Abstract
A series of eight copper (II) complexes with 3-(4-chloro-3-nitrophenyl)thiourea were designed and synthesized. The cytotoxic activity of all compounds was assessed in three human cancer cell lines (SW480, SW620, PC3) and human normal keratinocytes (HaCaT). The complexes 1, 3, 5, 7 and 8 were cytotoxic to the studied tumor cells in the low micromolar range, without affecting the normal cells. The complexes 1, 3, 7 and 8 induced lactate dehydrogenase (LDH) release in all cancer cell lines, but not in the HaCaT cells. They provoked early apoptosis in pathological cells, especially in SW480 and PC3 cells. The ability of compounds 1, 3, 7 and 8 to diminish interleukin-6 (IL-6) concentration in a cell was established. For the first time, the influence of the most promising Cu (II) complexes on intensities of detoxifying and reactive oxygen species (ROS) scavenging the enzymes of tumor cells was studied. The cytotoxic effect of all copper (II) conjugates against standard and hospital bacterial strains was also proved.
1. Introduction
Enzymes incorporate transition metal cofactors to perform a wide range of metabolic reactions. Among metal cations, copper (II) ions play an essential function in the human organism, being a structural component of several enzymatic proteins, such as ascorbate oxidase, ceruloplasmin, amine oxidase, cytochrome C oxidase, nitrite reductase and superoxide dismutase Cu–Zn (SODC) [1,2]. Complexes of organic chelators with Cu (II) have been receiving increased attention due to their potential biomedical applications, such as cytotoxic or antibacterial activities. Within a group of acylthiourea ligands, those containing electron-withdrawing chloro- and nitro-substituents were effective against adenocarcinoma cell lines [3]. Intercalating properties of N-(2-hydroxyethyl)-N′-benzoylthiourea-Cu (II) complexes with strong deoxyribonucleic acid (DNA) binding and cleavage abilities were denoted [4]. Chelates with orthophenanthroline units studied by Pivetta et al. exerted high cytotoxic affect against acute leukemia and other carcinomas [5]. Moreover, a wide antibacterial profile of copper-based thiourea coordinates of 4-azatricyclo-3,5-dione [6] and 3-(trifluoromethyl)aniline was also studied. Their inhibitory effect on bacterial topoisomerases was also established [7].
Requirements for safer, more active and more selective chemotherapeutics have stimulated the search for other metal-based antitumor candidates. First of all, investigations in the treatment of cancer diseases are oriented to synthesize new analogues of cisplatin, a platinum (II) coordinate and chemotherapeutic drug effective against various types of tumors. Recently published Pd (II) and Pt (II) complexes of N-allylthioureas containing morpholine and tert-butylamine moieties showed proapoptotic and cytostatic influence on HeLa (Henrietta Lacks) cell culture stronger or comparable with cisplatin [8]. Antitumor properties of structurally similar coordinates based on pyrazol-3-ylpyridine fragments were connected with their inducing effect on cell accumulations in the S phase (HeLa cells) and a reduction of cell subpopulations in G2/M and G0/G1 phases (pancreatic cancer cells) [9]. Among tested palladium complexes of N, N-disubstituted thioureas, those endowed with furoyl and ethyl groups were the most effective against human breast cancer cells, as compared to the platinum-derived standard drug [10]. On the other hand, Marverti et al. [11] introduced a series of thiourea-Pd (II) compounds that acted as productive growth inhibitors of cisplatin-sensitive ovarian cancer cell lines and their resistant counterparts. Coordinates altered malignant cell metabolism via inhibition of thymidylate synthase and dihydrofolate reductase expression. Interestingly, photoactivated platinum (IV) azide dipyridine complexes bonded strongly to the nuclear DNA of tumor cells and blocked ribonucleic acid (RNA) polymerase II more efficiently than conventional adducts of cisplatin [12]. Platinum (II)-acridine hybrids linked to the amidine group [13] have shown inhibition of lung cancer cell proliferation and a promising antitumor potency in mouse xenograft models. In contrast, cationic gold (I) complexes of thiourea containing an acridine core are devoid of cytotoxic properties. However, they exert remarkable activity against M. tuberculosis [14]. Gold (I) complexes of cyclic disubstituted 4-chlorophenylthiourea ligand severely affected mitochondrial respiration by inhibition of complexes II and IV of the respiratory chain and induced mitochondrial swelling, resulting in enhanced permeability of a membrane and its declined fluidity [15]. An alternative class of anticancer drugs has been identified within ruthenium (II)–arene compounds. The Ru (II)-acylthiourea organometallic complexes were found to be significantly cytotoxic towards lung [16,17,18], prostate [16] and breast [18] tumor cell cultures, inducing their apoptosis and reducing migration by interaction with human serum albumin. What is more, other results showed that silver (I) chelates of 2-benzimidazolylurea mediate a strong cytotoxic response to the tested breast cancer cell lines (MCF-7) and normal cells, but with better parameters than metallodrug cisplatin [19].
Reactive oxygen species (ROS), produced in the mitochondria, peroxisomes and endoplasmic reticulum, include highly reactive species such as hydroxyl and superoxide free radicals, singlet oxygen or less reactive hydrogen peroxide molecules. Regulation of ROS levels is crucial for cellular life, cell proliferation and differentiation. Under oxidative stress states, accumulation of ROS causes the damage of proteins, lipids and DNA, which contributes to carcinogenesis [20]. The expanded scavenging system of a cell is based on superoxide dismutases (SODs), glutathione peroxidase (GPX), glutathione reductase (GR), peroxiredoxins (PRDXs), thioredoxin and catalase (CAT), which neutralize reactive species or recover antioxidants to their reduced state [21]. In comparison with normal cells, an extent of ROS in cancer counterparts is higher, as they keep an improved antioxidant system [22]. What is more, expanded levels of GPX, CAT, glutathione and thioredoxin proteins are correlated with tumor aggression and its resistance to chemotherapy [22]. It was proved that a decline of ROS-scavenging capacity leads to apoptosis of malignant cells, especially when it is used in combination with ROS-increasing chemotherapeutics, such as 5-fluorouracil (therapy of colon cancer), celecoxib (prostate tumor) or doxorubicin (breast, liver cancers) [21,23,24,25,26]. Numerous organic complexes with transition metals are reported to induce autophagy and/or apoptosis by regulation of ROS. This group includes complexes of gold (I) with thiourea [15,27], ruthenium (II) coordinates with β-carboline [28], copper (II) complexes with 2-hydroxy-1-naphthaldehyde [29] and with the derivative of bipyridine-acetylacetonate (Casiopeina III-ia) [30]. The antioxidant abilities of coordinates evaluated by the diphenyl-2,2-picrylhydrazyl/ N, N-diethyl-p-phenylenediamine (DPPH/DPD) method were also confirmed for ruthenium (III) chelates with 1-ethyl-3-phenylthiourea ligands [31] and copper (II) complexes with biuret or urea [32], as well as with auxin [33] or N-ethylpiperazine [34].
As a part of our research on developing biologically active 1,3-disubstituted thioureas [7,35,36,37], we have synthesized a series of halogenated copper (II) complexes of (4-chloro-3-nitrophenyl)thiourea and reported their cytotoxic, prooxidant and antimicrobial properties.
2. Results and Discussion
2.1. Synthesis of Complexes
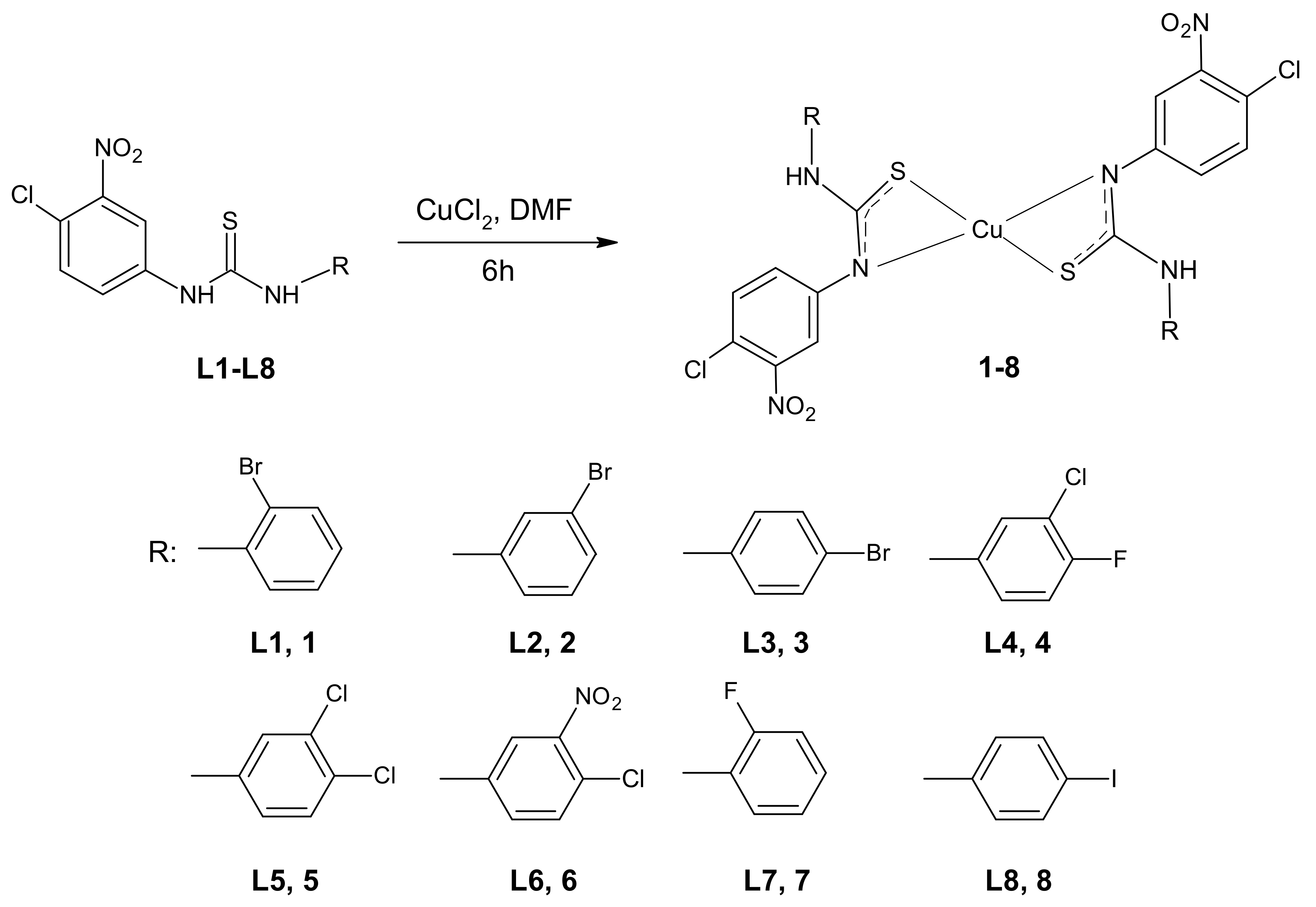
Thiourea–Cu (II) coordination compounds 1–8 were prepared by the reaction of copper (II) chloride with 3-(4-chloro-3-nitrophenyl)thiourea compounds (Figure 1). The complexes were obtained with satisfactory yields (45–61%). The identification of the parent ligands L1–L8, using nuclear magnetic resonance (NMR) and mass spectroscopy, was described previously [37,38]. The derivatives chosen for complexation reactions came from a series of mono- (L4–L6) or di-halogenated (L1–L3, L7, L8) 1-phenylthioureas with the most promising bioactivity. The presented selection of the phenyl ring substituents in their structures allow for the analysis of the influence of the substitution isomerism, as well as the impact of electron-withdrawing elements on the biological properties of newly synthesized metal–organic compounds.
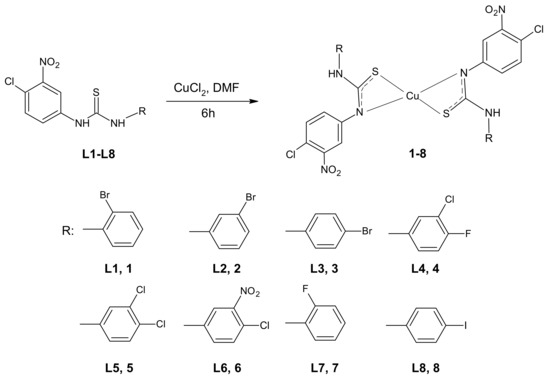
Figure 1.
Scheme of the complexation reaction of the respective arylthiourea ligands L1–L8 with proposed molecular structures of complexes 1–8 (DMF—dimethylformamide).
2.2. Structural Characterization of Complexes
The complexes were initially characterized by elemental analysis and infrared spectroscopy. During all complexation reactions, disubstituted thiourea acts as bidentate ligand (L), forming a hydrated complex of the type CuL2. In order to determine the geometry of metal–organic ligand complexes, ultraviolet–visible (UV–Vis) and electron paramagnetic resonance (EPR) spectroscopies have been applied.
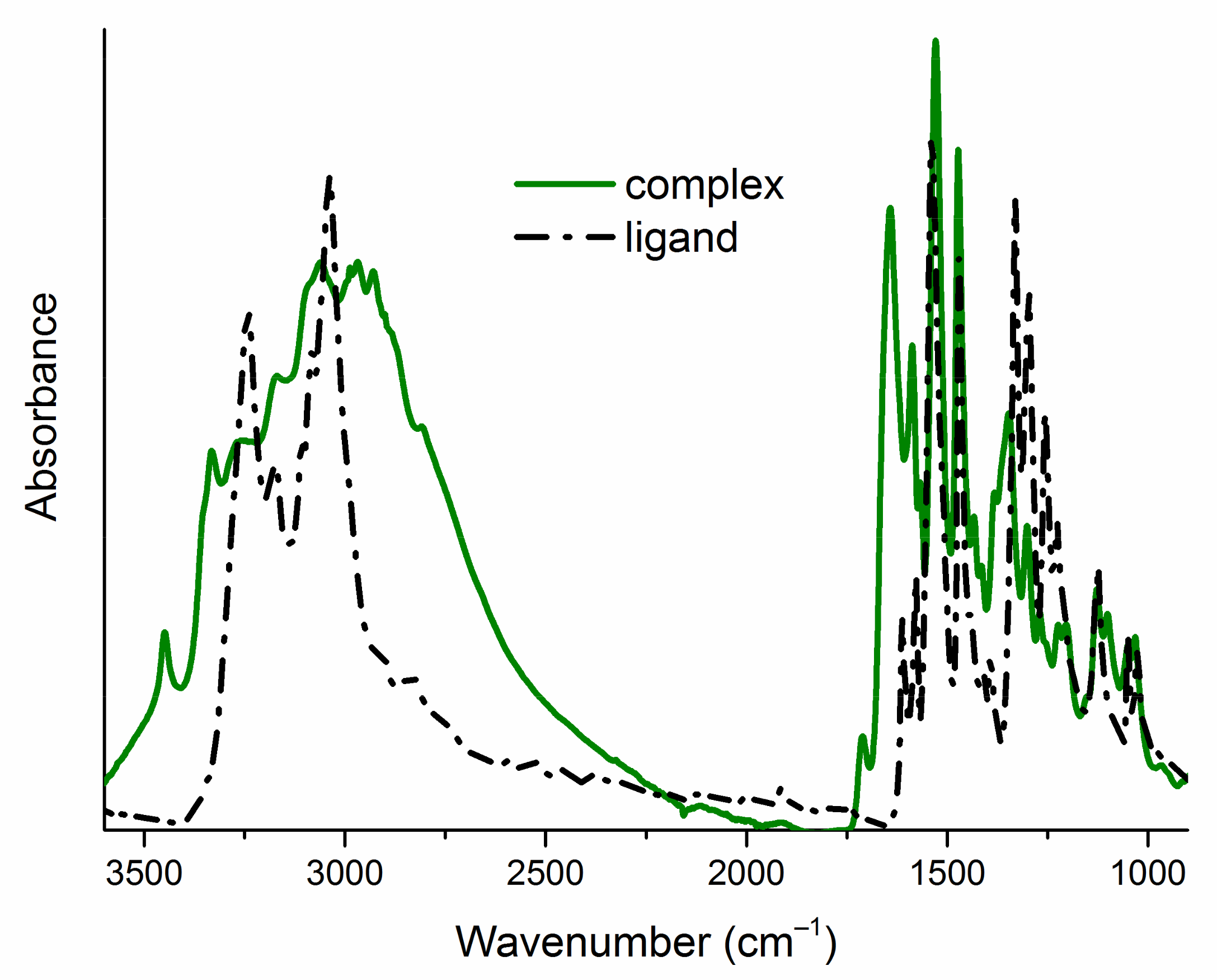
The infrared spectra of ligands (L1–L8) exhibit a broad band with several maxima in the high-energy part (Figure 2), corresponding to the stretching vibrations of the N–H and C–H groups. In the spectra of Cu (II) complexes (1–8), this band widens (Figure 2), which confirms the presence of water molecules in the product of the complexation reactions. The molecules of the initial ligands (L1–L8) and their complexes (1–8) contain two N–H groups (Figure 1), which are reflected in two infrared absorption bands. These bands are in different positions in the spectra of a pair of free and bonded ligands (Figure 2, Table 1). This confirms deprotonation of one N-atom from the ligand’s molecules and suggests different intermolecular interactions (N–H... X; X=O, Br, Cl, F, I hydrogen bonding patterns) in the crystal structure of free and bonded thioureas. The region of 1600–1400 cm−1 in the spectra of ligands (L1–L8) is dominated by the stretching vibrations of the C–C and C–N bonds. In the spectra of complexes (1–8), this feature is extended towards the higher frequencies (around 1700 cm−1, Figure 2) as the consequence of increased intensities of asymmetric ring stretching modes within the molecules of complexes [7]. The spectral ranges: 1550–1500 cm−1 (NO2 asymmetric stretching vibrations), 1360–1330 cm−1 (NO2 symmetric stretching vibrations) and 1240–1040 cm−1 (C–halogen stretching modes) are quite similar for parent ligands and their Cu (II) complexes (Table 1), which excludes the interaction of the nitro group or the halogen atom with the metal cation. The bands corresponding to the C=S stretching vibrations are found around 1333 cm−1 and in the 854–832 cm−1 range in the uncomplexed ligand’s spectra (Table 1). In the spectra of complexes, they are red shifted and observed at 1313–1300 cm−1 and 785–771 cm−1 regions (Table 1). This indicates that the ligands coordinate to the Cu (II) through the sulfur atom [39,40,41].
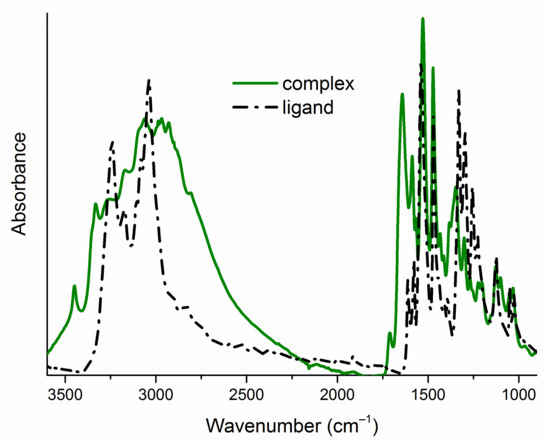
Figure 2.
Attenuated total reflection infrared (ATR-IR) spectra of one exemplary pair of a free ligand (L5) and a complex (5) in the range of 3600–900 cm–1. ATR-IR spectra of all studied compounds are gathered in the Supplementary Materials (Figure S1).

Table 1.
Characteristic infrared absorption bands of N−H, NO2, C=S and C−X (X = Br, Cl, F, I) groups. Band position in cm−1, intensity: w is weak, m is medium, s is strong, vs is very strong, sh is shoulder, as is asymmetric, s is symmetric, * is broad band.
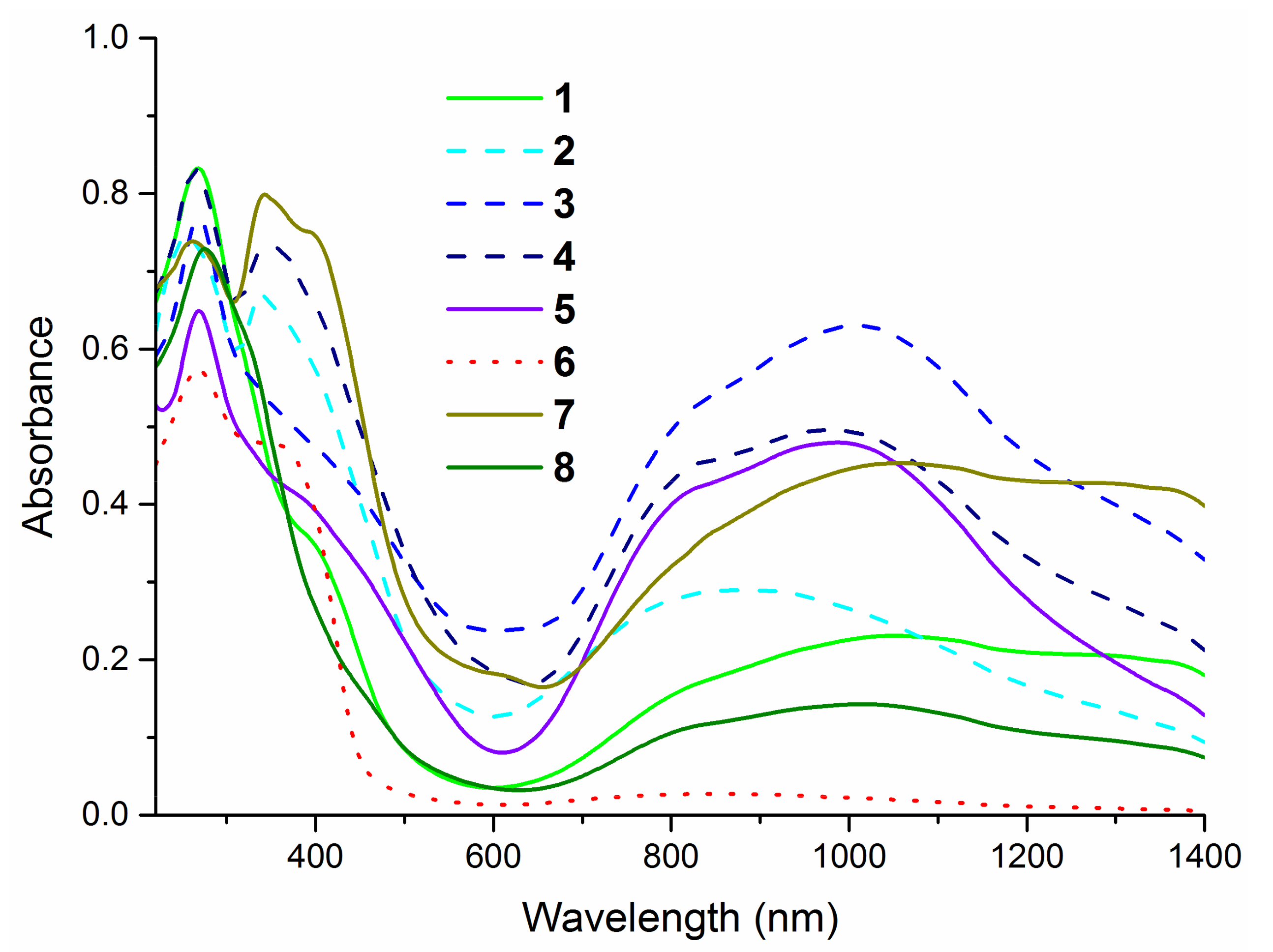
The electronic spectra of the complexes (Figure 3) exhibit the intense absorption bands in the region 230–300 nm corresponding to the π→π* and n→π* intra-ligand transitions. The presence of S→Cu (II) ligand-to-metal charge transfer (LMCT band) around 400 nm is additional evidence of the coordination of thiourea ligands to the metal cation via sulfur atom [39,40,42]. All presented compounds show the d-d bands in a wide region 650–1400 nm, which confirms the formation of four-coordinate complexes with copper on +2 oxidation state [41].
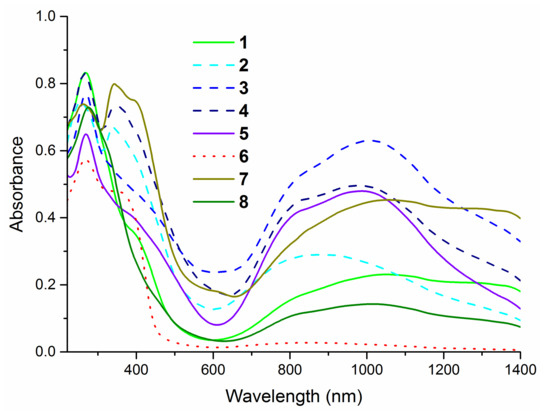
Figure 3.
Solid state ultraviolet–visible (UV–Vis) spectra of complexes in the range of 220–1400 nm.
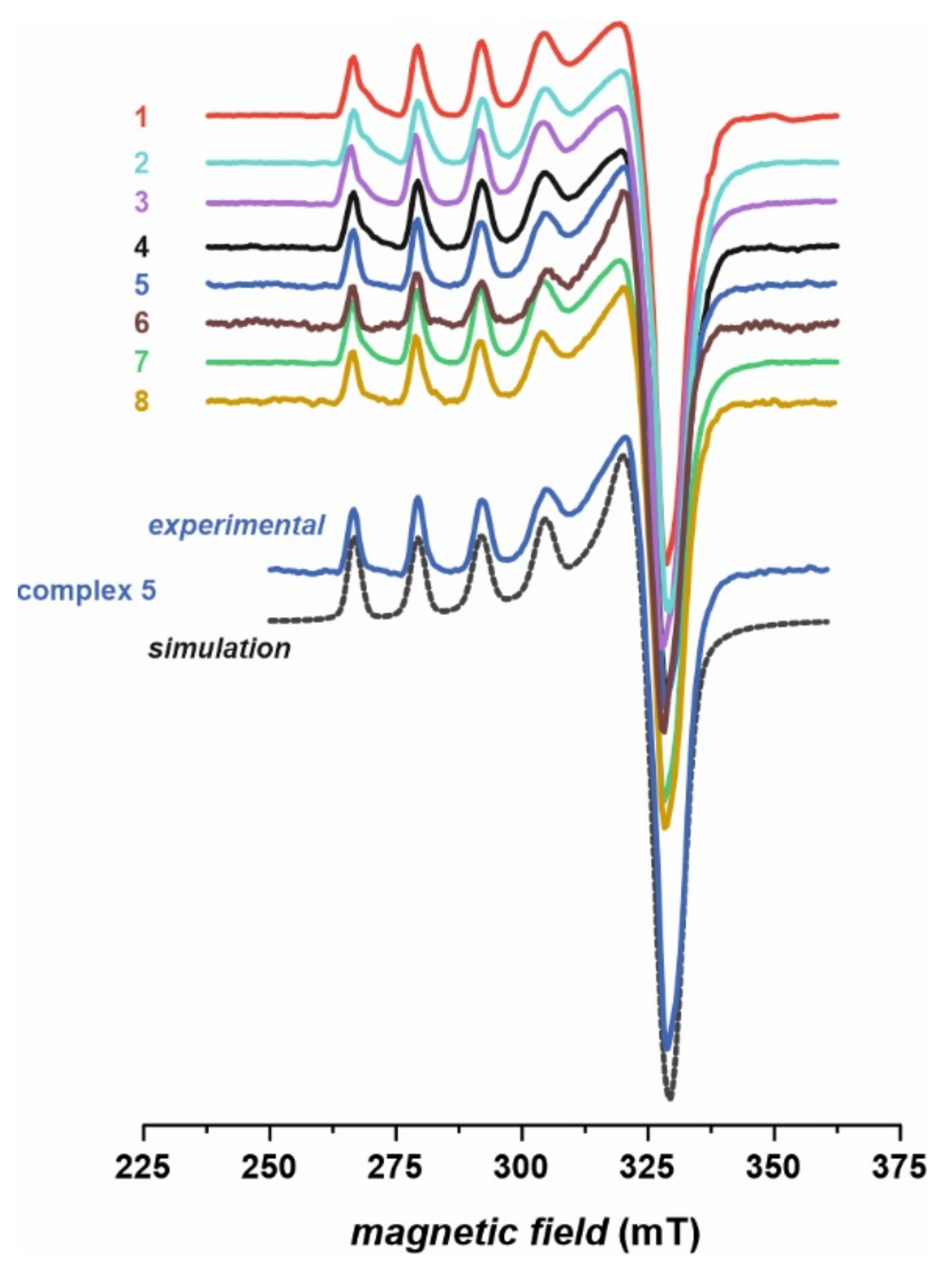
EPR spectra of the investigated complexes 1–8 (Figure 4) are in agreement with the conjectures drawn based on the infrared (IR) and UV–Vis results. The obtained spectra indicate clearly the divalent state of copper upon complexation and are very characteristic of typical Cu (II) complexes. The signals were simulated assuming rhombic symmetry with the z component (parallel) split into four lines due to the hyperfine interaction between the unpaired electron and nuclear magnetic moment of copper (I = 3/2, 63,65Cu). The splitting of the perpendicular line (x and y components) remains unresolved, yet it was necessary for obtaining the correct shape of the simulated spectra. Slight rhombicity of the spectra with gzz >> gxx > gyy confirms dominant contribution of a single d-orbital (3dx2–y2) in the semi-occupied molecular orbital. Exemplary parameters obtained for compound 5 are: gxx = 2.086, gyy = 2.054, gzz = 2.357, Axx and Ayy unresolved, |Azz| = 12.8 mT. Similar parameters were recently reported for Cu (II) thiourea complexes [6,7]. The EPR parameters obtained for other complexes are very similar (12.6 mT < |Azz| < 12.8 mT, 2.353 < gzz < 2.358), which strongly indicates very similar coordination around the copper center. It is in agreement with the molecular structure of the complexes (with two thiourea ligands chelated to the Cu (II) cation via deprotonated N and thiocarbonyl S atoms), which differ only in the substituents of thiourea ligands (Figure 1). Such modification has only a minute effect on the first coordination sphere of copper, to which continuous wave EPR is sensitive. No features characteristic of Cu dimer in solution were observed, e.g., a forbidden magnetic dipolar transition at half-field with g = 4 value.
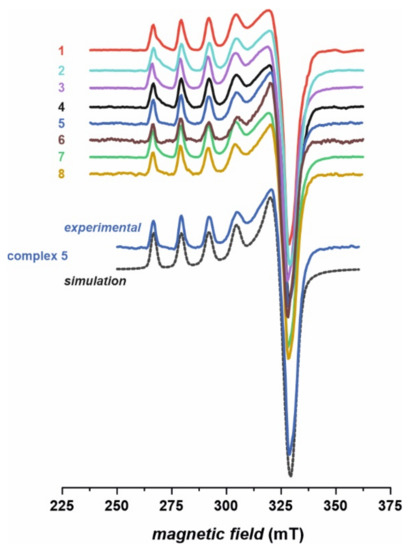
Figure 4.
Frozen solution (77 K) X-band electron paramagnetic resonance (EPR) spectra of complexes 1–8, along with exemplary computer simulation of spectrum for the complex 5.
The molecular structure proposed for compounds 1–8, in which the Cu (II) cation is the part of two four-membered rings (Cu-S-C-N, see Figure 1), is quite unusual for copper. However, this type of coordination was reported for one crystal structure determined by Singh et al. [43]. They found Cu (II) ions being chelated by two thiourea ligands in a trans manner and the distorted square-planar geometry of the complex was stabilized by electron delocalization in chelate rings. Moreover, such coordination was postulated by some of the authors of the current study for another series of Cu (II) complexes, namely for 3-(trifluoromethyl)phenylthiourea derivatives [7] and thioureas containing tricyclic imide’s part [6,41]. Structural characterization of those complexes, possibly owing to the combination of several experimental laboratory and synchrotron techniques (including X-ray absorption spectroscopy) and molecular modelling, indicated that 1,3-disubstituted thiourea ligands coordinate to Cu (II) cation in bidentate fashion through S and N atoms, forming a four membered 1,3-N, S chelate ring (as presented in Figure 1 for compounds 1–8).
2.3. Biological Studies
2.3.1. Anticancer Activity
MTT Assay
In vitro cytotoxicity studies of synthesized thiourea complexes 1–8 were assessed by standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) bioassay in different cancer cells at 72 h of drug exposure (Table 2). The ability of compounds to inhibit cell proliferation was established by means of their half-maximal inhibitory concentration (IC50) values towards the human tumor cell lines, such as SW480 (primary colon cancer), SW620 (metastatic colon cancer) and PC3 (metastatic prostate cancer), as well as against the non-tumor cell line HaCaT (immortal keratinocytes).

Table 2.
Cytotoxic activity (IC50, µM) of studied compounds estimated by the MTT assay a.
The 1,3-disubstituted arylthiourea complexes 1, 3, 5, 7 and 8 were cytotoxic to studied cancer cells at concentration ≤ 10 µM. The monohalogen derivatives 1 and 8 showed cytotoxic activity in the low micromolar range in all tested pathological cell lines. Their IC50 values ranged from 3.3 ± 0.2 to 9.7 ± 0.1 µM (compound 1) and between 3.9 ± 0.8 and 17.8 ± 1.3 µM for compound 8. In particular, they both were strongly potent towards SW480 cells (at 3.9 ± 0.8 and 4.7 ± 0.3 µM), whereas derivative 1 was against SW620 (at 3.3 ± 0.2 µM) and derivative 8 towards the PC3 cell line (with IC50 of 4.3 ± 0.5 µM).
The 4-bromophenyl compound (3) exerted remarkable growth-inhibiting activity mainly against PC3 cells (8.8 ± 0.8 µM). On the other hand, the 2-fluorophenyl-containing molecule 7 exerted the highest IC50 values for SW480 and PC3 cell lines (9.1 ± 0.8 and 10.8 ± 1.3 µM, respectively). Dichlorophenyl derivative 5 was potent against SW620 cancer cells at 10.8 ± 2.6 µM.
In general, the PC3 cell line was the most sensitive to the presence of new complexes, and substances 1, 3 and 8 appeared the most effective in the inhibition of their rise. The same set of compounds acted the strongest against primary cancerous SW480 cells. In contrast, for substances 1, 5 and 7, the lowest IC50 indexes for metastatic SW620 cells were denoted. The cytotoxicity of other tested derivatives (2, 4, 6) towards pathological cell lines was moderate, with IC50 values from 19.2 ± 1.1 to 26.8 ± 2.3 µM.
The studied thiourea complexes were non-cytotoxic against normal HaCaT cell lines (IC50 ˃ 100 µM). The highest selectivity indexes (SI) were observed for the most promising derivatives: 1 (33.2 vs. SW620; 23.3 for SW480) and 8 (26.3 vs. SW480; 23.8 vs. PC3 cells). The selectivity factors of other complexes 2–7 were in the range of 4.6–8.9 (SW480), 5.3–15.2 (SW620) and 5.2–14.4 (PC3 cell line). These indexes were incomparably greater than those of the reference doxorubicin and cisplatin (SI between 0.4 and 1.1), which proves a low toxicity of tested complexes towards health cells.
As compared to cisplatin, the cytotoxic properties of the synthesized diarylthiourea derivatives were considerably stronger. Complexes 1, 3, 5, 7 and 8 were 1.2–3.1-fold more active against human PC3 cell lines. Additionally, both studied colon cancer cell lines appeared to be 2–2.7 times more sensitive to the presence of coordination compounds 1 and 8 than cisplatin, one of the most frequently used chemotherapy drugs.
The present study reveals that monohalogen substituted phenylthiourea complexes (1–3, 7, 8) possess stronger cytotoxic properties in human cancer cell lines, when compared to disubstituted derivatives (4–6). The moderately electronegative atom, such as bromine or iodide at the ortho (1,7) or para (3, 8) position of the aromatic ring, was responsible for enhanced cytotoxicity in the MTT model. The location of an element at meta position (2) resulted in a severalfold reduction in anticancer activity. Among disubstituted halogen coordinates, the 3,4-dichlorophenyl derivative (5) appeared to be the most effective towards SW620 and PC3 cells. The substitution of the benzene with chlorine and nitro group together provided a less promising drug candidate (6), with IC50 at least 20.3 ± 3.1 µM against all tested cancer cells. Comparing pairs 7 (2-fluorophenyl-) and 4 (4-fluoro-3-chlorophenylthiourea), an introduction of the second halogen to the molecule led to the twofold decrease in cytotoxic activity vs. metastatic SW620 and PC3 cell lines. In fact, the location of two different groups in both third and fourth position of the ring was not beneficial on biological activity (4,6). Only 3,4-dichlorophenyl complex (5) restrained the potency in the low micromolar range, as compared to its monohalogen analogs 3 and 7. To sum up, the phenyl ring substituents can be arranged in order of their decreasing impact on bioactivity as follows: 2-bromo > 4-iodo > 2-fluoro > 4-bromo > 3,4-dichloro >> 3-chloro-4-fluoro > 3-bromo > 3-nitro-4-chloro. It is worth noting that all of them were at least 350 times less toxic for normal HaCaT cell lines than reference doxorubicin.
As reported, the cytotoxicity of standard copper (II) chloride towards cancer cells was negligible in comparison with thiourea derivatives. Its C50 ranged between 96.3 and 109.4 µM, and the salt was nontoxic for normal cells (IC50 >100 µM). DMF, an organic solvent that contaminates synthesized complexes, has already been tested on various cells and appears to be nontoxic at the concentrations used in our synthesis (see in the Supplementary Material section). Considering an influence of starting ligands L1–L8 on studied cell lines, the derivative L5, at the concentration 8 µM, caused an approximately 20% decrease in HaCaT cell viability, which indicated its weak cytotoxic impact against normal cells [36]. In contrast, new complexes are devoid of visible influence on keratinocytes. In preliminary tests published previously, ligands L1, L3–L5 and L8 influenced the growth of other cancer cells, such as human leukemia and solid (melanoma, prostate) tumors, as well as normal tissue foreskin fibroblasts [36]. These results encouraged us to proceed with deeper investigations on the cytotoxicity mechanism of complexes 1–8.
LDH Assay
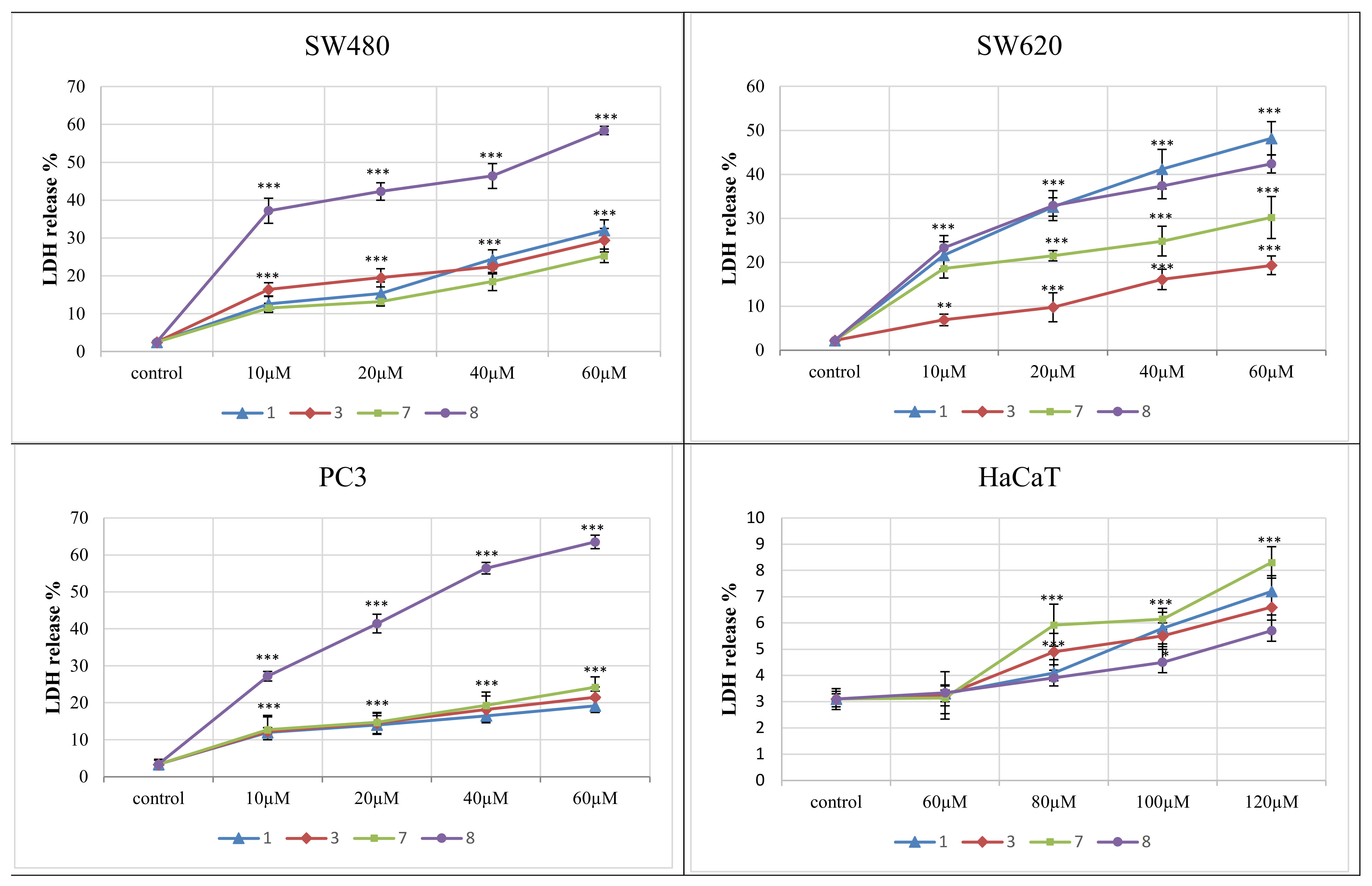
The lactate dehydrogenase (LDH) release assay was used to determine the level of plasma membrane damage of the most distinctive derivatives 1, 3, 7, 8. They were studied at concentrations of 10–60 μM against cancer cell lines and 60–120 μM towards normal HaCaT cells.
The LDH release curve results (Figure 5) demonstrated that the applied amounts of complexes did not affect the normal keratinocytes viability, as the secretion of LDH in their presence varied from 1.3% to 5.7%. The 4-iodophenylthiourea complex 8 exerted the highest cytotoxicity against all three tumor cell lines. This effect was the most evident in PC3 and SW480 cells, compared to SW620. When used at 60 μM, the derivative 8 achieved 64% LDH release in PC3 cells and 58% in SW480 cell lines. LDH secretion in these cells accounted for other complexes, used at the highest concentrations, ranging from 19% to 32%. The compound 8, applied in a dose of 40 and 20 μM, also expressed an increased response against SW480 cells (LDH release was 46.4% and 42%, respectively). Similarly, the LDH leakage induced in PC3 cells by this derivative used at lower concentrations differed from 56.4% (40 μM) to 27.2 % (10 μM). On the other hand, the number of lysed SW620 cells evoked by the presence of the 2-bromophenylthiourea complex 1 ranged from 48% (at 60 μM) to 22% (10 μM applied), and incubation of these cell lines with the derivative 8 expressed similar enzyme releases of 42.4% and 23%, respectively. 2-fluorophenyl compound 7, studied at its highest dose, was less cytotoxic and achieved 30.2% LDH secretion.
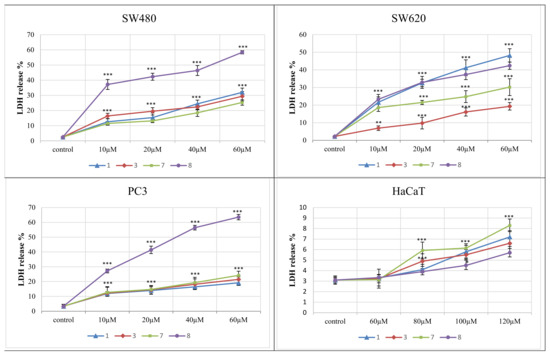
Figure 5.
Lactate dehydrogenase (LDH) release as a marker of cell death in the SW480, SW620, PC3 and HaCaT cells, treated for 72 h with different concentrations of compounds 1, 3, 7 and 8. LDH release in the HaCaT was analyzed after treatment with higher doses of compounds. *** p ≤ 0.001 as compared to the control.
The reference cancer chemotherapeutic doxorubicin showed very high LDH release in all studied cancer cell lines, even when applied in lower doses. The LDH percentage for this compound at 1.5 μM varied from 70% to 98%. However, its toxicity against normal HaCaT cells was significantly higher than examined complexes and accounted for 80%.
The data gained by the LDH activity assay are in an agreement with the results obtained for derivatives 1, 3, 7 and 8 by the MTT method.
Apoptotic Activity
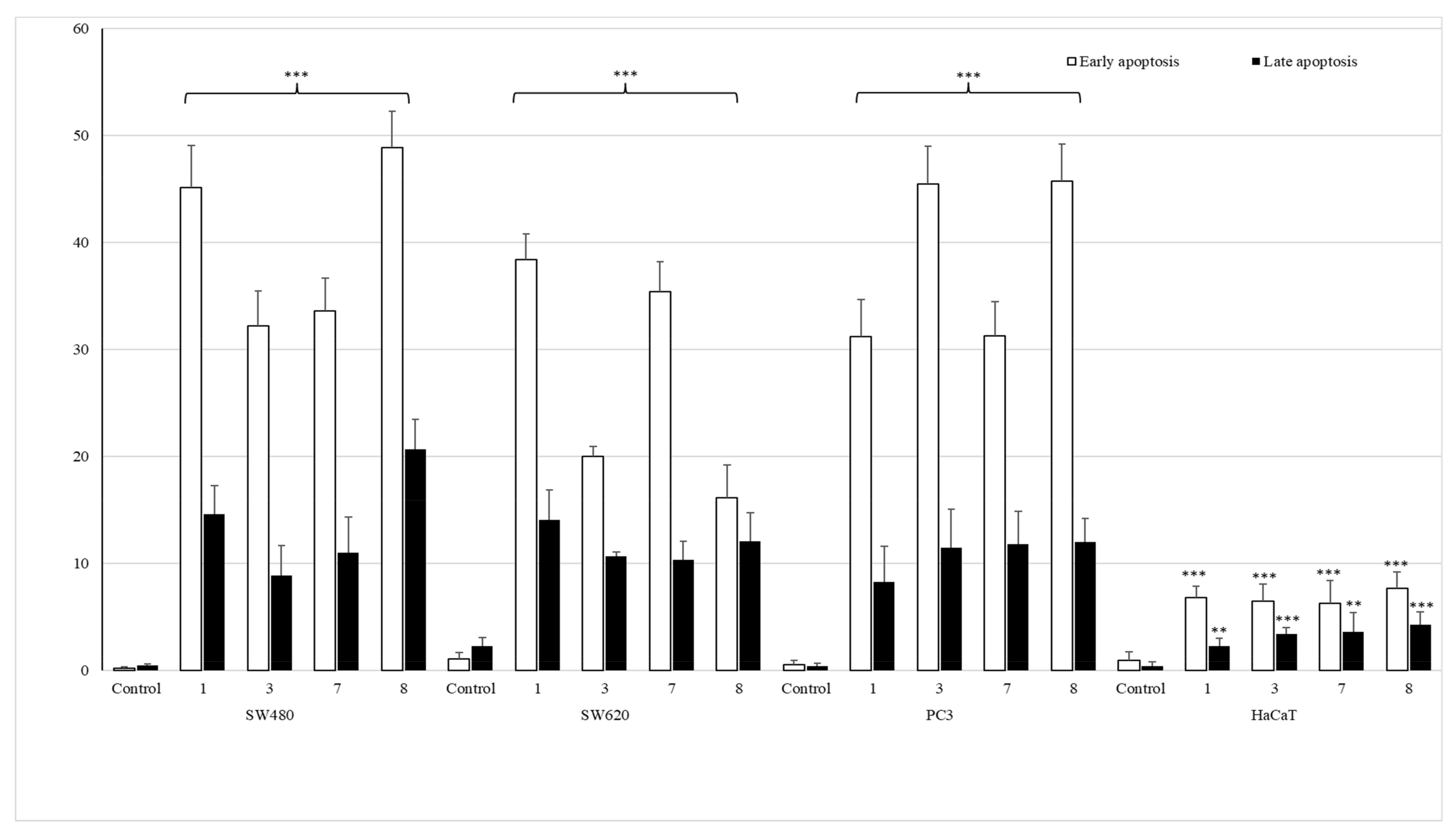
To establish the anticancer mechanism of activity, SW480, SW620 and PC3 cells were incubated for 72 h in the presence of the most promising monohalogeno complexes 1, 3, 7 and 8, after which annexin content was measured by flow cytometry analysis. The apoptotic effect is shown in Figure 6 and Figure S2.
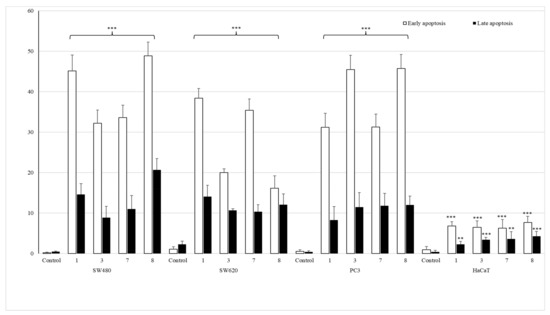
Figure 6.
The effect of complexes 1, 3, 7 and 8 on early and late apoptosis, or necrosis in SW480, SW620, PC3 and HaCaT cells. Cells were incubated for 72 h with tested compounds used in their IC50 concentrations. Then, cells were harvested, stained with annexin V-FITC and PI (fluorescein isothiocyanate and propidium iodide) and analyzed using flow cytometry. *** p ≤ 0.001, ** p ≤ 0.01 as compared to the control.
All studied compounds, applied in their IC50 doses, induced early apoptosis in pathological cells, especially in SW480 and PC3 cell lines, as compared to untreated cancerous controls. Derivatives of 2-bromophenyl- (1) and 4-iodophenylthiourea (8) revealed the most significant early-apoptosis-activating effect in primary colon cancer cell lines (45.18% ± 3.9% and 48.9% ± 3.4%, respectively). The influence of complexes bearing 4-bromophenyl (3) and 2-fluorophenyl (7) moieties in these cells was not so evident and did not exceed 40%. The most potent activators of apoptosis in metastatic PC3 cells were both para-substituted coordination compounds 3 and 8, as they promoted the process in 45.5% ± 3.5% and 45.7% ± 3.3% of cells. On the other hand, complexes containing a halogen atom in ortho position of the benzene ring (1 and 7) induced the early apoptosis in one third of tested PC3 cells. The similar pro-apoptotic inducing effect of these two compounds was observed in metastatic colon cancer cells (38.4% ± 2.4% and 35.4% ± 2.8%). The incubation of these cells with derivatives 3 or 8 increased early apoptosis in only approximately 20%.
The studied set of thiourea complexes did not considerably promote the process of late apoptosis of cells, as compared to controls. Only the most cytotoxic derivative 8 had the late apoptosis activating property, in 20.7% ± 2.8% of SW480 cells. It is also worth noting that tested coordination compounds 1, 3, 7 and 8 did not affect the level of early/late apoptosis in normal human keratinocytes (HaCaT), giving the result from 1.6% to 9.2% only.
In addition, conducted studies revealed that the referential doxorubicin was only a necrosis inducer. The number of SW480, SW620 and PC3 cells at the necrotic stage was 86%, 51% and 50%, respectively. Only in the case of PC3 cells, it induced late apoptosis as well (23%). HaCaT cells were considerably sensitive to doxorubicin, which produced a necrotic stage in 42% of cells, without affecting early and late apoptosis.
The obtained data comply with IC50 results found for mentioned cancer cells, particularly indicating the pro-apoptotic promoting role of complexes 1 and 8 in SW480 cells, as well as substances 3 and 8 in the prostate cancer cell line.
IL-6 Assay
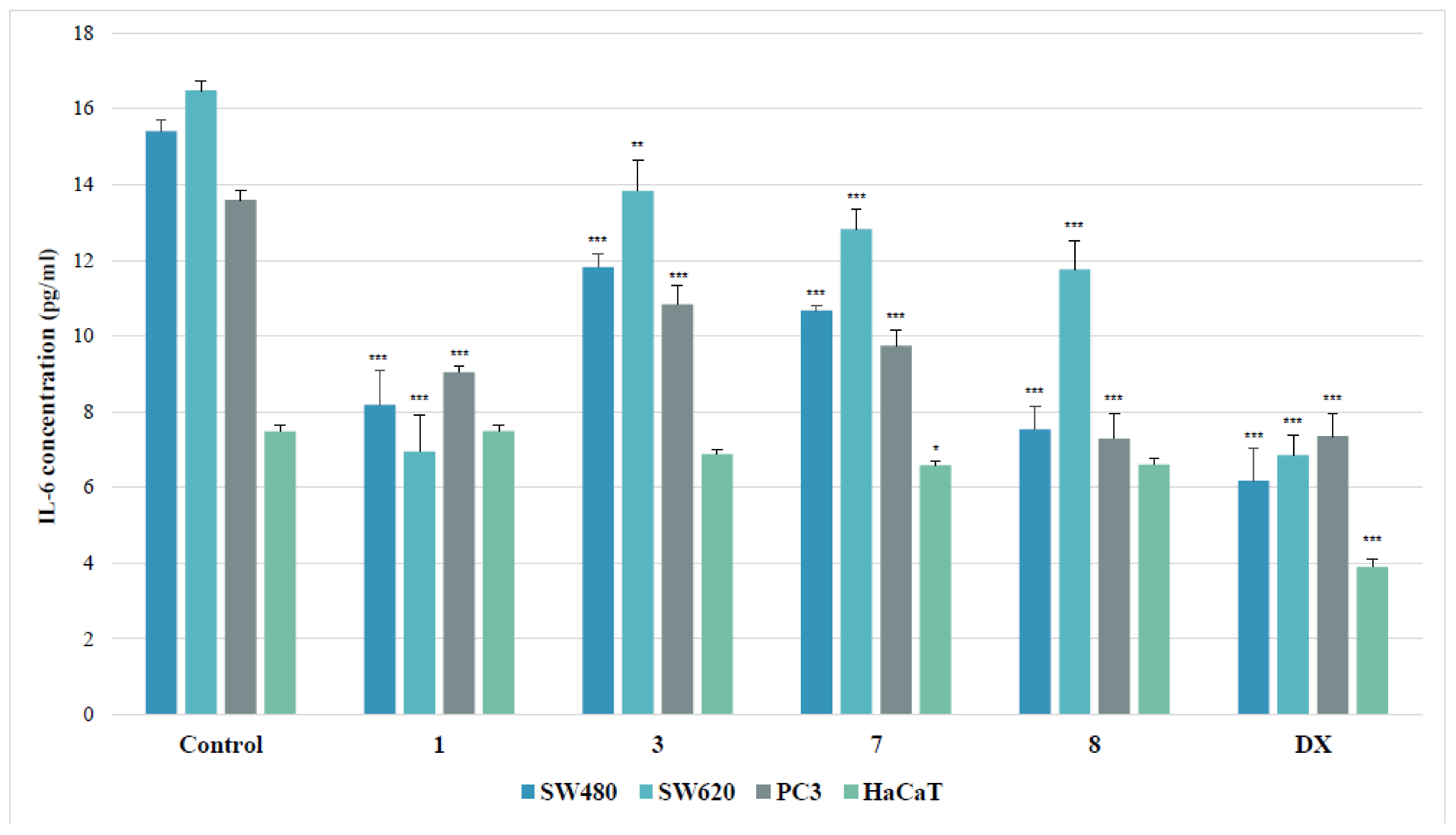
Human interleukin-6 (IL-6) is a pro-inflammatory cytokine, involved in numerous biological processes such as inflammation, cell growth, apoptosis, aging or bone remodeling. The ability of compounds to diminish IL-6 concentration in a cell, as a measure of their anti-inflammatory properties, was established for the most cytotoxic copper (II) complexes of thiourea derivatives (1, 3, 7, 8).
As shown in Figure 7, tested derivatives applied in their IC50 doses inhibited interleukin release in all evaluated cancer cell lines. Among them, the SW480 line was the most susceptible for the inhibition of the IL-6 secretion, elicited by the presence of evaluated substances. The 4-iodophenyl-containing complex (8) diminished the IL-6 level in these cells twofold. Similarly, the treatment of the primary colon cancer cells with 2-bromophenylyhiourea coordinate (1) reduced the cytokine amount 1.9 times. However, the other complexes (3 and 7) decreased that level not more than by 30%, as compared to the controls. The effectiveness of the complex 1 in metastatic SW620 cells equaled to the referential doxorubicin. Both substances inhibited the concentration of IL-6 2.4 times, while other tested derivatives diminished its secretion by an average of 23%. The strongest effect in PC3 cells was denoted for the 4-iodophenylthiourea complex (8), which, just as doxorubicin, diminished the IL-6 secretion almost twofold. On the other hand, the incubation of PC3 cells with the 2-bromophenyl compound (1) lowered the IL-6 concentration 1.5 times, with weaker effect of other complexes (3 and 7).
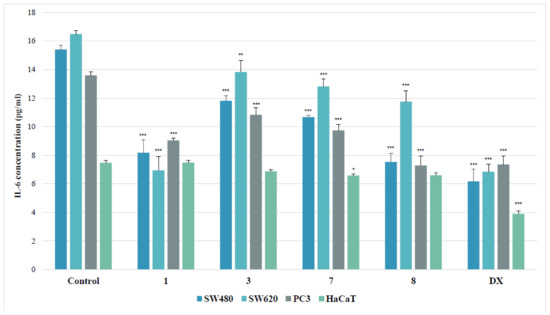
Figure 7.
Effects of thiourea complexes 1, 3, 7 and 8 on IL-6 levels, measured by enzyme-linked immunosorbent assay (ELISA) test. Data are expressed as the mean ± SD from three independent experiments performed in triplicate. *** p ≤ 0.001, ** p ≤ 0.01, * p ≤ 0.05, as compared to the control. DX (doxorubicin) is the reference compound.
Proteomic Analysis of Antioxidant and Detoxifying Enzymes
To evaluate the impact of the tested compounds on antioxidant status, they were screened for their influence on intensities of selected detoxifying and ROS-scavenging enzymes, occurring in human cancer cell lines such as glutathione S-transferase (GST), glutathione reductase (GR), superoxide dismutase (SOD) and peroxiredoxin (PRDX) (Table 3 and Table S1).

Table 3.
Liquid chromatography–mass spectrometry (LC–MS) proteome analysis provided in the SW480, SW620 and PC3 cells treated for 24 h with IC50 concentrations of complexes 1, 3, 7 and 8. Protein intensities were expressed as percentage of a control.
All tested complex compounds, applied in their IC50 concentrations, reduced intensities of colon and prostate cancer enzymatic proteins, in most cases by at least 30% of a control. The types of studied thiourea terminal moieties can be arranged in order of their decreasing influence, as shown: 2-fluoro- (7), 4-bromo- (3), 4-iodo- (8) and 2-bromophenyl (1). The most susceptible for the presence of tested coordinates were the proteins of metastatic PC3 cells. The level of all tested enzymes in the PC3 cell was diminished, while simultaneously superoxide dismutase [Cu-Zn] (SODC), mitochondrial superoxide dismutase (Mn) (SODM) and the majority of PRDXs belonged to the most sensitive. The 2-fluorophenyl derivative (7) reduced the intensity of 12 prostatic enzymes by 20.8-87.6%. Among them, both types of SOD and mitochondrial PRDX3 were the most vulnerable; their intensities were reduced by at least 75%. The compound 3 diminished the amount of all tested proteins in the PC3 cells by 29.4–69.1%, affecting most significantly the levels of both SODs. Similarly, after treatment of the cells with the complex 8, the amount of 12 enzymes was reduced by 31.5–75.2%, including glutathione S-transferase A1 (GSTA1), SODs, PRDX3 and PRDX4, which intensities equaled about 30% of a control. After incubation with the coordinate 1, quantities of SODM and PRDX5 were diminished up to 29% of the initial level.
On the other hand, GSTA1, glutathione S-transferase P (GSTP1) and PRDX3 of metastatic SW620 cells were the most susceptible to the treatment with copper (II) complexes. The derivative 7 appeared to be the most powerful. It reduced intensities of all tested colon proteins by 32.5–63.9%, most heavily both GSTs (up to 63.6%) and SODs (55.3%). Other analogs (1, 3, 8) affected considerably mainly on GSTA1, leading to a 45.7% (compound 8) and 54.8% (complex 3) decrease in the amount of this protein, respectively.
The impact of the tested compounds on enzymes of the primary SW480 cells was also evident, but weaker than that observed for both metastatic lines. The quantitative changes affected mainly GSTA1, SODC and PRDX4. The derivative 7 exerted the strongest effect, reducing their intensities by 54.9%, 59.4% and 41.7%, respectively. Additionally, it diminished the amount of five other proteins by 28.4% or more. Complexes 1 and 3 acted similarly to their analogs, but they also affected smaller group of enzymes, most significantly mitochondrial glutathione reductase (GSHR) (compound 1) and PRDX4 (compound 3). The thiourea coordination compound 8 had the greatest relevance for GSTA1, giving 47.6% inhibition of its intensity.
Obtained results confirmed the cytotoxic effect of tested complexes by the decreasing of antioxidant influence on cytoplasmic and mitochondrial proteins of malignant cells, especially observed for derivatives 7 and 3.
According to the literature data, there are few evidences that suggest GSTs function in the cancer development and resistance of colon and prostate tumor cells to anticancer agents. Colon cancers reflected elevated levels of GST expression compared with normal mucosa [44]. It was observed that GST overexpression was related to a doxorubicin-resistance phenomenon [45]. Similarly, the high level of glutathione-S-transferase pi (GST-pi) was detected in prostate cancer cells, and it contributed to the development of drug resistance and a significant increase in the cell proliferation rate of androgen independent PC3 cells [46].
Likewise, an increased GR expression and activity was shown in colon tumors, where it was involved in cellular defense against ROS by producing a reduced form of GSH. This activity may favor tumor development [47]. It was also shown that this enzyme may protect PC3 cells under persistent ROS production induced by radiotherapy and chemotherapy. Therefore, GR activity depletion could lead to the intensified H2O2 toxicity in metastatic prostate cancer cells [48].
There are contradictory reports indicating the function of SOD enzymes on primary tumor proliferation and metastasis activity. It was shown that overexpression of SOD could increase cell differentiation, decrease proliferation and turn back a malignant phenotype [49]. Other studies demonstrated that high SOD expression correlates with colon tumor aggressiveness and with resistance to cytotoxic drugs and radiotherapy [50]. The loss of SOD1 expression by siRNA knockdown significantly increased prostate cell sensitivity to cytotoxic agents, confirming SOD1 participation in cellular response and resistance [51]. Some evidence has indicated SOD1 overexpression in cancers maintaining cellular ROS below the crucial threshold [52]. Suddenly, down-regulation of SOD2 itself was reported in breast cancer cell lines [53].
Beyond the PRDX family protein’s regulatory function on cytokine-induced hydrogen peroxide concentration, some members can act independently during their peroxidase activity on cell proliferation, differentiation, apoptosis and gene expression [54]. Recent studies highlight dual catalytic activities of PRDX1, because besides antioxidant activity it has a physiologically significant overoxidation site [55,56]. The positive PRDXI immunohistochemical staining was strongly associated with a poor response to neoadjuvant chemoradiotherapy and a worse prognosis in rectal cancer patients [57]. The PRDX3 protein in antiandrogen resistant prostate cancer cell lines is responsible for increased tolerance to oxidative stress and a lack of activation of pro-apoptotic pathways. Therefore, the knockdown of PRDX3 leads to an increased tendency to oxidative stress [58]. In vivo studies showed the up-regulation of both PRDX3 and PRDX4 in prostate tumors. It could be an attempt at adaptation of the cancer cells to the microenvironment in a way favorable for survival and proliferation rate, while maintaining their tumor’s aggressiveness [59].
2.3.2. In Vitro Antimicrobial Activity
To assess the antimicrobial profile of coordination compounds 1–8, we assembled a panel of isolates, which included Gram-positive and Gram-negative organisms of clinical importance, as well as strains of fungal pathogens. The synthesized derivatives displayed significant to weak inhibitory effects towards standard staphylococci, as shown in Table 4. Derivatives of 3-chloro-4-fluorophenyl- (4) and 4-iodophenylthiourea (8) appeared to be the most active, with MIC values of 4 µg/mL. Bromophenyl compounds (1 and 3) and 3-nitro-4-chlorophenyl-containing complex (6) moderately inhibited the growth of S. aureus isolates, at concentrations from 4 to 8 µg/mL. The least potent monohalogen derivatives 2, 7 and dichlorophenyl complex 5 in doses of 16–32 µg/mL suppress the rise of staphylococcal rods. On the other hand, Gram-negative strains were weakly susceptible to the presence of tested thiourea coordinates (minimal inhibitory concentration, MIC ≥ 128 µg/mL). No evident antifungal properties were also observed; compounds 2, 3, 6 and 8, applied only at a dose of 64 µg/mL, exerted growth inhibitory properties towards studied Candida species

Table 4.
In vitro activity of complexes 1–8 against standard bacterial and fungal strains—minimal inhibitory concentrations (MIC, μg/mL).
On extended testing, a series of copper (II) complexes showed activity against 30 clinical methicillin-resistant strains of S. aureus (MRSA) and S. epidermidis (MRSE) (Table 5), and their potency against most of above-mentioned cocci was severalfold higher than the reference ciprofloxacin. The derivative 3, possessing 4-bromophenyl substituent at the thiourea branch, exhibited its growth-inhibitory effect towards all evaluated hospital S. aureus rods with MIC of 4 µg/mL, whereas against most of S. epidermidis species at a dose of 4–8 µg/mL. The similar sensitivity of clinical isolates of S. aureus to the presence of 3-chloro-4-fluorophenyl complex (4) was observed. Monosubstituted compounds, containing bromine (1, 2), fluorine (7) and iodide (8) on the phenyl ring, also possessed a moderate antibacterial potency (MIC 4–8 µg/mL). However, complexes 3, 4, 7 and 8 were 64 times more active towards S. aureus 572 and 481 strains than ciprofloxacin, while the bioactivity of coordinates 1–4, 7 and 8 against six clinical S. aureus rods was 16–64 times stronger than the reference drug. Disubstituted derivatives 5 and 6 acted with lesser strength than their analogues incorporating one halogen in a molecule, but they were still 4–16 times more potent as compared to ciprofloxacin.

Table 5.
In vitro activity of complexes 1–8 against hospital methicillin-resistant strains of Staphylococcus aureus (MRSA) and Staphylococcus epidermidis (MRSE)—minimal inhibitory concentrations (MIC, µg/mL).
Most of the clinical strains of S. epidermidis were less susceptible to the presence of the synthesized complexes. However, derivatives 1, 2, 4, 7 and 8 appeared to be 2–16 times more effective against S. epidermidis 424, 471, 511, 515 and 431–433 isolates than the reference chemotherapeutic. Similarly, weak antimicrobials, such as disubstituted compounds 5 and 6, were still equally or more potent towards these cocci, as compared to ciprofloxacin.
To sum up, thiourea complexes incorporating a copper (II) ion expressed higher inhibitory growth properties against hospital than standard bacterial rods. What is more, their efficiency depended on the type of electronegative functionalities attached to the benzene ring. Derivatives incorporating 3-chloro-4-fluorophenyl- (4) and 4-iodophenyl substituents (8) showed the strongest effect towards both types of strains, while their 4-bromosubstituted analog (3) was powerful distinctly against clinical isolates. Ortho-substituted compounds, bearing bromine (1) or fluorine (7) atoms, shared a similar inhibitory activity when incubated with Staphylococci, but were weaker than the mentioned para-substituted. Comparing dihalogeno derivatives 4 and 5, the replacement of the fluorine atom by chlorine led to a severalfold decrease of the antimicrobial potency. On the other hand, the compound possessing both chlorine and nitro group (6) was much more active against standard than hospital S. aureus species.
As proved by Bacillus subtilis rec-assay test, newly synthesized complexes 1–8 exerted no mutagenic and carcinogenic activities (Table S2). Thus, their antimicrobial effects were not linked with DNA-damaging potency.
As compared to results obtained for complexes, the antimicrobial activities of parental ligands were considerably higher [37]. MIC values of the most active ligands L1–L5, L7 and L8 against standard Staphylococcal strains ranged from 0.5 to 2 µg/mL. As expected, new copper (II) complexes were much more potent than copper (II) chloride itself (MIC ≥ 128 µg/mL for all strains). Although the complexation of halogen-containing 4-chloro-3-nitrophenylthioureas with copper (II) ion diminished their antibacterial properties [36,37], it incomparably increased their cytotoxicity towards various cancer cell lines, without influencing on normal keratinocytes.
Antimicrobial activity of the close analogs of the title complexes was previously described [6,7]. The formerly tested copper (II) complex of 1-(3-chloro-4-fluorophenyl)-3-[3-trifluoromethyl)phenylthiourea, an analog of the complex 4 described in this paper, exerted comparable growth-inhibitory activity towards standard bacterial strains (MIC 4–256 μg/mL) [7]. The same level of activities of both compounds was observed for clinical strains (MIC 4–8 μg/mL). However, for the derivative of 3,4-dichlorophenylthiourea, the observed differences in bioactivity were considerable. The complex of 3-(trifluoromethyl)phenylthiourea inhibited the growth of Staphylococcal strains at 2 μg, whereas the coordinate of 1-(4-chloro-3-nitrophenyl)-3-(3,4-dichlorophenyl)thiourea (5) was only at 32 μg/mL. Variabilities in antibacterial actions were even higher when hospital strains were considered. The 3-(trifluoromethyl)phenylthiourea analog was 16-64 times more potent than its 4-chloro-3-nitrophenylthiourea counterpart (5). On the other hand, copper (II) complexes with 2-bromo- and 3-bromophenylthiourea derivatives of 4-azatricyclo[5.2.1.0 2,6]dec-8-ene-3,5-dione were poorly active in comparison with the 4-chloro-3-nitrophenyl thioureas published in this paper [6]. The only exception was the 4-bromophenylthiourea with an azatricyclodione terminal fragment, which was four times more active against standard Staphylococcal isolates than its 3-chloro-4-fluorophenylthiourea analog (3). To conclude, derivatives of substituted phenylthioureas, mainly 3-trifluoromethylphenylthiourea complexes, revealed higher antimicrobial activity in comparison with coordinates of cyclic imides.
3. Materials and Methods
3.1. General Procedure
All chemicals were of analytical grade and were purchased from Sigma-Aldrich. The melting point (m.p.) was determined on a Boetius (HMK65/ 1360) microscope. Elemental analysis of all complexes was carried out using elemental analyzer CHNS (Vario Micro Cube) with an electronic microbalance. The copper content for complex 5 was determined using the energy dispersive X-ray fluorescence spectrometer EDX-7000 from Shimadzu.
3.1.1. Synthesis of Cu (II) Complexes of 3-(4-Chloro-3-nitrophenyl)thiourea (1‒8)
The appropriate 3-(4-chloro-3-nitrophenyl)thiourea L1–L8 (1 mmol) was stirred in dimethylformamide (DMF) (2 mL) until its dissolution. Next, anhydrous copper (II) chloride (1 mmol) was added to the solution. After stirring for 6 h, the solvent was evaporated. The solid residue was collected, washed several times with cold distilled water and dried in vacuo over anhydrous calcium chloride at room temperature to yield complexes 1–8.
- Copper (II) complex with 1-(2-bromophenyl)-3-(4-chloro-3-nitrophenyl)thiourea.
Yield 53%; dark brown solid; m.p. 93–95 °C; Anal. Calc for Cu(L1)2·0.75DMF·0.5H2O; Calc. C 37.76, H 2.50, N 10.52, Found C 37.92, H 2.28, N 10.57 (%).
- 2.
- Copper (II) complex with 1-(3-bromophenyl)-3-(4-chloro-3-nitrophenyl)thiourea.
Yield 61%; dark brown solid; m.p. 80–82 °C; Anal. Calc for Cu(L2)2·0.25 DMF·1.25 H2O, Calc. C 36.69, H 2.23, N 10.00, Found C 36.28, H 2.39, N 10.40 (%).
- 3.
- Copper (II) complex with 1-(4-bromophenyl)-3-(4-chloro-3-nitrophenyl)thiourea.
Yield 58%; dark brown solid; m.p. 88–90 °C; Anal. Calc for Cu(L3)2·0.75 DMF·0.5 H2O, Calc. C 37.76, H 2.50, N 10.52, Found C 38.04, H 2.24, N 10.30 (%).
- 4.
- Copper (II) complex with 1-(3-chloro-4-fluorophenyl)-3-(4-chloro-3-nitrophenyl)thiourea.
Yield 55%; dark brown solid; m.p. 95–97 °C; Anal. Calc for Cu(L4)2·0.75 DMF·0.5 H2O, Calc. C 40.12, H 2.41, N 11.18, Found C 40.26, H 2.18, N 11.30 (%).
- 5.
- Copper (II) complex with 1-(4-chloro-3-nitrophenyl)-3-(3,4-dichlorophenyl)thiourea.
Yield 57%; brown solid; m.p. 76–78 °C; Anal. Calc for Cu(L5)2·3.25H2O, Calc. C 35.76, H 2.37, N 9.62, Cu 7.28, Found C 35.76, H 2.20, N 9.88, Cu 7.74 (%).
- 6.
- Copper (II) complex with 1,3-bis(4-chloro-3-nitrophenyl)thiourea.
Yield 45%; light yellow solid; m.p. 107–108 °C; Anal. Calc for Cu(L6)2 ·0.75 DMF·0.75 H2O, Calc. C 37.52, H 2.31, N 13.55, Found C 37.41, H 2.22, N 13.41 (%).
- 7.
- Copper (II) complex with 1-(2-fluorophenyl)-3-(4-chloro-3-nitrophenyl)thiourea.
Yield 57%; yellow-green solid; m.p. 97–99 °C; Anal. Calc for Cu(L7)2·0.5 DMF·0.25 H2O, Calc. C 43.8, H 2.67, N 12.07, Found C 43.82, H 2.47, N 12.22 (%).
- 8.
- Copper (II) complex with 1-(4-iodophenyl)-3-(4-chloro-3-nitrophenyl)thiourea.
Yield 47%; orange brown solid; m.p. 99–101 °C; Anal. Calc for Cu(L8)2 ·3.5H2O, Calc. C 31.48, H 2.32, N 8.47, Found C 31.20, H 2.08, N 8.50 (%).
3.1.2. Instrumentation
Infrared spectra were performed on Nicolet iS5 FTIR spectrometer (Thermo Scientific) with diamond ATR sample accessory. The complexes 1–8 as well as organic ligands L1–L8 were recorded in the range of 400–4000 cm−1. The solid state electronic reflectance spectra of complexes 1–8 were collected on SHIMADZU UV-2600 spectrophotometer with UV-2600Plus Integrating Sphere in the range of 220–1400 nm. Electron paramagnetic resonance (EPR) spectra were recorded with a Bruker ELEXSYS-E580 X-band spectrometer (100 kHz field modulation). The microwave power of 5 mW and the modulation amplitude of 0.1–0.5 mT were applied. Prior, the measurements of the samples were dissolved in a mixture of ethanol–toluene solvent (1:1 v/v) to form solution of ca. 0.1 mM. The EPR measurements of frozen solutions were carried out at 77 K. The EPR parameters of the copper complexes were determined by computer simulation of the experimental spectra using the EPRsim32 package [60].
3.2. Cell Culture
The human primary (SW480), metastatic (SW620) colon cancer, metastatic prostate cancer (PC3) and human immortal keratinocyte (HaCaT) cell lines were purchased from the American Type Culture Collection (ATCC, Rockville, USA). The cells were cultured in medium according to protocols (MEM for SW480 and SW620, RPMI 1640 for PC3 and DMEM for HaCaT cells) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL) and streptomycin (100 μg/mL) and cultured in 37 °C/ 5% CO2 humidified incubator. The cells were cultured until appropriate confluence was achieved (80–90%). Next, they were harvested by treatment with 0.25% trypsin (Gibco Life Technologies, USA) and used for studies.
3.3. MTT Assay
To determine IC50 of the thiourea complexes, cells were seeded in 96-well plates (1 × 104 cells per well) and treated for 72 h with different concentrations of compounds. Cells without studied compounds in medium were used as a control.
The cell viability was assessed by determination of MTT salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) conversion by mitochondrial dehydrogenase. MTT assay was performed as previously described [61]. Experiments were repeated three times. Cell viability was presented as a percentage of MTT reduction in the treated cells versus the control cells. Number of viable cells cultured without studied compounds was assumed to be 100%. Decreased relative MTT level means decreased cell viability. Thiourea complexes with the highest cytotoxic potential assessed by MTT determination (with the lowest IC50) were chosen for subsequent assessments of cytotoxicity mechanisms.
3.4. LDH Assay
The presence of lactate dehydrogenase (LDH) in culture medium indicates a disturbance of the integrity of the cellular membrane. The LDH activity was performed after 72 h incubation of cells (1 × 104 cells per well) in 96-well plates with selected compounds according to manufacturer’s protocol (Roche Diagnostics, Germany), as was described by Chrzanowska et al. [61]. Compound mediated cytotoxicity was determined using equation: ((A test sample−A low control)/ (A high control−A low control)) × 100% (A-absorbance), where “low control” means cells in medium with 2% FBS without tested compounds, and “high control” means cells incubated in medium with 2% FBS and 1% Triton X-100 (100% LDH release). The cytotoxicity was expressed as percentage of LDH release as compared with the maximum release of LDH from Triton-X-100-treated cells.
3.5. Annexin V Binding Assay
The cells were cultured and harvested under the conditions described in the cell culture section. Then, they were seeded in six-well plates (2 × 105 cells per well) and treated with selected thioureas complexes at their IC50 concentration for 72 h. The effect of these compounds on the process of early and late apoptosis and necrosis was determined, as described previously [61], by dual staining with annexin V-FITC and propidium iodide according to manufacturer’s protocol (Becton Dickinson). The cells that were annexin V-FITC positive and PI-negative were identified as early apoptotic and annexin V-FITC and PI-positive as late apoptotic or necrotic.
3.6. Il-6 Level Assay
IL-6 concentration at all studied cancer cells and normal HaCaT cell lines was measured by ELISA kit (Diaclon SAS Besancon Cedex, France). Cells were seeded in twelve-well plates (1 × 105 cells per well) and treated with IC50 concentration of selected studied complexes for 72 h. IL-6 in cell culture supernatant was measured using enzyme-linked immunosorbent assay in accordance with the manufacturer’s protocol.
3.7. LC-MS Proteome Analysis
Enzymes involved in oxidoreductive potential were analyzed in the cell lysates obtained after treatment cells with selected conjugates for 24 h. Cells were washed with phosphate-buffered saline (PBS) and harvested, then centrifuged at 1000× g for 10 min. Then, lysis buffer (containing protease inhibitor, 1% RIPA Lysis and Extraction Buffer (ThermoFisher)) and cold PBS were added, and samples were sonicated three times in ice bath. Next, the cell lysates were centrifuged at 14,000× g at 4 °C for 15 min, and then supernatants were stored at 70 °C before use. Protein concentration was measured by the Bradford method.
Normalized protein concentrations (5 µg) from cell lysate were precipitated by ice cold (–20 °C) acetonitrile (ACN, Merck, in ratio 1:4). Then, samples were centrifuged (–9 °C, 30 min., 18,000× g), the supernatant was discarded and ACN excess was evaporated using a vacuum centrifuge (5 min., room temp.). Protein pellet was dissolved in 40 mM ammonium bicarbonate. An amount of 500 mM dithiothreitol (DTT, with final concentration 20 mM) and 1 M iodoacetamide (IAA, with final concentration 40 mM) were used for reduction and alkylation processes. After 16 h of incubation in 37 °C with Trypsin Gold (Promega), digested protein samples were diluted with 0.1% formic acid (ThermoFisher) and centrifuged (+2 °C, 30 min, 18,000× g).
LC–MS analysis was carried out with the use of nanoUHPLC (nanoElute, Bruker) coupled by CaptiveSpray (Bruker) to ESI-Q-TOF mass spectrometer (Compact, Bruker). Two-column separation method was used, i.e., pre-column (300 µm × 5 mm, C18 PepMap 100, 5 µm, 100Å, Thermo Scientific) and Aurora separation column with CSI fitting (75 µm × 250 mm, C18 1.6 µm) in gradient 2% B to 35% B in 90 min with the 300 nL/min flow rate. Mobile phases (A) 0.1% formic acid in water and (B) 0.1% formic acid in ACN were used.
Sample ionizations were performed at a gas flow of 3.0 L/min, temperature of 150 °C and voltage of the capillary at 1600 V. The quadrupole energy was fixed to 5.0 eV and collision chamber energy 7.0 eV, with an ion transfer time of 90 µs. The ions were analyzed in the positive polarity mode in the range 150–2200 m/z, with the acquisition frequency of the 1 Hz spectrum, as well as with the autoMS/MS system.
The collected spectra were analyzed and calibrated using DataAnalysis software (Bruker) and then identified in ProteinScape (Bruker) by the MASCOT server. Protein identification was conducted using the online SwissProt and NCBIprot databases, and their references and biological significance were identified using Reactome.org, String.org and KEGG.
3.8. In Vitro Evaluation of Antimicrobial Activity
The antimicrobial activity of the compounds was tested on Gram-positive bacteria (Staphylococcus aureus NCTC 4163, Staphylococcus aureus ATCC 25923, Staphylococcus aureus ATCC 6538, Staphylococcus aureus ATCC 29213, Staphylococcus epidermidis ATCC 12228, Staphylococcus epidermidis ATCC 35984), Gram-negative rods (Escherichia coli ATCC 10538, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 15442, Pseudomonas aeruginosa ATCC 27863) and yeasts (Candida albicans ATCC 10231, Candida albicans ATCC 90028, Candida parapsilosis ATCC 22019). Hospital methicillin-resistant strains of Staphylococcus aureus and Staphylococcus epidermidis were obtained from the collection of the Department of Pharmaceutical Microbiology, Medical University of Warsaw, Poland.
Antibacterial activity was examined by the disc-diffusion method under standard conditions using Mueller–Hinton II agar medium (Becton Dickinson) according to CLSI (previously NCCLS) guidelines [62]. Antifungal activities were assessed using Mueller–Hinton agar + 2% glucose and 0.5 µg/mL methylene blue dye medium [63]. Sterile filter paper discs (9 mm diameter, Whatman Number 3 chromatography paper) were dripped with tested compound solutions (in dimethylsulfoxide, DMSO) to load 400 μg of a given compound per disc. Dry discs were placed on the surface of appropriate agar medium. The results (diameter of the growth inhibition zone) were read after 18 h of incubation at 35 °C. Minimal inhibitory concentration (MIC) was tested by the twofold serial microdilution method (in 96-well microtiter plates) using Mueller–Hinton broth medium (Beckton Dickinson) for bacteria or RPMI-1640 medium for Candida species, according to CLSI guidelines [64,65]. The stock solution of tested agent was prepared in DMSO and diluted in sterile water. Concentrations of tested agents ranged from 0.125 to 512 µg/mL. The final inoculum of all studied microorganisms was 105 CFU/ mL−1 (colony forming units per ml). Minimal inhibitory concentrations (the lowest concentration of a tested agent that prevents visible growth of a microorganism) were read after 18 h (bacteria) or 24 h (yeasts) of incubation at 35 °C.
3.9. Genotoxicity Studies
DNA-damaging activity of compounds was tested by rec-assay using two genetically modified Bacillus subtilis strains: M45 (rec−) and H17 (rec+) [66,67]. Tested compounds were dissolved in DMSO, and 10 µL of each solution was dripped onto sterile cotton discs (Rotilabo) to load 256 µg of a given compound per 9 mm disc. Discs were placed on the surface nutrient agar plates (Difco), inoculated with 100 µL of bacterial overnight culture and incubated 24 h at 35 °C. After incubation, the growth inhibition zones were measured. 4-nitroquinoline N-oxide (NOQ) was used as reference genotoxin (concentration 2 µg per disc). Results of the genotoxicity test were estimated after 18 h of incubation at 35 °C by comparing the diameter of the inhibition zone on the B. subtilis M45 (rec−) strain with that on the B. subtilis H17 (rec+) strain.
3.10. Statistical Analyses
Statistical analyses were performed using GraphPad Prism 9 software (GraphPad Software). Statistical significance was assessed by ANOVA with Dunnett’s post hoc test. P values below 0.05 were considered statistically significant. Data were presented as the mean ± SD from at least three independent experiments.
4. Conclusions
To conclude, we have presented the cytotoxic influences of four-coordinate Cu (II) complexes with 3-(4-chloro-3-nitrophenyl)thiourea derivatives on a panel of human cancer and normal cell lines, as well as on bacterial isolates. In contrast to initial ligands, the complexation with metal ions has revealed the cytotoxic profile of synthesized compounds towards tumor cells and to a lesser extent to bacterial strains. Diversity of a type, a position and quantity of halogen substituents on the phenyl ring of the thiourea branch allowed for the examination of the impact of the structure of synthesized complexes on their bioactivity. Studied coordination compounds did not express cytotoxic effects in normal cells (HaCaT) at clinically achievable concentrations, and they were also proven to be non-genotoxic. The most active halogen phenylthiourea complexes (1, 3, 7, 8) showed stronger anticancer potential against PC3 compared to colon cancer cell lines. They were also more effective than the tested disubstituted derivatives (4–6). Compound 8 achieved the highest percentage of LDH release from PC3 and SW480 cells. Studied complexes, especially 3 and 8, induced early apoptosis in the above-mentioned pathological cells. Additionally, all coordination compounds, with emphasis on derivatives 1 and 8, reduced the secretion of IL-6 by tumor cell lines. Their interleukin-inhibitory properties in the selected cells were equally as strong as doxorubicin. Moreover, new compounds were active against selected strains of Staphylococci of clinical importance, being up to 2–64 times more potent in comparison to the reference antibiotic, ciprofloxacin. Our preliminary studies also showed the general tendency of tested compounds to disturb the antioxidant and detoxifying systems in cancer cells, especially evident for complexes 3, 7 and 8. This is an initial signal that this may be one of the cytotoxicity mechanisms. Such a diminishing effect of compounds on the antioxidant defense of cancer cells may support the action of other pro-oxidative agents, including drug resistance as well as support radiotherapy treatment.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222111415/s1.
Author Contributions
Conceptualization, A.B.; methodology, A.C. and A.D.-A.; software, P.P.; validation, A.C.; formal analysis, J.S.; investigation, A.C., A.D.-A., K.D. and P.P.; resources, A.B.; data curation, A.C.; writing—original draft preparation, A.B.; writing—review and editing, M.S.; visualization, A.D.-A. and K.D.; supervision, J.S.; project administration, A.B.; funding acquisition, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that have or could be perceived to have influenced the work reported in this article.
References
- Solomon, E.I.; Heppner, D.E.; Johnston, E.M.; Ginsbach, J.W.; Cirera, J.; Qayyum, M.; Kieber-Emmons, M.T.; Kjaergaard, C.H.; Hadt, R.G.; Tian, L. Copper active sites in biology. Chem. Rev. 2014, 114, 3659–3853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacPherson, I.S.; Murphy, M.E.P. Type-2 copper-containing enzymes. Cell. Mol. Life Sci. 2007, 64, 2887–2899. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Gorritti, W.-R.; Spodine-Spirinova, E.; Beyer, L.; Schröder, U.; Richter, R.; Ferreira, J.; Pavani, M. Synthesis, characterization and antitumor activity of copper (II) complexes, [CuL2] [HL1-3 = N,N-Diethyl-N′-(R-Benzoyl)Thiourea (R = H, o-Cl and p-NO2)]. Bioinorg. Chem. Appl. 2005, 3, 299–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, B.; Gao, Z.; Li, X.; Li, T.; Chen, G.; Zhou, M.; Zhang, J. DNA binding, DNA cleavage and HSA interaction of several metal complexes containing N-(2-hydroxyethyl)-N′-benzoylthiourea and 1,10-phenanthroline ligands. JBIC J. Biol. Inorg. Chem. 2016, 21, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Pivetta, T.; Isaia, F.; Verani, G.; Cannas, C.; Serra, L.; Castellano, C.; Demartin, F.; Pilla, F.; Manca, M.; Pani, A. Mixed-1,10-phenanthroline–Cu(II) complexes: Synthesis, cytotoxic activity versus hematological and solid tumor cells and complex formation equilibria with glutathione. J. Inorg. Biochem. 2012, 114, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Drzewiecka-Antonik, A.; Rejmak, P.; Klepka, M.; Wolska, A.; Pietrzyk, P.; Stępień, K.; Sanna, G.; Struga, M. Synthesis, structural studies and biological activity of novel Cu (II) complexes with thiourea derivatives of 4-azatricyclo [5.2.1.0 2,6 ] dec-8-ene-3,5-dione. J. Inorg. Biochem. 2017, 176, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Bielenica, A.; Drzewiecka-Antonik, A.; Rejmak, P.; Stefańska, J.; Koliński, M.; Kmiecik, S.; Lesyng, B.; Włodarczyk, M.; Pietrzyk, P.; Struga, M. Synthesis, structural and antimicrobial studies of type II topoisomerase-targeted copper (II) complexes of 1,3-disubstituted thiourea ligands. J. Inorg. Biochem. 2018, 182, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Repich, H.; Orysyk, V.; Palchykovska, L.; Orysyk, S.; Zborovskii, Y.L.; Vasylchenko, O.; Storozhuk, O.; Biluk, A.; Nikulina, V.; Garmanchuk, L.; et al. Synthesis, spectral characterization of novel Pd (II), Pt (II) π-coordination compounds based on N-allylthioureas. Cytotoxic properties and DNA binding ability. J. Inorg. Biochem. 2017, 168, 98–106. [Google Scholar] [CrossRef]
- Ćoćić, D.; Jovanovic, S.; Radisavljević, S.; Korzekwa, J.; Scheurer, A.; Puchta, R.; Baskić, D.; Todorovic, D.; Popovic, S.; Matić, S.; et al. New monofunctional platinum (II) and palladium (II) complexes: Studies of the nucleophilic substitution reactions, DNA/BSA interaction, and cytotoxic activity. J. Inorg. Biochem. 2018, 189, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Plutín, A.M.; Mocelo, R.; Alvarez, A.; Ramos, R.; Castellano, E.E.; Cominetti, M.R.; Graminha, A.E.; Ferreira, A.G.; Batista, A.A. On the cytotoxic activity of Pd (II) complexes of N,N-disubstituted-N′-acyl thioureas. J. Inorg. Biochem. 2014, 134, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Marverti, G.; Gozzi, G.; Lauriola, A.; Ponterini, G.; Belluti, S.; Imbriano, C.; Costi, M.P.; D’Arca, D. The 1,10-phenanthroline ligand enhances the antiproliferative activity of DNA-intercalating thiourea-Pd (II) and -Pt (II) complexes against cisplatin-sensitive and-resistant human ovarian cancer cell lines. Int. J. Mol. Sci. 2019, 20, 6122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pracharova, J.; Zerzankova, L.; Stepankova, J.; Novakova, O.; Farrer, N.J.; Sadler, P.J.; Brabec, V.; Kasparkova, J. Interactions of DNA with a new platinum (IV) azide dipyridine complex activated by UVA and visible light: Relationship to toxicity in tumor cells. Chem. Res. Toxicol. 2012, 25, 1099–1111. [Google Scholar] [CrossRef]
- Ma, Z.; Choudhury, J.R.; Wright, M.W.; Day, C.S.; Saluta, G.; Kucera, G.L.; Bierbach, U. A non-cross-linking platinum−acridine agent with potent activity in non-small-cell lung cancer. J. Med. Chem. 2008, 51, 7574–7580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eiter, L.C.; Hall, N.W.; Day, C.S.; Saluta, G.; Kucera, G.L.; Bierbach, U. Gold (I) analogues of a platinum−acridine antitumor agent are only moderately cytotoxic but show potent activity against mycobacterium tuberculosis. J. Med. Chem. 2009, 52, 6519–6522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, B.; Ma, L.; Jin, J.; Jiang, F.; Zhou, G.; Yan, K.; Liu, Y. Mitochondrial toxicity induced by a thiourea gold (I) complex: Mitochondrial permeability transition and respiratory deficit. Toxicol. Res. 2018, 7, 1081–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correa, R.; de Oliveira, K.M.; Delolo, F.G.; Alvarez, A.; Mocelo, R.; Plutin, A.M.; Cominetti, M.R.; Castellano, E.E.; Batista, A.A. Ru (II)-based complexes with N-(acyl)-N′,N′-(disubstituted)thiourea ligands: Synthesis, characterization, BSA- and DNA-binding studies of new cytotoxic agents against lung and prostate tumour cells. J. Inorg. Biochem. 2015, 150, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Cunha, B.N.; Colina-Vegas, L.; Plutín, A.M.; da Silveira, R.G.; Honorato, J.; de Oliveira, K.M.; Cominetti, M.R.; Ferreira, A.G.; Castellano, E.E.; Batista, A.A. Hydrolysis reaction promotes changes in coordination mode of Ru (II)/acylthiourea organometallic complexes with cytotoxicity against human lung tumor cell lines. J. Inorg. Biochem. 2018, 186, 147–156. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, T.D.; Plutín, A.M.; Luna-Dulcey, L.; Castellano, E.E.; Cominetti, M.R.; Batista, A.A. Cytotoxicity of ruthenium-N,N-disubstituted-N′-acylthioureas complexes. Mater. Sci. Eng. C 2020, 115, 111106. [Google Scholar] [CrossRef] [PubMed]
- Poyraz, M.; Berber, H.; Banti, C.N.; Kourkoumelis, N.; Manos, M.J.; Hadjikakou, S.K. Synthesis characterization and biological activity of mixed ligand silver (I) complex of 2-benzimidazolylurea and triphenylphosphine. Polyhedron 2017, 128, 95–103. [Google Scholar] [CrossRef]
- Perillo, B.; di Donato, M.; Pezone, A.; di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Arfin, S.; Jha, N.; Jha, S.; Kesari, K.; Ruokolainen, J.; Roychoudhury, S.; Rathi, B.; Kumar, D. Oxidative stress in cancer cell metabolism. Antioxidants 2021, 10, 642. [Google Scholar] [CrossRef]
- Bułdak, R.; Buldak, L.; Kukla, M.; Gabriel, A.; Żwirska-Korczala, K. Significance of selected antioxidant enzymes in cancer cell progression. Pol. J. Pathol. 2014, 3, 167–175. [Google Scholar] [CrossRef] [Green Version]
- Hwang, P.M.; Bunz, F.; Yu, J.; Rago, C.; Chan, T.A.; Murphy, M.P.; Kelso, G.F.; Smith, R.A.J.; Kinzler, K.W.; Vogelstein, B. Ferredoxin reductase affects p53-dependent, 5-fluorouracil–induced apoptosis in colorectal cancer cells. Nat. Med. 2001, 7, 1111–1117. [Google Scholar] [CrossRef] [Green Version]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Using cyclooxygenase-2 inhibitors as molecular platforms to develop a new class of apoptosis-inducing agents. J. Natl. Cancer Inst. 2002, 94, 1745–1757. [Google Scholar] [CrossRef]
- Zhou, W.; Liotta, L.A.; Petricoin, E.F. Cancer metabolism: What we can learn from proteomic analysis by mass spectrometry. Cancer Genom.-Proteom. 2012, 9, 373–381. [Google Scholar]
- Yu, B.; Liu, Y.; Peng, X.; Hua, S.; Zhou, G.; Yan, K.; Liu, Y. Synthesis, characterization, and antitumor properties of Au (I)–thiourea complexes. Metallomics 2020, 12, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.-P.; Lai, S.; Wu, S.; Hu, S.; Zhou, L.; Chen, Y.; Wang, M.; Zhu, Y.; Lian, W.; Peng, W.; et al. Nuclear permeable ruthenium (II) β-carboline complexes induce autophagy to antagonize mitochondrial-mediated apoptosis. J. Med. Chem. 2010, 53, 7613–7624. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.H.; Cai, M.; Deng, J.; Yu, P.; Liang, H.; Yang, F. Anticancer function and ROS-mediated multi-targeting anticancer mechanisms of copper (II) 2-hydroxy-1-naphthaldehyde complexes. Molecules 2019, 24, 2544. [Google Scholar] [CrossRef] [Green Version]
- Trejo-Solís, C.; Jimenez-Farfan, D.; Rodriguez-Enriquez, S.; Fernandez-Valverde, F.; Cruz-Salgado, A.; Ruiz-Azuara, L.; Sotelo, J. Copper compound induces autophagy and apoptosis of glioma cells by reactive oxygen species and JNK activation. BMC Cancer 2012, 12, 156. [Google Scholar] [CrossRef] [Green Version]
- Terbouche, A.; Ramdane-Terbouche, C.A.; Bendjilali, Z.; Berriah, H.; Lakhdri, H.; Lerari, D.; Bachari, K.; Mezaoui, D.; Bensiradj, N.E.H.; Guegan, J.-P.; et al. Synthesis, spectral characterization, molecular modeling, antibacterial and antioxidant activities and stability study of binuclear Pd (II) and Ru (III) complexes with novel bis-[1-(2-[(2-hydroxynaphthalen-1-yl)methylidene]amino}ethyl)-1-ethyl-3-phenylthiourea] ligand: Application to detection of cholesterol. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 205, 146–159. [Google Scholar] [CrossRef]
- Esmaeili, L.; Perez, M.G.; Jafari, M.; Paquin, J.; Ispas-Szabo, P.; Pop, V.; Andruh, M.; Byers, J.; Mateescu, M.A. Copper complexes for biomedical applications: Structural insights, antioxidant activity and neuron compatibility. J. Inorg. Biochem. 2019, 192, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Inci, D.; Aydın, R.; Zorlu, Y. Cu (II) complex with auxin (3-indoleacetic acid) and an aromatic planar ligand: Synthesis, crystal structure, biomolecular interactions and radical scavenging activity. Eur. Biophys. J. 2021, 50, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Karagoz, Z.; Genc, M.; Yılmaz, E.; Keser, S. Synthesis and antitumor, antioxidant effects studies of N-ethylpiperazine substitute thiourea ligands and their copper (II) complexes. Spectrosc. Lett. 2013, 46, 182–190. [Google Scholar] [CrossRef]
- Bielenica, A.; Stefańska, J.; Stępień, K.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Sanna, G.; Madeddu, S.; Boi, S.; Giliberti, G.; Wrzosek, M.; et al. Synthesis, cytotoxicity and antimicrobial activity of thiourea derivatives incorporating 3-(trifluoromethyl)phenyl moiety. Eur. J. Med. Chem. 2015, 101, 111–125. [Google Scholar] [CrossRef]
- Bielenica, A.; Sanna, G.; Madeddu, S.; Giliberti, G.; Stefańska, J.; Kozioł, A.E.; Savchenko, O.; Strzyga-Łach, P.; Chrzanowska, A.; Kubiak-Tomaszewska, G.; et al. Disubstituted 4-chloro-3-nitrophenylthiourea derivatives: Antimicrobial and cytotoxic studies. Molecules 2018, 23, 2428. [Google Scholar] [CrossRef] [Green Version]
- Bielenica, A.; Stępień, K.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Krukowski, S.; Włodarczyk, M.; Struga, M. Synthesis and antimicrobial activity of 4-chloro-3-nitrophenylthiourea derivatives targeting bacterial type II topoisomerases. Chem. Biol. Drug Des. 2016, 87, 905–917. [Google Scholar] [CrossRef]
- Stefańska, J.; Stepien, K.; Bielenica, A.; Wrzosek, M.; Struga, M. Antistaphylococcal activity of selected thiourea derivatives. Pol. J. Microbiol. 2016, 65, 451–460. [Google Scholar] [CrossRef] [Green Version]
- John, R.P.; Sreekanth, A.; Kurup, M.R.P.; Usman, A.; Ibrahim, A.R.; Fun, H.K. Spectral studies and structure of a 2-hydroxyacetophenone 3-hexamethyleneiminyl thiosemicarbazonate (−2) copper (II) complex containing 1,10-phenanthroline. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2003, 59, 1349–1358. [Google Scholar] [CrossRef]
- Sreekanth, A.; Kurup, M.R.P. Structural and spectral studies on four coordinate copper (II) complexes of 2-benzoylpyridine N(4), N(4)-(butane-1,4-diyl) thiosemicarbazone. Polyhedron 2003, 22, 3321–3332. [Google Scholar] [CrossRef]
- Drzewiecka-Antonik, A.; Rejmak, P.; Klepka, M.; Wolska, A.; Chrzanowska, A.; Struga, M. Structure and anticancer activity of Cu (II) complexes with (bromophenyl) thiourea moiety attached to the polycyclic imide. J. Inorg. Biochem. 2020, 212, 111234. [Google Scholar] [CrossRef] [PubMed]
- Low, M.L.; Maigre, L.; Tahir, M.I.M.; Tiekink, E.R.; Dorlet, P.; Guillot, R.; Ravoof, T.B.; Rosli, R.; Pagès, J.-M.; Policar, C.; et al. New insight into the structural, electrochemical and biological aspects of macroacyclic Cu (II) complexes derived from S-substituted dithiocarbazate schiff bases. Eur. J. Med. Chem. 2016, 120, 1–12. [Google Scholar] [CrossRef]
- Singh, D.P.; Pratap, S.; Shukla, M. Solvent induced geometry transformation of trigonal planar Cu (I) complexes of N-((2/4-methyoxy carbonyl) phenyl)-N′-(ethoxy/methoxy carbonyl) thiocarbamides to square-planar Cu (II) complexes: Synthesis, spectral, single crystal, DFT and in vitro cytotoxic study. Inorg. Chim. Acta 2014, 423, 386–396. [Google Scholar] [CrossRef]
- Singh, S. Cytoprotective and regulatory functions of glutathione S-transferases in cancer cell proliferation and cell death. Cancer Chemother. Pharmacol. 2014, 75, 1–15. [Google Scholar] [CrossRef]
- Beaumont, P.; Moore, M.J.; Ahmad, K.; Payne, M.M.; Lee, C.; Riddick, D.S. Role of glutathione S-transferases in the resistance of human colon cancer cell lines to doxorubicin. Cancer Res. 1998, 58, 947–955. [Google Scholar] [PubMed]
- Hasegawa, N.; Mizutani, K.; Suzuki, T.; Deguchi, T.; Nozawa, Y. A comparative study of protein profiling by proteomic analysis in camptothecin-resistant PC3 and camptothecin-sensitive LNCaP human prostate cancer cells. Urol. Int. 2006, 77, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Lorestani, S.; Hashemy, S.I.; Mojarad, M.; Shahrestanaki, M.K.; Bahari, A.; Asadi, M.; Avval, F.Z. Increased glutathione reductase expression and activity in colorectal cancer tissue samples: An investigational study in Mashhad, Iran. Middle East J. Cancer 2018, 9, 99–104. [Google Scholar]
- Freitas, M.; Baldeiras, I.; Proença, T.; Alves, V.; Mota-Pinto, A.; Sarmento-Ribeiro, A. Oxidative stress adaptation in aggressive prostate cancer may be counteracted by the reduction of glutathione reductase. FEBS Open Bio 2012, 2, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhao, W.; Zhang, H.J.; Domann, F.E.; Oberley, L.W. Overexpression of copper zinc superoxide dismutase suppresses human glioma cell growth. Cancer Res. 2002, 62, 1205–1212. [Google Scholar] [PubMed]
- Satomi, A.; Murakami, S.; Hashimoto, T.; Ishida, K.; Matsuki, M.; Sonoda, M. Significance of superoxide dismutase (SOD) in human colorectal cancer tissue: Correlation with malignant intensity. J. Gastroenterol. 1995, 30, 177–182. [Google Scholar] [CrossRef]
- Martino, T.; Kudrolli, T.A.; Kumar, B.; Salviano, I.; Mencalha, A.L.; Coelho, M.G.P.; Justo, G.; Costa, P.R.R.; Sabino, K.C.C.; Lupold, S.E. The orally active pterocarpanquinone LQB-118 exhibits cytotoxicity in prostate cancer cell and tumor models through cellular redox stress. Prostate 2018, 78, 140–151. [Google Scholar] [CrossRef]
- Papa, L.; Manfredi, G.; Germain, D. SOD1, an unexpected novel target for cancer therapy. Genes Cancer 2014, 5, 15–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papa, L.; Hahn, M.; Marsh, E.L.; Evans, B.S.; Germain, D. SOD2 to SOD1 switch in breast cancer. J. Biol. Chem. 2014, 289, 5412–5416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Wang, Y.; Su, Y. Peroxiredoxins, a novel target in cancer radiotherapy. Cancer Lett. 2009, 286, 154–160. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Lee, W.-S.; Ip, C.; Chae, H.-Z.; Park, E.-M.; Park, Y.-M. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Res. 2006, 66, 7136–7142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, H.H.; Kim, S.Y.; Park, S.K.; Jeon, H.S.; Lee, Y.M.; Jung, J.H.; Lee, S.Y.; Chae, H.B.; Jung, Y.J.; Lee, K.O.; et al. Phosphorylation and concomitant structural changes in human 2-Cys peroxiredoxin isotype I differentially regulate its peroxidase and molecular chaperone functions. FEBS Lett. 2005, 580, 351–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.-F.; Lee, K.-D.; Yeh, C.-H.; Chen, W.-C.; Huang, W.-S.; Chin, C.-C.; Lin, P.-Y.; Wang, J.-Y. Role of peroxiredoxin I in rectal cancer and related to p53 status. Int. J. Radiat. Oncol. 2010, 78, 868–878. [Google Scholar] [CrossRef]
- Whitaker, H.C.; Patel, D.J.; Howat, W.J.; Warren, A.Y.; Kay, J.; Sangan, T.; Marioni, J.; Mitchell, J.F.B.; Aldridge, S.; Luxton, H.J.; et al. Peroxiredoxin-3 is overexpressed in prostate cancer and promotes cancer cell survival by protecting cells from oxidative stress. Br. J. Cancer 2013, 109, 983–993. [Google Scholar] [CrossRef] [Green Version]
- Basu, A.; Banerjee, H.; Rojas, H.; Martinez, S.R.; Roy, S.; Jia, Z.; Lilly, M.B.; de Leon, M.; Casiano, C.A. Differential expression of peroxiredoxins in prostate cancer: Consistent upregulation of PRDX3 and PRDX4. Prostate 2011, 71, 755–765. [Google Scholar] [CrossRef] [Green Version]
- Spałek, T.; Pietrzyk, P.; Sojka, Z. Application of the genetic algorithm joint with the powell method to nonlinear least-squares fitting of powder EPR spectra. J. Chem. Inf. Model. 2005, 45, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard M7-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Determining Bactericidal Activity of Antimicrobial Agents. Approved Guideline M26-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard M7-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2006. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. In CLSI Document M27-A3, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008; pp. 1–25. [Google Scholar]
- Kada, T.; Hirano, K.; Shirasu, Y. Bacillus subtilis rec-assay test. In Chemical Mutagens; de Sevres, F.E., Hollaende, A., Eds.; Plenum Press: New York, NY, USA, 1980; Volume 6, p. 149. [Google Scholar]
- Sadaie, Y.; Kada, T. Recombination-deficient mutants of Bacillus subtilis. J. Bacteriol. 1976, 125, 489–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chrzanowska, A.; Roszkowski, P.; Bielenica, A.; Olejarz, W.; Stępień, K.; Struga, M. Anticancer and antimicrobial effects of novel ciprofloxacin fatty acids conjugates. Eur. J. Med. Chem. 2020, 185, 111810. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).