Structural and Functional Analysis of Female Sex Hormones against SARS-CoV-2 Cell Entry
Abstract
:1. Introduction
2. Results
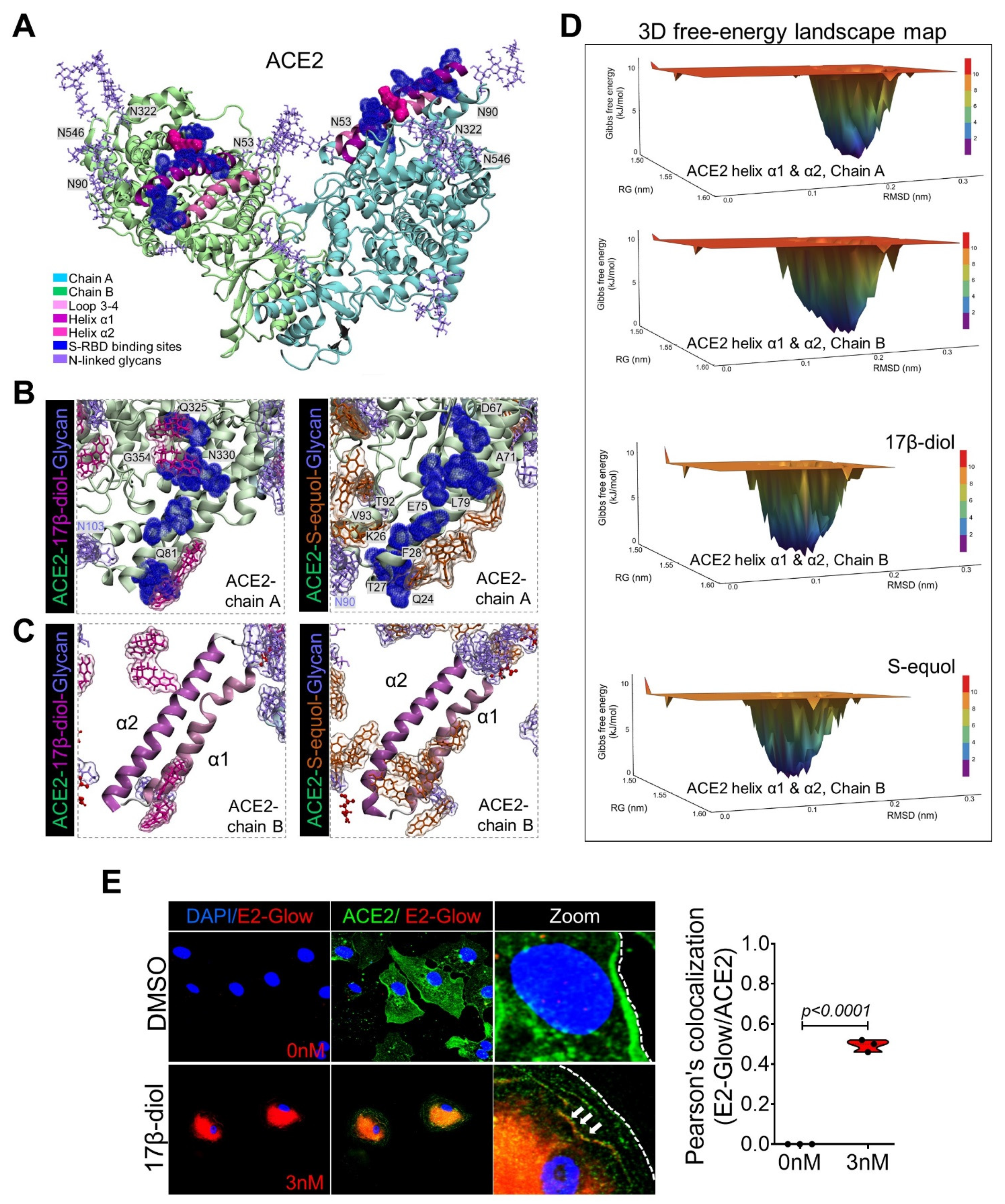
2.1. Glycosylation Site-Mapping of Human ACE2 and SARS-CoV-2 Spike Interactions
2.2. Estrogens Bind to ACE2 and Stimulate Its Stabilization and Internalization
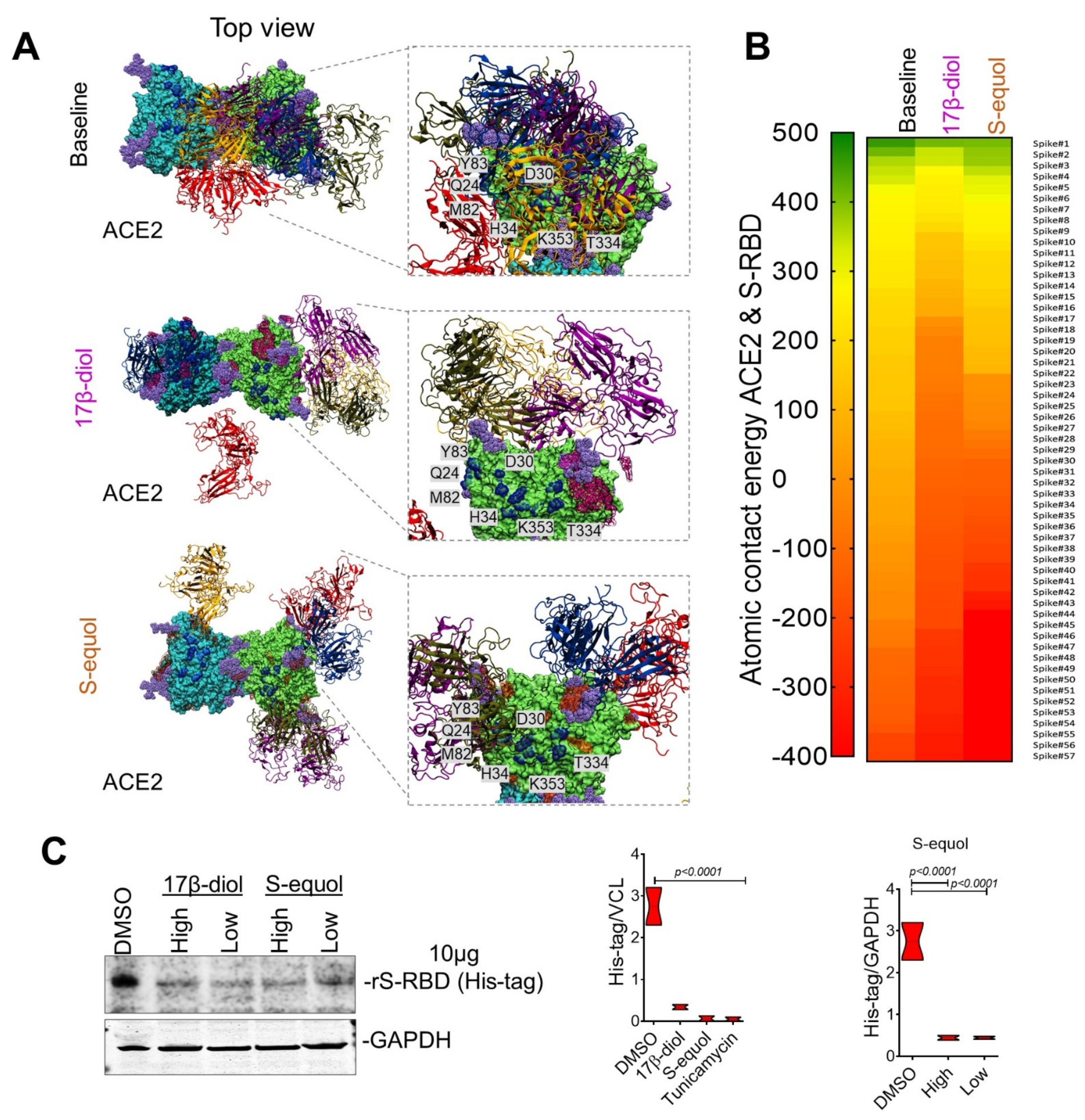
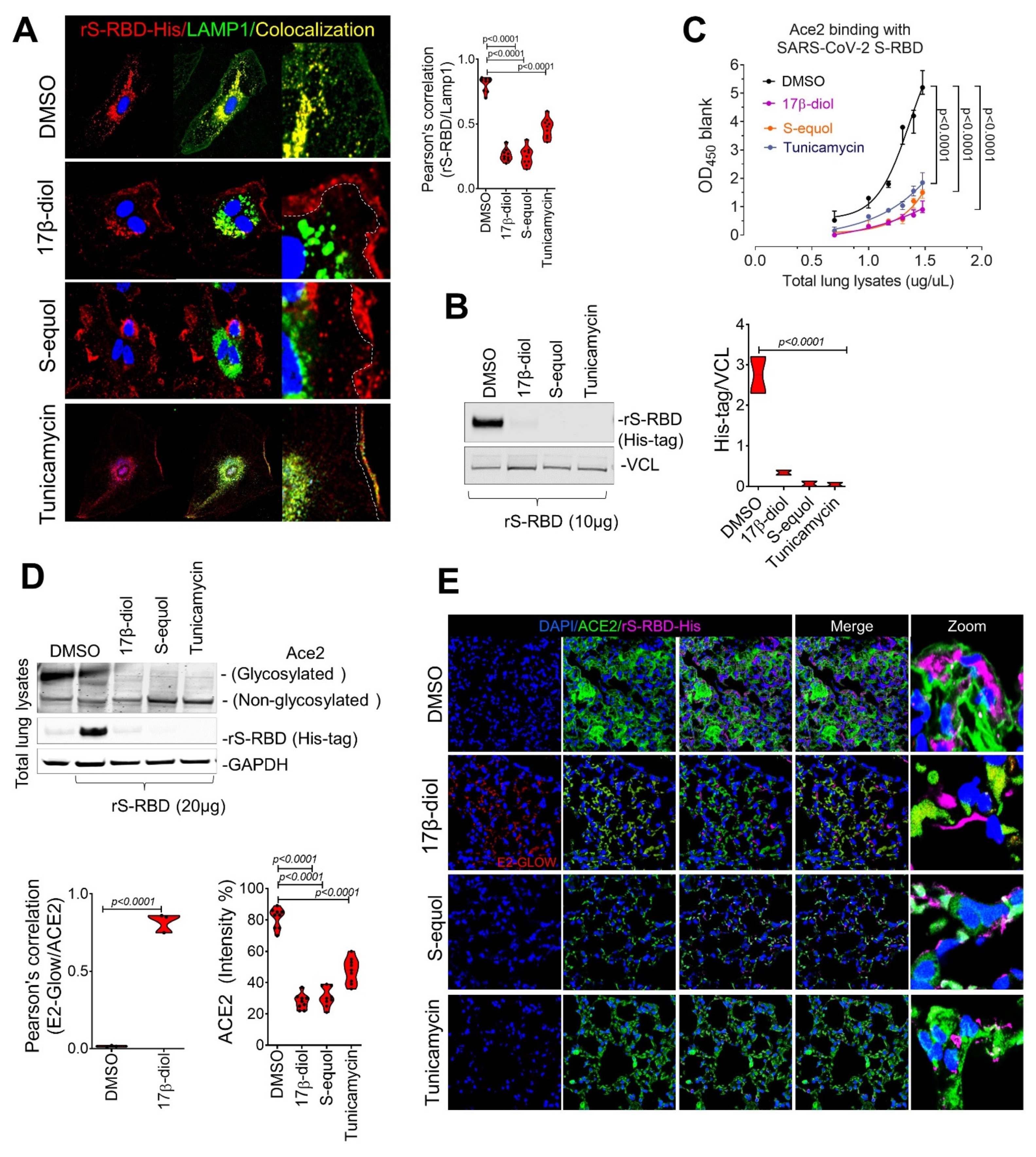
2.3. Estrogens Interfere with SARS-CoV-2 Receptor Binding and Block Entry into the Cell
2.4. Estrogens Block SARS-CoV-2 Infection of the Respiratory Tract in an Animal Model of COVID-19
3. Discussion
4. Methods
4.1. Immunofluorescence Microscopy
4.2. Protein Extraction and Immunoblot
4.3. Animal Treatment
4.4. In Vitro Treatment
4.5. rS-RBD-ACE2 Binding Assay
4.6. Statistics
4.7. Model Building and Glycosylation Process
4.8. Estrogens Solvated System
4.9. Simulation Details
4.10. Structure and Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glinsky, G.V. Tripartite combination of candidate pandemic mitigation agents: Vitamin D, quercetin, and estradiol manifest properties of medicinal agents for targeted mitigation of the COVID-19 pandemic defined by genomics-guided tracing of SARS-CoV-2 targets in human cells. Biomedicines 2020, 8, 129. [Google Scholar]
- Sama, I.; Ravera, A.; Santema, B.T.; Van Goor, H.; Ter Maaten, J.M.; Cleland, J.G.F.; Rienstra, M.; Friedrich, A.W.; Samani, N.J.; Ng, L.L.; et al. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin–angiotensin–aldosterone inhibitors. Eur. Heart J. 2020, 41, 1810–1817. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Jiang, W.; Yao, J.; Nicholson, C.J.; Li, R.; Sigurslid, H.; Wooster, L.; Rotter, J.I.; Guo, X.; Malhotra, R. Predictors of mortality in hospitalized COVID-19 patients: A systematic review and meta-analysis. J. Med. Virol. 2020, 92, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, C.; Regitz-Zagrosek, V.; Neuhauser, H.K.; Morgan, R.; Klein, S.L. Impact of sex and gender on COVID-19 outcomes in Europe. Biol. Sex Differ. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Peckham, H.; de Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020, 11, 6317. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Rasool, R.U.; Russell, R.M.; Natesan, R.; Asangani, I.A. Targeting androgen regulation of TMPRSS2 and ACE2 as a therapeutic strategy to combat COVID-19. iScience 2021, 24, 102254. [Google Scholar] [CrossRef]
- Strope, J.; Chau, C.H.; Figg, W.D. Are sex discordant outcomes in COVID-19 related to sex hormones? Semin. Oncol. 2020, 47, 335–340. [Google Scholar] [CrossRef]
- Dutta, S.; Sengupta, P. SARS-CoV-2 and Male Infertility: Possible Multifaceted Pathology. Reprod. Sci. 2021, 28, 23–26. [Google Scholar] [CrossRef]
- A Bhowmick, N.; Oft, J.; Dorff, T.; Pal, S.; Agarwal, N.; A Figlin, R.; Posadas, E.M.; Freedland, S.J.; Gong, J. COVID-19 and androgen-targeted therapy for prostate cancer patients. Endocr.-Relat. Cancer 2020, 27, R281–R292. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef] [PubMed]
- Samavati, L.; Uhal, B.D. ACE2, Much More Than Just a Receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 2020, 10, 317. [Google Scholar] [CrossRef]
- Libert, C.; Dejager, L.; Pinheiro, I. The X chromosome in immune functions: When a chromosome makes the difference. Nat. Rev. Immunol. 2010, 10, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Fish, E.N. The X-files in immunity: Sex-based differences predispose immune responses. Nat. Rev. Immunol. 2008, 8, 737–744. [Google Scholar] [CrossRef]
- Scully, E.P.; Haverfield, J.; Ursin, R.L.; Tannenbaum, C.; Klein, S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020, 20, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Dalpiaz, E.L.M.; Lamas, A.Z.; Caliman, I.F.; Ribeiro, R.F.; Abreu, G.R.; Moyses, M.R.; Andrade, T.; Gouvea, S.A.; Alves, M.F.; Carmona, A.; et al. Correction: Sex Hormones Promote Opposite Effects on ACE and ACE2 Activity, Hypertrophy and Cardiac Contractility in Spontaneously Hypertensive Rats. PLoS ONE 2015, 10, e0133225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, H.; Park, S.-J.; Choi, Y.K.; Park, T.; Tanveer, M.; Cao, Y.; Kern, N.R.; Lee, J.; Yeom, M.S.; Croll, T.I.; et al. Developing a Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein Model in a Viral Membrane. J. Phys. Chem. B 2020, 124, 7128–7137. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Xia, Y.L.L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Vankadari, N.; Wilce, J.A. Emerging COVID-19 coronavirus: Glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020, 9, 601–604. [Google Scholar] [CrossRef]
- Heckmann, B.; Teubner, B.J.; Tummers, B.; Boada-Romero, E.; Harris, L.; Yang, M.; Guy, C.S.; Zakharenko, S.S.; Green, D.R. LC3-Associated Endocytosis Facilitates β-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 2019, 178, 536–551.e14. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Barrientos, R.; Shiota, T.; Madigan, V.; Misumi, I.; McKnight, K.L.; Sun, L.; Li, Z.; Meganck, R.M.; Li, Y.; et al. Gangliosides are essential endosomal receptors for quasi-enveloped and naked hepatitis A virus. Nat. Microbiol. 2020, 5, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fett, C.; Mack, M.; Eyck, P.P.T.; Meyerholz, D.; Perlman, S. Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J. Immunol. 2017, 198, 4046–4053. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.-Y.; Li, J.-L.; Yang, X.-L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shoemaker, R.; Thatcher, S.; Batifoulier-Yiannikouris, F.; English, V.L.; Cassis, L.A. Administration of 17β-estradiol to ovariectomized obese female mice reverses obesity-hypertension through an ACE2-dependent mechanism. Am. J. Physiol. Metab. 2015, 308, E1066–E1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, K.E.; Thomas, E.; Leaver, M.; Wells, D. Coronavirus disease-19 and fertility: Viral host entry protein expression in male and female reproductive tissues. Fertil. Steril. 2020, 114, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Goad, J.; Rudolph, J.; Rajkovic, A. Female reproductive tract has low concentration of SARS-CoV-2 receptors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Goodman, W.A.; Bedoyan, S.M.; Havran, H.L.; Richardson, B.; Cameron, M.J.; Pizarro, T.T. Impaired estrogen signaling underlies regulatory T cell loss-of-function in the chronically inflamed intestine. Proc. Natl. Acad. Sci. USA 2020, 117, 17166–17176. [Google Scholar] [CrossRef]
- Li, Y.; Jerkic, M.; Slutsky, A.S.; Zhang, H. Molecular mechanisms of sex bias differences in COVID-19 mortality. Crit. Care 2020, 24, 1–6. [Google Scholar] [CrossRef]
- Sreekumar, S.; Levine, K.M.; Sikora, M.J.; Chen, J.; Tasdemir, N.; Carter, D.; Dabbs, D.J.; Meier, C.; Basudan, A.; Boone, D.; et al. Differential Regulation and Targeting of Estrogen Receptor α Turnover in Invasive Lobular Breast Carcinoma. Endocrinology 2020, 161, bqaa109. [Google Scholar] [CrossRef]
- Cavounidis, A.; Mann, E.H. SARS-CoV-2 has a sweet tooth. Nat. Rev. Immunol. 2020, 20, 460. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Ding, T.; Zhang, J.; Wang, T.; Cui, P.; Chen, Z.; Jiang, J.; Zhou, S.; Dai, J.; Wang, B.; Yuan, S.; et al. Potential influence of menstrual status and sex hormones on female SARS-CoV-2 infection: A cross-sectional study from multicentre in Wuhan, China. Clin. Infect. Dis. 2020, 72, e240–e248. [Google Scholar] [CrossRef]
- De Francesco, E.M.; Vella, V.; Belfiore, A. COVID-19 and Diabetes: The Importance of Controlling RAGE. Front. Endocrinol. 2020, 11, 526. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; She, Z.-G.; Cheng, X.; Qin, J.-J.; Zhang, X.-J.; Cai, J.; Lei, F.; Wang, H.; Xie, J.; Wang, W.; et al. Association of Blood Glucose Control and Outcomes in Patients with COVID-19 and Pre-existing Type 2 Diabetes. Cell Metab. 2020, 31, 1068–1077.e3. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Simons, J.P.; Stanca-Kaposta, E.C.; Cocinero, E.J.; Liu, B.; Davis, B.G.; Gamblin, D.P.; Kroemer, R.T. Probing the glycosidic linkage: Secondary structures in the gas phase. Phys. Scr. 2008, 78, 058124. [Google Scholar] [CrossRef]
- Stanley, P.; Schachter, H.; Taniguchi, N. Essentials of Glycobiology: N-Glycans, Ch. 3; CSH Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Danne, R.; Poojari, C.; Martinez-Seara, H.; Rissanen, S.; Lolicato, F.; Rog, T.; Vattulainen, I. doGlycans-tools for preparing carbohydrate structures for atomistic simulations of glycoproteins, glycolipids, and carbohydrate polymers for GROMACS. J. Chem. Inf. Model. 2017, 57, 2401–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Tirado-Rives, J. Potential energy functions for atomic-level simulations of water and organic and bio-molecular systems. Proc. Natl. Acad. Sci. USA 2005, 102, 6665–6670. [Google Scholar] [CrossRef] [Green Version]
- Dodda, L.S.; Vilseck, J.Z.; Tirado-Rives, J.; Jorgensen, W.L. 1.14*CM1A-LBCC: Localized Bond-Charge Corrected CM1A Charges for Condensed-Phase Simulations. J. Phys. Chem. B 2017, 121, 3864–3870. [Google Scholar] [CrossRef] [Green Version]
- Dodda, L.; De Vaca, I.C.; Tirado-Rives, J.; Jorgensen, W.L. LigParGen web server: An automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017, 45, W331–W336. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Madura, J.D. Temperature and size dependence for Monte Carlo simulations of TIP4P water. Mol. Phys. 1985, 56, 1381–1392. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Parr, R.G.; Yang, W. International Series of Monographs on Chemistry 16; Oxford University Press: Oxford, UK, 1989. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. IV. A new dynamical correlation functional and implications for exact-exchange mixing. J. Chem. Phys. 1996, 104, 1040–1046. [Google Scholar] [CrossRef]
- Schäfer, A.; Horn, H.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J. Chem. Phys. 1992, 97, 2571–2577. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Dennington, R.; Keith, T.A.; Millam, J.M. GaussView, Version 6; Semichem Inc.: Shawnee Mission, KS, USA, 2016. [Google Scholar]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Hockney, R.; Goel, S.; Eastwood, J. Quiet high-resolution computer models of a plasma. J. Comput. Phys. 1974, 14, 148–158. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewald, P.P. Die Berechnung optischer und elektrostatischerGitterpotentiale. Ann. Phys. 1921, 369, 253–287. [Google Scholar] [CrossRef] [Green Version]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Vasmatzis, G.; Cornette, J.L.; DeLisi, C. Determination of atomic desolvation energies from the structures of crystallized proteins. J. Mol. Biol. 1997, 267, 707–726. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. Science forum: SARS-CoV-2 (COVID-19) by the numbers. eLife 2020, 9, e57309. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Turner, P.J. XM Grace, Version 5.1.19; Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology: Beaverton, OR, USA, 2005. [Google Scholar]
- Wolfram Research, Inc. Mathematica, Version 12.1; Wolfram Research, Inc.: Champaign, IL, USA, 2020. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilar-Pineda, J.A.; Albaghdadi, M.; Jiang, W.; Vera-Lopez, K.J.; Nieto-Montesinos, R.; Alvarez, K.L.F.; Davila Del-Carpio, G.; Gómez, B.; Lindsay, M.E.; Malhotra, R.; et al. Structural and Functional Analysis of Female Sex Hormones against SARS-CoV-2 Cell Entry. Int. J. Mol. Sci. 2021, 22, 11508. https://doi.org/10.3390/ijms222111508
Aguilar-Pineda JA, Albaghdadi M, Jiang W, Vera-Lopez KJ, Nieto-Montesinos R, Alvarez KLF, Davila Del-Carpio G, Gómez B, Lindsay ME, Malhotra R, et al. Structural and Functional Analysis of Female Sex Hormones against SARS-CoV-2 Cell Entry. International Journal of Molecular Sciences. 2021; 22(21):11508. https://doi.org/10.3390/ijms222111508
Chicago/Turabian StyleAguilar-Pineda, Jorge Alberto, Mazen Albaghdadi, Wanlin Jiang, Karin J. Vera-Lopez, Rita Nieto-Montesinos, Karla Lucia F. Alvarez, Gonzalo Davila Del-Carpio, Badhin Gómez, Mark E. Lindsay, Rajeev Malhotra, and et al. 2021. "Structural and Functional Analysis of Female Sex Hormones against SARS-CoV-2 Cell Entry" International Journal of Molecular Sciences 22, no. 21: 11508. https://doi.org/10.3390/ijms222111508
APA StyleAguilar-Pineda, J. A., Albaghdadi, M., Jiang, W., Vera-Lopez, K. J., Nieto-Montesinos, R., Alvarez, K. L. F., Davila Del-Carpio, G., Gómez, B., Lindsay, M. E., Malhotra, R., & Lino Cardenas, C. L. (2021). Structural and Functional Analysis of Female Sex Hormones against SARS-CoV-2 Cell Entry. International Journal of Molecular Sciences, 22(21), 11508. https://doi.org/10.3390/ijms222111508






