LegumeSSRdb: A Comprehensive Microsatellite Marker Database of Legumes for Germplasm Characterization and Crop Improvement
Abstract
1. Introduction
2. Results
2.1. Cross-Species Comparison of Legume Species SSRs
2.2. Characterization of the Perfect SSRs
2.3. Characterization of SSRs by Motif Type
2.4. Functional Annotations of Predicted SSRs
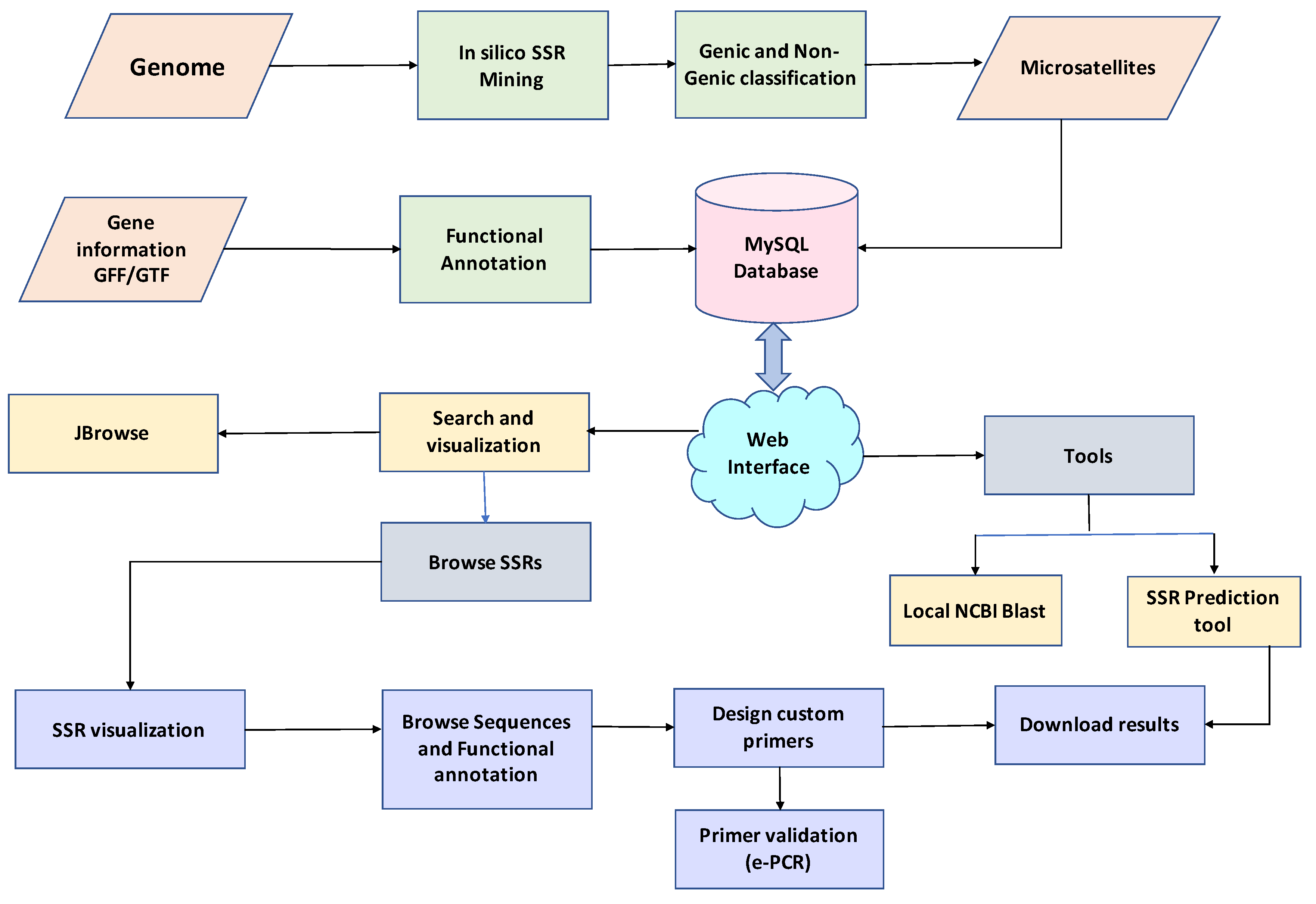
2.5. Web Genomic Resource: legumeSSRdb
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. In Silico Simple Sequence Repeat Mining and Functional Annotation
4.3. Webserver Development and Web Interface
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stagnari, F.; Maggio, A.; Galieni, A.; Pisante, M. Multiple benefits of legumes for agriculture sustainability: An overview. Chem. Biol. Technol. Agric. 2017, 4, 1–13. [Google Scholar] [CrossRef]
- van Loon, M.P.; Deng, N.; Grassini, P.; Rattalino Edreira, J.I.; Wolde-meskel, E.; Baijukya, F.; Marrou, H.; van Ittersum, M.K. Prospect for increasing grain legume crop production in East Africa. Eur. J. Agron. 2018, 101, 140–148. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Septiningsih, E.M.; Das, S.R.; Carandang, J.J.; Pamplona, A.M.; Sanchez, D.L.; Kato, Y.; Ye, G.; Reddy, J.N.; Singh, U.S.; et al. Developing new flood-tolerant varieties at the international rice research institute (IRRI). Sabrao J. Breed. Genet. 2013, 45, 42–56. [Google Scholar]
- Xu, Y.; Crouch, J.H. Marker-assisted selection in plant breeding: From publications to practice. Crop Sci. 2008, 48, 391–407. [Google Scholar] [CrossRef]
- Gupta, H.S.; Agrawal, P.K.; Mahajan, V.; Bisht, G.S.; Kumar, A.; Verma, P.; Srivastava, A.; Saha, S.; Babu, R.; Pant, M.C.; et al. Quality protein maize for nutritional security: Rapid development of short duration hybrids through molecular marker assisted breeding. Curr. Sci. 2009, 96, 230–237. [Google Scholar]
- Cuenca, J.; Aleza, P.; Garcia-Lor, A.; Ollitrault, P.; Navarro, L. Fine mapping for identification of citrus alternaria brown spot candidate resistance genes and development of new SNP markers for marker-assisted selection. Front. Plant Sci. 2016, 7, 1948. [Google Scholar] [CrossRef]
- Omura, M.; Shimada, T. Citrus breeding, genetics and genomics in Japan. Breed. Sci. 2016, 66, 3–17. [Google Scholar] [CrossRef]
- Yu, J.; Dossa, K.; Wang, L.; Zhang, Y.; Wei, X.; Liao, B.; Zhang, X. PMDBase: A database for studying microsatellite DNA and marker development in plants. Nucleic Acids Res. 2017, 45, D1046–D1053. [Google Scholar] [CrossRef]
- Xu, X.; Peng, M.; Fang, Z.; Xu, X. The direction of microsatellite mutations is dependent upon allele length. Nat. Genet. 2000, 24, 396–399. [Google Scholar] [CrossRef]
- Wierdl, M.; Dominska, M.; Petes, T.D. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics 1997, 146, 769–779. [Google Scholar] [CrossRef]
- Akemi, A.; Pereira, J.; Macedo, P.; Alessandra, K. Microsatellites as tools for genetic diversity analysis. In Genetic Diversity in Microorganisms; InTechOpen: London, UK, 2012; Available online: https://www.intechopen.com/chapters/28891 (accessed on 13 October 2021). [CrossRef]
- Senan, S.; Kizhakayil, D.; Sasikumar, B.; Sheeja, T.E. Methods for development of microsatellite markers: An overview. Not. Sci. Biol. 2014, 6, 1–13. [Google Scholar] [CrossRef]
- Sharma, P.C.; Grover, A.; Kahl, G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007, 25, 490–498. [Google Scholar] [CrossRef]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tian, Y.; Yang, R.; Feng, H.; Ouyang, Q.; Tian, Y.; Tan, Z.; Li, M.; Niu, Y.; Jiang, J.; et al. Coevolution between simple sequence repeats (SSRs) and virus genome size. BMC Genom. 2012, 13, 435. [Google Scholar] [CrossRef] [PubMed]
- Portis, E.; Lanteri, S.; Barchi, L.; Portis, F.; Valente, L.; Toppino, L.; Rotino, G.L.; Acquadro, A. Comprehensive characterization of simple sequence repeats in eggplant (Solanum melongena L.) genome and construction of a web resource. Front. Plant Sci. 2018, 9, 401. [Google Scholar] [CrossRef] [PubMed]
- Haseneyer, G.; Schmutzer, T.; Seidel, M.; Zhou, R.; Mascher, M.; Schön, C.C.; Taudien, S.; Scholz, U.; Stein, N.; Mayer, K.F.X.; et al. From RNA-seq to large-scale genotyping - genomics resources for rye (Secale cereale L.). BMC Plant Biol. 2011, 11, 131. [Google Scholar] [CrossRef] [PubMed]
- Portis, E.; Portis, F.; Valente, L.; Moglia, A.; Barchi, L.; Lanteri, S.; Acquadro, A. A genome-wide survey of the microsatellite content of the globe artichoke genome and the development of a web-based database. PLoS ONE 2016, 11, e0162841. [Google Scholar] [CrossRef] [PubMed]
- Kariin, S.; Burge, C. Dinucleotide relative abundance extremes: A genomic signature. Trends Genet. 1995, 11, 283–290. [Google Scholar] [CrossRef]
- Shioiri, C.; Takahata, N. Skew of mononucleotide frequencies, relative abundance of dinucleotides, and DNA strand asymmetry. J. Mol. Evol. 2001, 53, 364–376. [Google Scholar] [CrossRef]
- Cheng, J.; Zhao, Z.; Li, B.; Qin, C.; Wu, Z.; Trejo-Saavedra, D.L.; Luo, X.; Cui, J.; Rivera-Bustamante, R.F.; Li, S.; et al. A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Xiao, Y.; Xia, W.; Ma, J.; Mason, A.S.; Fan, H.; Shi, P.; Lei, X.; Ma, Z.; Peng, M. Genome-wide identification and transferability of microsatellite markers between palmae species. Front. Plant Sci. 2016, 7, 1578. [Google Scholar] [CrossRef]
- Guo, W.J.; Ling, J.; Li, P. Consensus features of microsatellite distribution: Microsatellite contents are universally correlated with recombination rates and are preferentially depressed by centromeres in multicellular eukaryotic genomes. Genomics 2009, 93, 323–331. [Google Scholar] [CrossRef]
- Eujayl, I.; Sledge, M.K.; Wang, L.; May, G.D.; Chekhovskiy, K.; Zwonitzer, J.C.; Mian, M.A.R. Medicago truncatula EST-SSRs reveal cross-species genetic markers for Medicago spp. Theor. Appl. Genet. 2004, 108, 414–422. [Google Scholar] [CrossRef]
- Gonthier, L.; Blassiau, C.; Mörchen, M.; Cadalen, T.; Poiret, M.; Hendriks, T.; Quillet, M.C. High-density genetic maps for loci involved in nuclear male sterility (NMS1) and sporophytic self-incompatibility (S-locus) in chicory (Cichorium intybus L., Asteraceae). Theor. Appl. Genet. 2013, 126, 2103–2121. [Google Scholar] [CrossRef]
- Würschum, T.; Langer, S.M.; Longin, C.F.H.; Korzun, V.; Akhunov, E.; Ebmeyer, E.; Schachschneider, R.; Schacht, J.; Kazman, E.; Reif, J.C. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theor. Appl. Genet. 2013, 126, 1477–1486. [Google Scholar] [CrossRef]
- Singh, A.; Knox, R.E.; DePauw, R.M.; Singh, A.K.; Cuthbert, R.D.; Campbell, H.L.; Shorter, S.; Bhavani, S. Stripe rust and leaf rust resistance QTL mapping, epistatic interactions, and co-localization with stem rust resistance loci in spring wheat evaluated over three continents. Theor. Appl. Genet. 2014, 127, 2465–2477. [Google Scholar] [CrossRef] [PubMed]
- Buerstmayr, M.; Huber, K.; Heckmann, J.; Steiner, B.; Nelson, J.C.; Buerstmayr, H. Mapping of QTL for Fusarium head blight resistance and morphological and developmental traits in three backcross populations derived from Triticum dicoccum × Triticum durum. Theor. Appl. Genet. 2012, 125, 1751–1765. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Zongo, A.; Khera, P.; Sawadogo, M.; Shasidhar, Y.; Sriswathi, M.; Vishwakarma, M.K.; Sankara, P.; Ntare, B.R.; Varshney, R.K.; Pandey, M.K.; et al. SSR markers associated to early leaf spot disease resistance through selective genotyping and single marker analysis in groundnut (Arachis hypogaea L.). Biotechnol. Rep. 2017, 15, 132–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, J.; Somta, P.; Chen, X.; Cui, X.; Yuan, X.; Srinives, P. Gene mapping of a mutant mungbean (Vigna radiata L.) using new molecular markers suggests a gene encoding a YUC4-like protein regulates the chasmogamous flower trait. Front. Plant Sci. 2016, 7, 830. [Google Scholar] [CrossRef] [PubMed]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Iquebal, M.A.; Sarika; Arora, V.; Verma, N.; Rai, A.; Kumar, D. First whole genome based microsatellite DNA marker database of tomato for mapping and variety identification. BMC Plant Biol. 2013, 13, 197. [Google Scholar] [CrossRef]
- Duhan, N.; Meshram, M.; Loaiza, C.D.; Kaundal, R. citSATdb: Genome-wide simple sequence repeat (SSR) marker database of citrus species for germplasm characterization and crop improvement. Genes 2020, 11, 1486. [Google Scholar] [CrossRef] [PubMed]
| Genome | Size MB | No. of Base Pairs | No. of SSRs | Freq/Mbp | Perfect SSRs (Repeat Units ≥ 15) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | % | Freq/Mbp | Genic | % | Non-Genic | % | |||||
| Glycine max | 974 | 973,419,153 | 475,123 | 488.1 | 150,682 | 31.7 | 154.8 | 35,588 | 23.6 | 115,094 | 76.4 |
| Cicer arietinum | 350 | 350,719,855 | 193,672 | 552.2 | 51,797 | 26.7 | 147.7 | 8014 | 15.5 | 43,783 | 84.5 |
| Medicago truncatula | 391 | 390,874,780 | 238,882 | 611.1 | 68,657 | 28.7 | 175.6 | 18,109 | 26.4 | 50,548 | 73.6 |
| Trifolium pratense | 192 | 192,330,821 | 105286 | 547.4 | 23,674 | 22.5 | 123.1 | 10,993 | 46.4 | 12,681 | 53.6 |
| Phaseolus vulgaris | 520 | 520,399,038 | 193,735 | 372.3 | 127,463 | 65.8 | 244.9 | 16,083 | 12.6 | 111,380 | 87.4 |
| Vigna unguiculata | 481 | 481,347,227 | 290,479 | 603.5 | 54,679 | 18.8 | 113.6 | 9510 | 17.4 | 45,169 | 82.6 |
| Arachis hypogaea | 2600 | 2,570,012,282 | 1,009,984 | 393.0 | 319,463 | 31.6 | 124.3 | 50,863 | 15.9 | 268,600 | 84.1 |
| Arachis ipaensis | 1400 | 1,359,188,642 | 437,350 | 321.8 | 99,538 | 22.8 | 73.2 | 21,942 | 22.0 | 77,596 | 78.0 |
| Cajanus cajun | 250 | 250,588,641 | 165,919 | 662.1 | 56,287 | 33.9 | 224.6 | 12,504 | 22.2 | 43,783 | 77.8 |
| Lupinus albus | 480 | 480,287,150 | 146,505 | 305.0 | 62,895 | 42.9 | 131.0 | 13,542 | 21.5 | 49,353 | 78.5 |
| Lupinus angustifolius | 476 | 476,300,322 | 132,282 | 277.7 | 54,187 | 41.0 | 113.8 | 12,508 | 23.1 | 41,679 | 76.9 |
| Vigna angularis | 377 | 377,395,406 | 140,751 | 373.0 | 38,517 | 27.4 | 102.1 | 8514 | 22.1 | 30,003 | 77.9 |
| Vigna radiata | 338 | 337,474,823 | 176,308 | 522.4 | 61,388 | 34.8 | 181.9 | 12,866 | 21.0 | 48,522 | 79.0 |
| Genome | Mono% | Di% | Tri% | Tetra% | Penta% | Hexa% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | P | All | P | All | P | All | P | All | P | All | P | |
| Glycine max | 51.1 | 15.9 | 39.3 | 53.8 | 8.6 | 27.3 | 0.8 | 2.5 | 0.2 | 0.5 | 0.1 | 0.2 |
| Cicer arietinum | 53.3 | 15.1 | 31.7 | 38.1 | 12.6 | 40.3 | 1.6 | 5.2 | 0.4 | 1.2 | 0.3 | 1.1 |
| Medicago truncatula | 67.6 | 41.8 | 25.5 | 34.1 | 6.1 | 21.4 | 0.6 | 2.3 | 0.1 | 0.4 | 0.1 | 0.2 |
| Trifolium pratense | 62.7 | 13 | 25.5 | 34.7 | 9.7 | 43.1 | 1.8 | 8.2 | 0.2 | 1.1 | 0.1 | 0.4 |
| Vigna unguiculata | 49.4 | 8.4 | 39.3 | 52.0 | 10.3 | 36.7 | 0.7 | 2.5 | 0.1 | 0.5 | 0.2 | 0.6 |
| Phaseolus vulgaris | 40.2 | 5.5 | 46.6 | 64.7 | 11.6 | 26.4 | 0.9 | 2.2 | 0.6 | 1.3 | 0.1 | 0.3 |
| Arachis hypogaea | 44.3 | 11.9 | 40.5 | 40.2 | 13.2 | 42.1 | 1.4 | 4.5 | 0.4 | 1.4 | 0.2 | 0.6 |
| Arachis ipaensis | 47.4 | 9.9 | 39.2 | 31.7 | 11.1 | 49.2 | 1.6 | 6.9 | 0.5 | 2.3 | 0.2 | 0.8 |
| Cajanus cajun | 46.3 | 17.5 | 44.5 | 56.0 | 7.7 | 22.7 | 1.1 | 3.3 | 0.2 | 0.5 | 0.2 | 0.7 |
| Lupinus albus | 25.0 | 5.3 | 49.4 | 35.4 | 8.0 | 18.7 | 1.0 | 2.3 | 16.4 | 38.3 | 0.3 | 0.6 |
| Lupinus angustifolius | 25.9 | 2.3 | 49.3 | 49.0 | 14.0 | 44.4 | 0.9 | 2.9 | 0.4 | 1.4 | 9.5 | 30.2 |
| Vigna angularis | 46.5 | 5.9 | 43.1 | 56.4 | 9.3 | 34.0 | 0.7 | 2.5 | 0.3 | 1.1 | 0.1 | 0.5 |
| Vigna radiata | 52.4 | 12.6 | 38.6 | 61.7 | 7.8 | 22.6 | 0.8 | 2.3 | 0.3 | 0.8 | 0.1 | 0.2 |
| Features | legumeSSRdb | CicArMiSatDB | Legumeinfo | LegumeIP | PMDbase |
|---|---|---|---|---|---|
| Number of species | 13 | 1 | 22 | 21 | 15 |
| Microsatellites | Yes | Yes | No | No | Yes |
| Microsatellite Search Criteria | Yes (Advanced) | Limited | No | No | Limited |
| Microsatellites results—Graphical visualization | Yes | No | No | No | No |
| Genic and non-genic classification of SSRs | Yes | No | No | No | No |
| Primer Designing | Yes (Custom) | Yes (Predesigned) | No | No | Yes (Predesigned) |
| Primer Validation using e-PCR | Yes | No | No | No | No |
| BLAST | Yes | Yes | Yes | Yes | Yes |
| Blast result graphical visualization | Yes | No | No | No | No |
| Genome Browse | Yes | Yes | No | No | Yes |
| SSR Predictor | Yes | No | No | No | Yes |
| Primer Designing for predicted SSRs | Yes | No | No | No | No |
| Functional Annotation | Yes | No | Yes | Yes (In-depth) | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duhan, N.; Kaundal, R. LegumeSSRdb: A Comprehensive Microsatellite Marker Database of Legumes for Germplasm Characterization and Crop Improvement. Int. J. Mol. Sci. 2021, 22, 11350. https://doi.org/10.3390/ijms222111350
Duhan N, Kaundal R. LegumeSSRdb: A Comprehensive Microsatellite Marker Database of Legumes for Germplasm Characterization and Crop Improvement. International Journal of Molecular Sciences. 2021; 22(21):11350. https://doi.org/10.3390/ijms222111350
Chicago/Turabian StyleDuhan, Naveen, and Rakesh Kaundal. 2021. "LegumeSSRdb: A Comprehensive Microsatellite Marker Database of Legumes for Germplasm Characterization and Crop Improvement" International Journal of Molecular Sciences 22, no. 21: 11350. https://doi.org/10.3390/ijms222111350
APA StyleDuhan, N., & Kaundal, R. (2021). LegumeSSRdb: A Comprehensive Microsatellite Marker Database of Legumes for Germplasm Characterization and Crop Improvement. International Journal of Molecular Sciences, 22(21), 11350. https://doi.org/10.3390/ijms222111350







