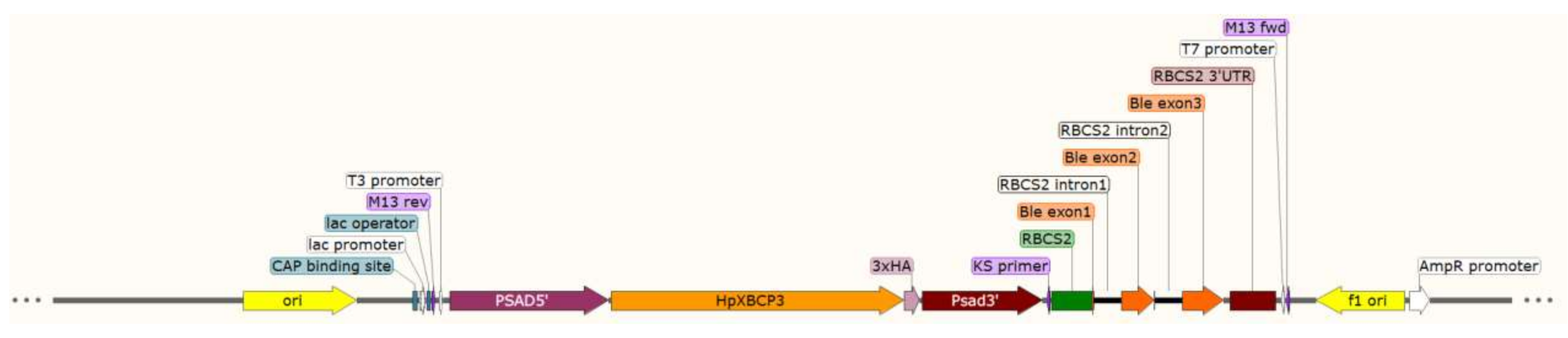
4.5. Plasmid Construction and Genetic Transformation
4.6. Determination of Chlorophyll and Carotenoids Content
4.7. Transcriptomic Analyses
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, P.; Gayen, D. Plant protease as regulator and signaling molecule for enhancing environmental stress-tolerance. Plant Cell Rep. 2021, 10, 2081–2095. [Google Scholar] [CrossRef]
- Zou, Y.; Bozhkov, P.V. Chlamydomonas proteases: Classification, phylogeny, and molecular mechanisms. J. Exp. Bot. 2021, erab383. [Google Scholar] [CrossRef]
- Ikui, A.E.; Ueki, N.; Pecani, K.; Cross, F.R. Control of pre-replicative complex during the division cycle in Chlamydomonas reinhardtii. PLoS Genet. 2021, 17, e1009471. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Shabab, M.; Strasser, R.; Wolters, P.J.; Shindo, T.; Niemer, M.; Kaschani, F.; Mach, L.; Van Der Hoorn, R.A.L. Post-Translational Regulation and Trafficking of the Granulin-Containing Protease RD21 of Arabidopsis thaliana. PLOS ONE 2012, 7, e32422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Hu, M.; Wang, Q.; Cheng, L.; Zhang, Z. Role of Papain-Like Cysteine Proteases in Plant Development. Front. Plant Sci. 2018, 9, 1717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richau, K.H.; Kaschani, F.; Verdoes, M.; Pansuriya, T.C.; Niessen, S.; Stüber, K.; Colby, T.; Overkleeft, H.S.; Bogyo, M.; Van der Hoorn, R.A. Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 2012, 158, 1583–1599. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Li, H.; Yin, B.; Zhang, Y.; Liu, Y.; Cheng, Z.; Liu, D.; Lu, H. The papain-like cysteine protease CEP1 is involved in programmed cell death and secondary wall thickening during xylem development in Arabidopsis. J. Exp. Bot. 2019, 70, 205–215. [Google Scholar] [CrossRef]
- Alomrani, S.; Kunert, K.J.; Foyer, C.H. Papain-like cysteine proteases are required for the regulation of photosynthetic gene expression and acclimation to high light stress. J. Exp. Bot. 2021, 72, 3441–3454. [Google Scholar] [CrossRef]
- Xiao, H.-J.; Yin, Y.-X.; Chai, W.-G.; Gong, Z.-H. Silencing of the CaCP Gene Delays Salt- and Osmotic-Induced Leaf Senescence in Capsicum annuum L. Int. J. Mol. Sci. 2014, 15, 8316–8334. [Google Scholar] [CrossRef] [Green Version]
- Velasco-Arroyo, B.; Diaz-Mendoza, M.; Gomez-Sanchez, A.; Moreno-Garcia, B.; Santamaria, M.E.; Torija-Bonilla, M.; Hensel, G.; Kumlehn, J.; Martinez, M.; Diaz, I. Silencing barley cystatins HvCPI-2 and HvCPI-4 specifically modifies leaf responses to drought stress. Plant Cell Environ. 2018, 41, 1776–1790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The Cysteine Protease CEP1, a Key Executor Involved in Tapetal Programmed Cell Death, Regulates Pollen Development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umen, J.G. Sizing up the cell cycle: Systems and quantitative approaches in Chlamydomonas. Curr. Opin. Plant Biol. 2018, 46, 96–103. [Google Scholar] [CrossRef]
- Schaller, G.E.; Street, I.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef]
- Jain, R.; Aggarwal, A.K.; Rechkoblit, O. Eukaryotic DNA polymerases. Curr. Opin. Struct. Biol. 2018, 53, 77–87. [Google Scholar] [CrossRef]
- Deegan, T.D.; Yeeles, J.; Diffley, J.F. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 2016, 35, 961–973. [Google Scholar] [CrossRef]
- Aklilu, B.B.; Culligan, K.M. Molecular Evolution and Functional Diversification of Replication Protein A1 in Plants. Front. Plant Sci. 2016, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross, F.R.; Umen, J.G. The Chlamydomonas cell cycle. Plant J. 2015, 82, 370–392. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.N.; Caputo, C.; Criado, M.V.; Funk, C. Senescence-associated proteases in plants. Physiol. Plant. 2012, 145, 130–139. [Google Scholar] [CrossRef]
- Díaz-Mendoza, M.; Velasco-Arroyo, B.; González-Melendi, P.; Martínez, M.; Díaz, I. C1A cysteine protease-cystatin interactions in leaf senescence. J. Exp. Bot. 2014, 65, 3825–3833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.K.; Ryu, M.Y.; Kim, J.H.; Hong, J.S.; Oh, T.R.; Kim, W.T.; Yang, S.W. RING E3 ligases: Key regulatory elements are involved in abiotic stress responses in plants. BMB Rep. 2017, 50, 393–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, K.; Yang, W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Melo, F.V.; Oliveira, M.M.; Saibo, N.J.M.; Lourenço, T.F. Modulation of Abiotic Stress Responses in Rice by E3-Ubiquitin Ligases: A Promising Way to Develop Stress-Tolerant Crops. Front. Plant Sci. 2021, 12, 640193. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.C.; Chapagain, S.; Jang, C.S. The microtubule-associated RING finger protein 1 (OsMAR1) acts as a negative regulator for salt-stress response through the regulation of OCPI2 (O. sativa chymotrypsin protease inhibitor 2). Planta 2018, 247, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Yi, D.; Yang, J.; Liu, X.; Pang, Y. Genome-Wide Identification, Expression Analysis and Functional Study of CCT Gene Family in Medicago truncatula. Plants 2020, 9, 513. [Google Scholar] [CrossRef]
- Sharma, M.; Pandey, G.K. Expansion and Function of Repeat Domain Proteins During Stress and Development in Plants. Front. Plant Sci. 2016, 6, 1218. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Wang, J.; Xia, X.; Zhang, Z.; He, J.; Nong, B.; Luo, T.; Feng, R.; Wu, Y.; Pan, Y.; et al. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021, 107, 198–214. [Google Scholar] [CrossRef]
- Hodul, M.; Ganji, R.; Dahlberg, C.L.; Raman, M.; Juo, P. The WD40-repeat protein WDR-48 promotes the stability of the deubiquitinating enzyme USP-46 by inhibiting its ubiquitination and degradation. J. Biol. Chem. 2020, 295, 11776–11788. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, Y.; Shen, J.; Yin, J.; Li, D.; Gao, Y.; Xu, W.; Liang, J. OsRACK1A, encodes a circadian clock-regulated WD40 protein, negatively affect salt tolerance in rice. Rice 2018, 11, 45. [Google Scholar] [CrossRef] [Green Version]
- Smythers, A.L.; McConnell, E.W.; Lewis, H.C.; Mubarek, S.N.; Hicks, L.M. Photosynthetic Metabolism and Nitrogen Reshuffling Are Regulated by Reversible Cysteine Thiol Oxidation Following Nitrogen Deprivation in Chlamydomonas. Plants 2020, 9, 784. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Masuda, T. Transcriptional Regulation of Tetrapyrrole Biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Stenbaek, A.; Jensen, P.E. Redox regulation of chlorophyll biosynthesis. Phytochemistry 2010, 71, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Czarnecki, O.; Grimm, B. Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 2012, 63, 1675–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopečná, J.; Cabeza de Vaca, I.; Adams, N.B.P.; Davison, P.A.; Brindley, A.A.; Hunter, C.N.; Guallar, V.; Sobotka, R. Porphyrin Binding to Gun4 Protein, Facilitated by a Flexible Loop, Controls Metabolite Flow through the Chlorophyll Biosynthetic Pathway. J. Biol. Chem. 2015, 290, 28477–28488. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Bian, C.; Tao, M.; Huang, Y.; Zheng, Y.; Lv, Y.; Li, J.; Wang, C.; You, X.; Jia, B.; et al. Genome and Transcriptome Sequencing of the Astaxanthin-Producing Green Microalga, Haematococcus pluvialis. Genome Biol. Evol. 2019, 11, 166–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Li, Z.; Tao, M.; Li, J.; Hu, Z. Effects of selenite on green microalga Haematococcus pluvialis: Bioaccumulation of selenium and enhancement of astaxanthin production. Aquat. Toxicol. 2017, 183, 21–27. [Google Scholar] [CrossRef]
- Sarada, R.; Tripathi, U.; Ravishankar, G. Influence of stress on astaxanthin production in Haematococcus pluvialis grown under different culture conditions. Process. Biochem. 2002, 37, 623–627. [Google Scholar] [CrossRef]
- Chen, X.; Tian, D.; Kong, X.; Chen, Q.; Abd Allah, E.F.; Hu, X.; Jia, A. The role of nitric oxide signalling in response to salt stress in Chlamydomonas reinhardtii. Planta 2016, 244, 651–669. [Google Scholar] [CrossRef]
- Zuo, Z.; Chen, Z.; Zhu, Y.; Bai, Y.; Wang, Y. Effects of NaCl and Na2CO3 stresses on photosynthetic ability of Chlamydomonas reinhardtii. Biologia 2014, 69, 1314–1322. [Google Scholar] [CrossRef]
- Wen, Z.; Liu, Z.; Hou, Y.; Liu, C.; Gao, F.; Zheng, Y.; Chen, F. Ethanol induced astaxanthin accumulation and transcriptional expression of carotenogenic genes in Haematococcus pluvialis. Enzym. Microb. Technol. 2015, 78, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Jiang, X.; Hu, C.; Sun, T.; Zeng, Z.; Cai, X.; Li, H.; Hu, Z. Optogenetic regulation of artificial microRNA improves H2 production in green alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2017, 10, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Einhaus, A.; Baier, T.; Rosenstengel, M.; Freudenberg, R.A.; Kruse, O. Rational Promoter Engineering Enables Robust Terpene Production in Microalgae. ACS Synth Biol. 2021, 10, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Neupert, J.; Shao, N.; Lu, Y.; Bock, R. Genetic Transformation of the Model Green Alga Chlamydomonas reinhardtii. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2012; Volume 847, pp. 35–47. [Google Scholar]
- Lin, T.-H.; Rao, M.-Y.; Lu, H.-W.; Chiou, C.-W.; Lin, S.-T.; Chao, H.-W.; Zheng, Z.-L.; Cheng, H.-C.; Lee, T.-M. A role for glutathione reductase and glutathione in the tolerance of Chlamydomonas reinhardtii to photo-oxidative stress. Physiol. Plant. 2017, 162, 35–48. [Google Scholar] [CrossRef]
- Hu, Q.; Huang, D.; Li, A.; Hu, Z.; Gao, Z.; Yang, Y.; Wang, C. Transcriptome-based analysis of the effects of salicylic acid and high light on lipid and astaxanthin accumulation in Haematococcus pluvialis. Biotechnol. Biofuels 2021, 14, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Sturn, A.; Quackenbush, J.; Trajanoski, Z. Genesis: Cluster analysis of microarray data. Bioinformatics 2002, 18, 207–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.; Huang, R.; Wang, C.; Hu, Q.; Li, H.; Li, X. Expression of Anti-Lipopolysaccharide Factor Isoform 3 in Chlamydomonas reinhardtii Showing High Antimicrobial Activity. Mar. Drugs 2021, 19, 239. [Google Scholar] [CrossRef] [PubMed]


| Online Analysis Software | Predicted Location(s) |
|---|---|
| YLOC | Cytoplasm |
| Plant-PLoc | Cytoplasm |
| PSORT | Cytoplasm/Chloroplast stroma |
| Gene ID | Forward Primer 5′-3′ | Reverse Primer 5′-3′ |
|---|---|---|
| CHLRE_01g019250v5 | GGGTCACTACATCGTGGCTT | TGATGACGGAGTGGTTGGAC |
| CHLRE_08g372550v5 | AGCAGCTATGGATGCCTACG | TTCAAGGCGGCACTTCTTCA |
| CHLRE_16g656700v5 | CGCAACTCCAAGCTCACCTA | TTTCGCACGCATTGACCTTG |
| CHLRE_13g603350v5 | GTCAGCGTCCCAACAAAAACA | ACCTTGTACACACCGAACGC |
| CHLRE_01g019250v5 | GTCCAACCACTCCGTCATCA | ATCCAGCTGCTTGTACTCGG |
| CHLRE_01g045200v5 | CGAGACCAACCAGACGAAGT | GTAGTTCAGCATCCAGCCGT |
| CHLRE_12g491050v5 | CACTCAGCTCATGTCTCCTCC | ATAGGGAACATGCAGAAGCGG |
| CHLRE_12g558100v5 | CCGACTTCGCCAACTACTTCT | CCACCGCGTACACCTTCTT |
| CrActin | ACCCCGTGCTGCTGACTG | ACGTTGAAGGTCTCGAACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Guo, C.; Huang, D.; Li, H.; Wang, C. The Papain-like Cysteine Protease HpXBCP3 from Haematococcus pluvialis Involved in the Regulation of Growth, Salt Stress Tolerance and Chlorophyll Synthesis in Microalgae. Int. J. Mol. Sci. 2021, 22, 11539. https://doi.org/10.3390/ijms222111539
Liu W, Guo C, Huang D, Li H, Wang C. The Papain-like Cysteine Protease HpXBCP3 from Haematococcus pluvialis Involved in the Regulation of Growth, Salt Stress Tolerance and Chlorophyll Synthesis in Microalgae. International Journal of Molecular Sciences. 2021; 22(21):11539. https://doi.org/10.3390/ijms222111539
Chicago/Turabian StyleLiu, Wenfu, Chunli Guo, Danqiong Huang, Hui Li, and Chaogang Wang. 2021. "The Papain-like Cysteine Protease HpXBCP3 from Haematococcus pluvialis Involved in the Regulation of Growth, Salt Stress Tolerance and Chlorophyll Synthesis in Microalgae" International Journal of Molecular Sciences 22, no. 21: 11539. https://doi.org/10.3390/ijms222111539
APA StyleLiu, W., Guo, C., Huang, D., Li, H., & Wang, C. (2021). The Papain-like Cysteine Protease HpXBCP3 from Haematococcus pluvialis Involved in the Regulation of Growth, Salt Stress Tolerance and Chlorophyll Synthesis in Microalgae. International Journal of Molecular Sciences, 22(21), 11539. https://doi.org/10.3390/ijms222111539






