Prediction of Protein–ATP Binding Residues Based on Ensemble of Deep Convolutional Neural Networks and LightGBM Algorithm
Abstract
1. Introduction
2. Results
2.1. Performance Comparison with other Sequence-Based Prediction Methods
| Method | ACC | Sen | Spe | MCC | AUC |
|---|---|---|---|---|---|
| Complementary template | 0.967 | 0.189 | 0.995 | 0.324 | -- |
| ATPint a | 0.665 | 0.512 | 0.660 | 0.066 | 0.606 |
| ATPsite a | 0.969 | 0.367 | 0.991 | 0.451 | 0.868 |
| NsitePred b | 0.967 | 0.460 | 0.985 | 0.476 | 0.875 |
| TargetATPsite c | 0.972 | 0.458 | 0.991 | 0.530 | 0.882 |
| TargetNUCs d | 0.975 | 0.516 | 0.992 | 0.584 | -- |
| Multi-Xception-based predictor | 0.977 | 0.565 | 0.993 | 0.630 | 0.909 |
| TargetATP a | 0.969 | 0.489 | 0.989 | 0.542 | 0.912 |
| Multi-IncepResNet-based predictor | 0.969 | 0.608 | 0.983 | 0.565 | 0.915 |
| LightGBM predictor | 0.977 | 0.556 | 0.992 | 0.618 | 0.917 |
| Ensemble without template | 0.978 | 0.569 | 0.993 | 0.638 | 0.925 |
| Ensemble predictor | 0.978 | 0.589 | 0.992 | 0.639 | 0.925 |
| Method | ACC | Sen | Spe | MCC | AUC |
|---|---|---|---|---|---|
| Complementary template | 0.964 | 0.239 | 0.998 | 0.451 | -- |
| NsitePred a | 0.954 | 0.467 | 0.977 | 0.456 | 0.852 |
| TargetATPsite a | 0.968 | 0.413 | 0.995 | 0.559 | 0.853 |
| TargetNUCs a | 0.972 | 0.469 | 0.997 | 0.627 | 0.856 |
| Multi-IncepResNet-based predictor | 0.969 | 0.441 | 0.995 | 0.589 | 0.875 |
| ATPseq a | 0.972 | 0.545 | 0.993 | 0.639 | 0.878 |
| Multi-Xception-based predictor | 0.968 | 0.480 | 0.992 | 0.585 | 0.889 |
| LightGBM predictor | 0.970 | 0.447 | 0.996 | 0.597 | 0.896 |
| Ensemble without template | 0.972 | 0.461 | 0.997 | 0.625 | 0.902 |
| Ensemble predictor | 0.973 | 0.497 | 0.996 | 0.642 | 0.902 |
2.2. Case Study
3. Discussions
3.1. Feature Importance Analysis
3.2. Proposed CNN-Based Models Showed Better Performance than Simple 2D CNN Model
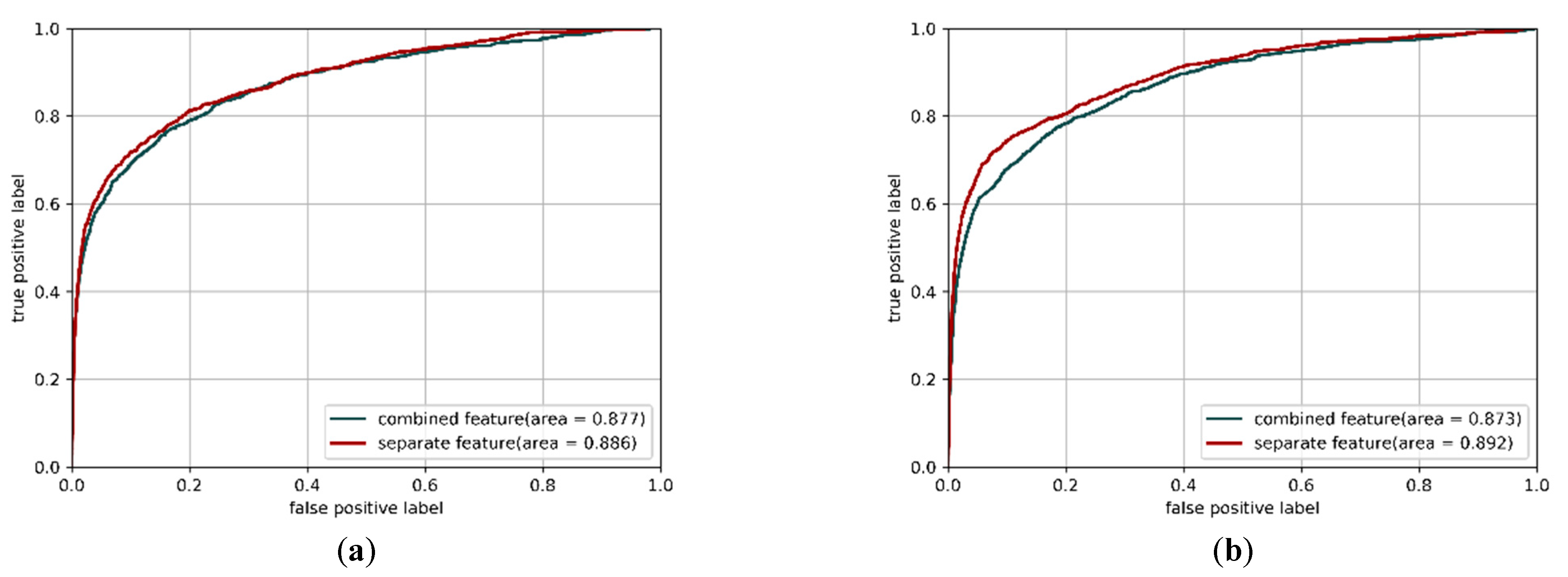
3.3. Applying Separate Features as Inputs in CNN Models can Improve Performance
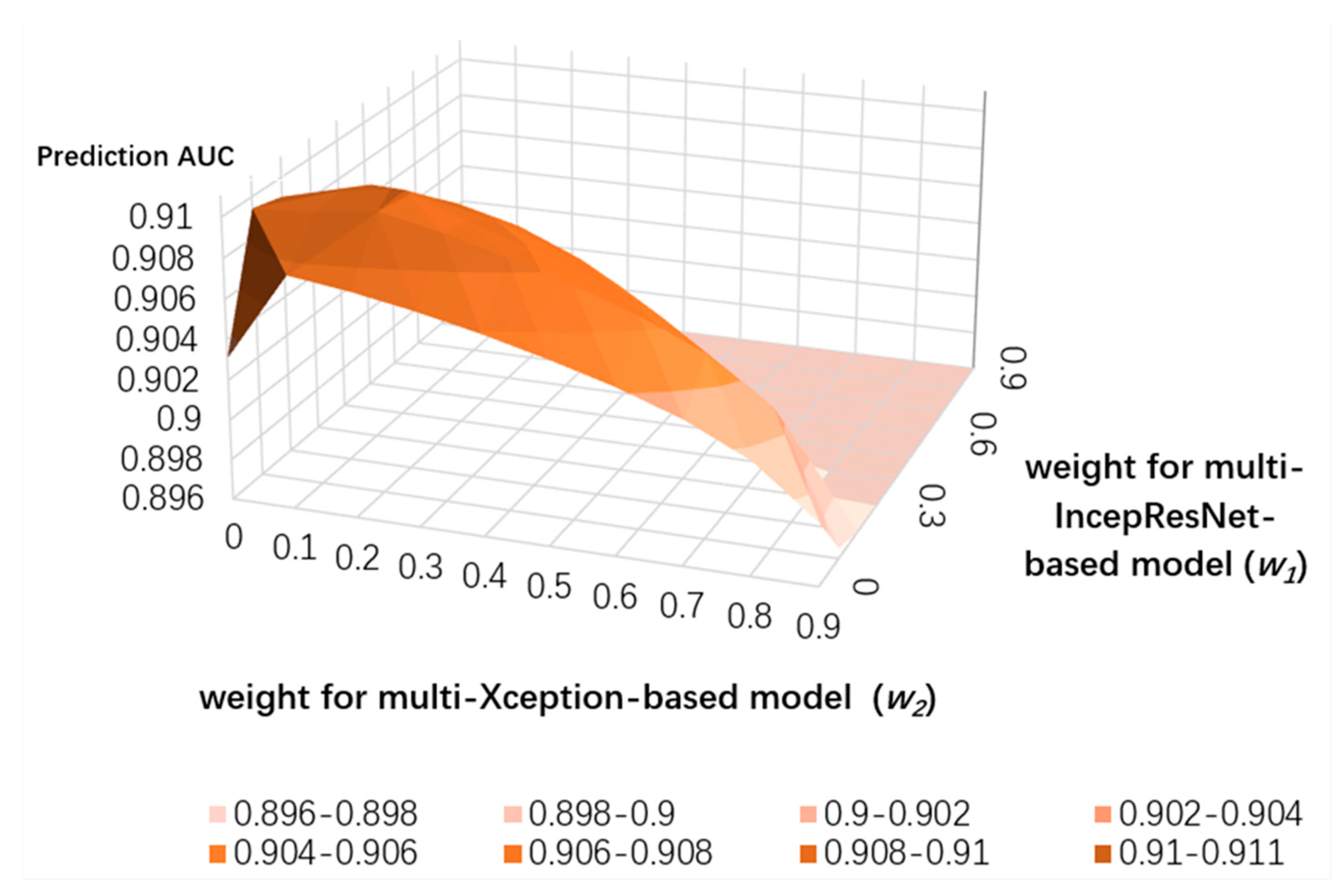
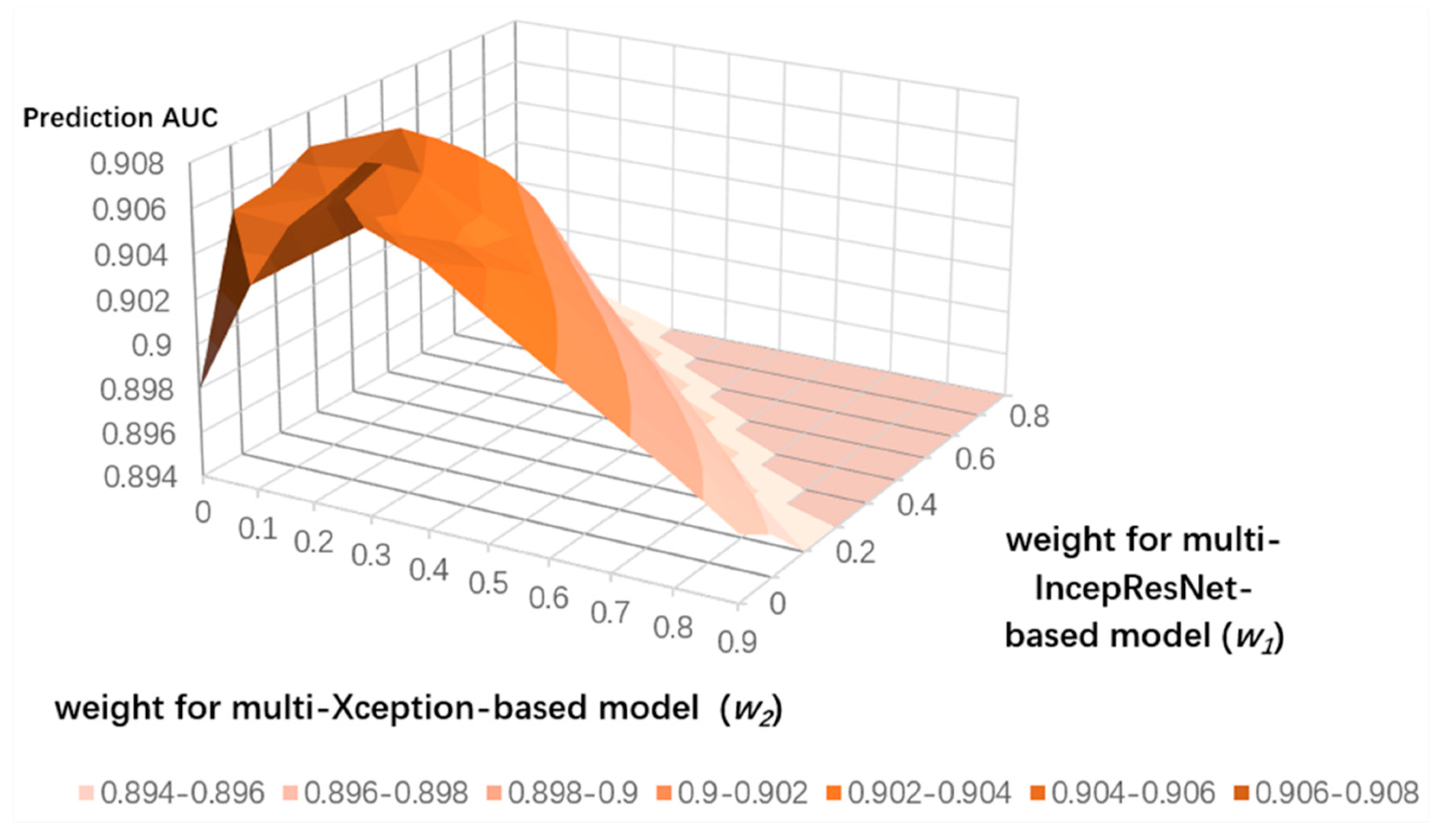
3.4. Ensemble Learning for CNN Predictors and the LightGBM Predictor
4. Materials and Methods
4.1. Datasets
4.1.1. ATP-227 and ATP-17
4.1.2. ATP-388 and ATP-41
4.2. Feature Representation
4.2.1. Position-Specific Scoring Matrix (PSSM)
4.2.2. Predicted Secondary Structure
4.2.3. Residue One-Hot Encoding
4.3. Deep Convolutional Neural Network
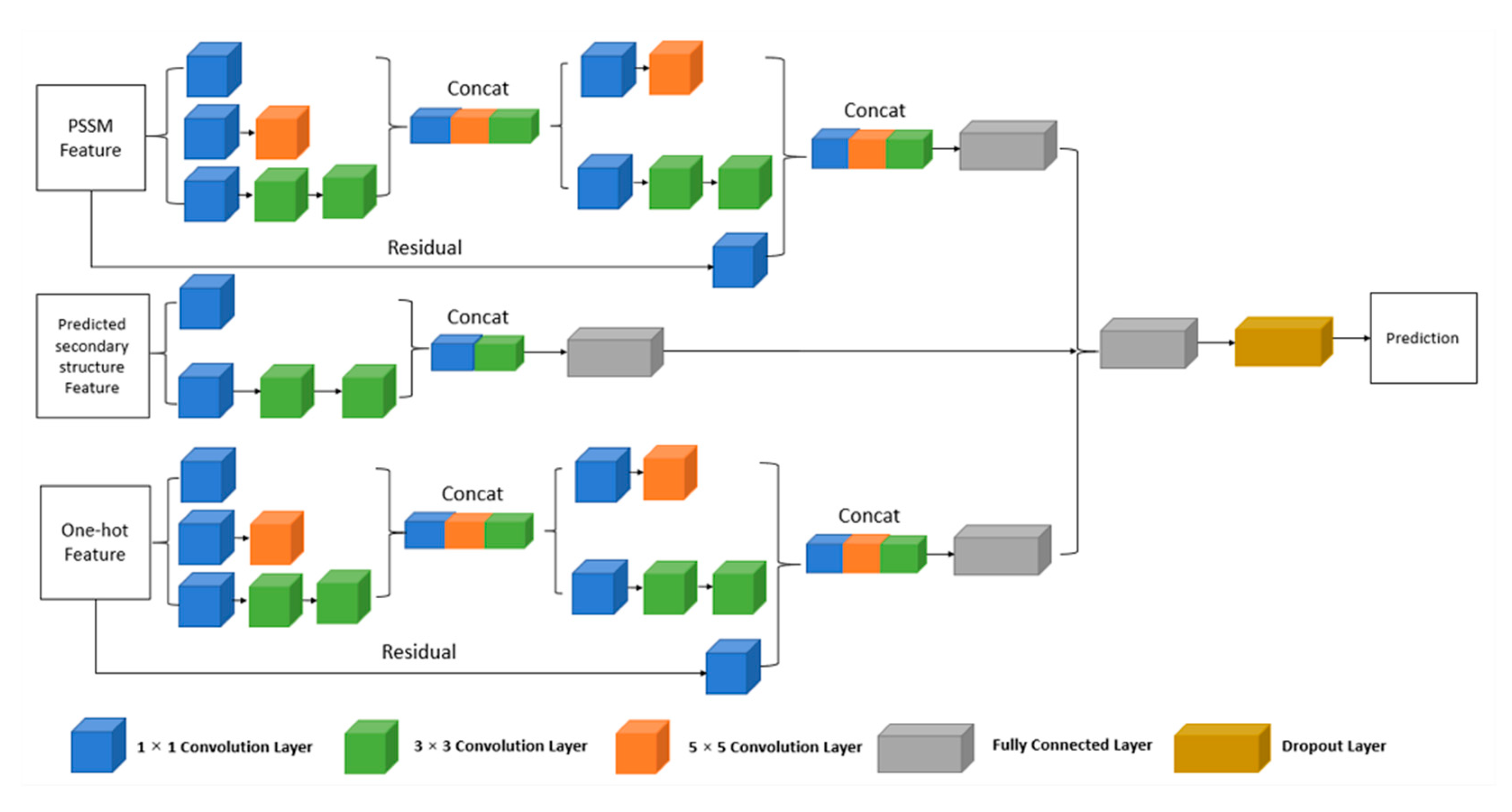
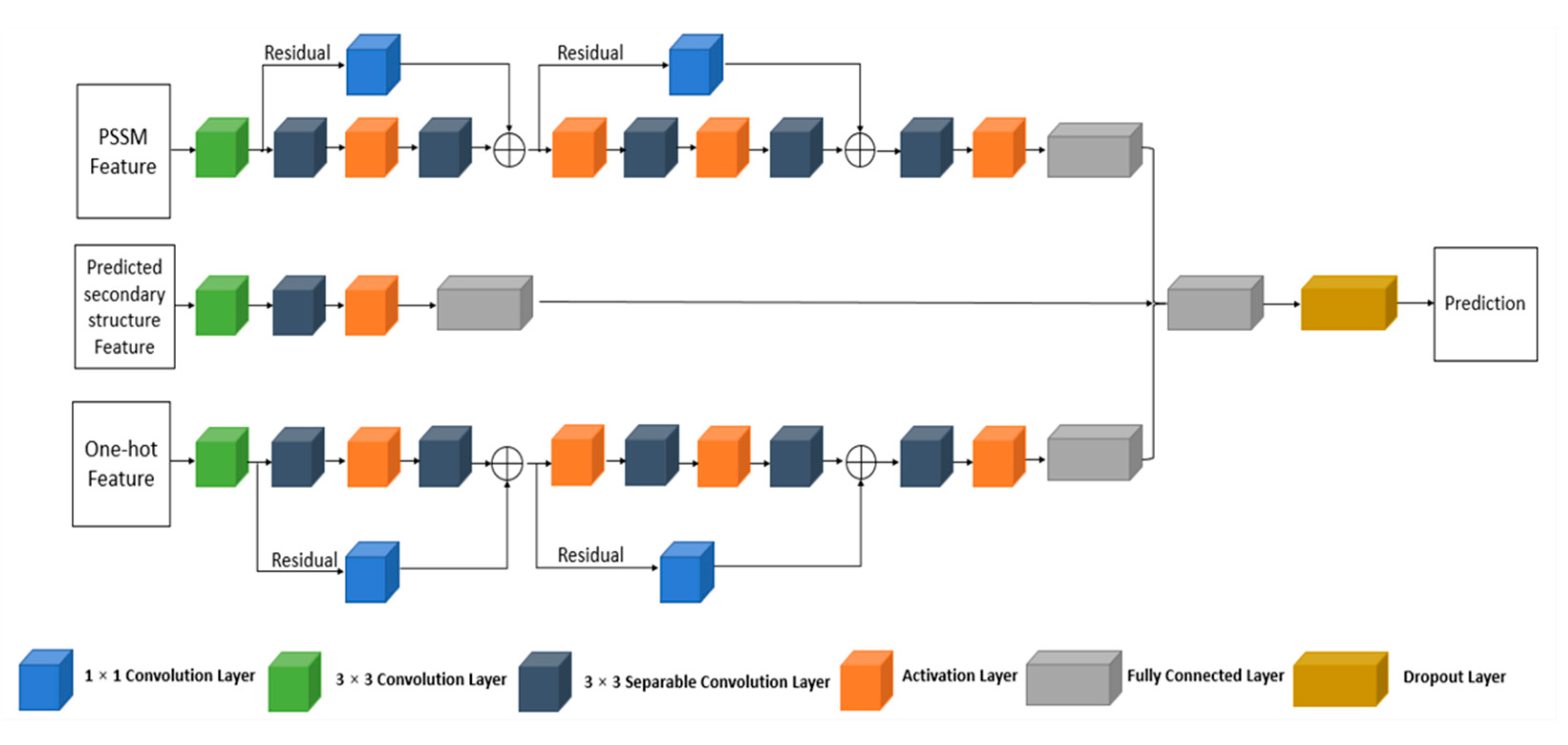
4.3.1. The Multi-IncepResNet-Based Predictor
4.3.2. The Multi-Xception-Based Predictor
4.4. LightGBM Predictor
4.5. Complementary Template-Based Prediction Method
4.6. Imbalanced Learning Problem
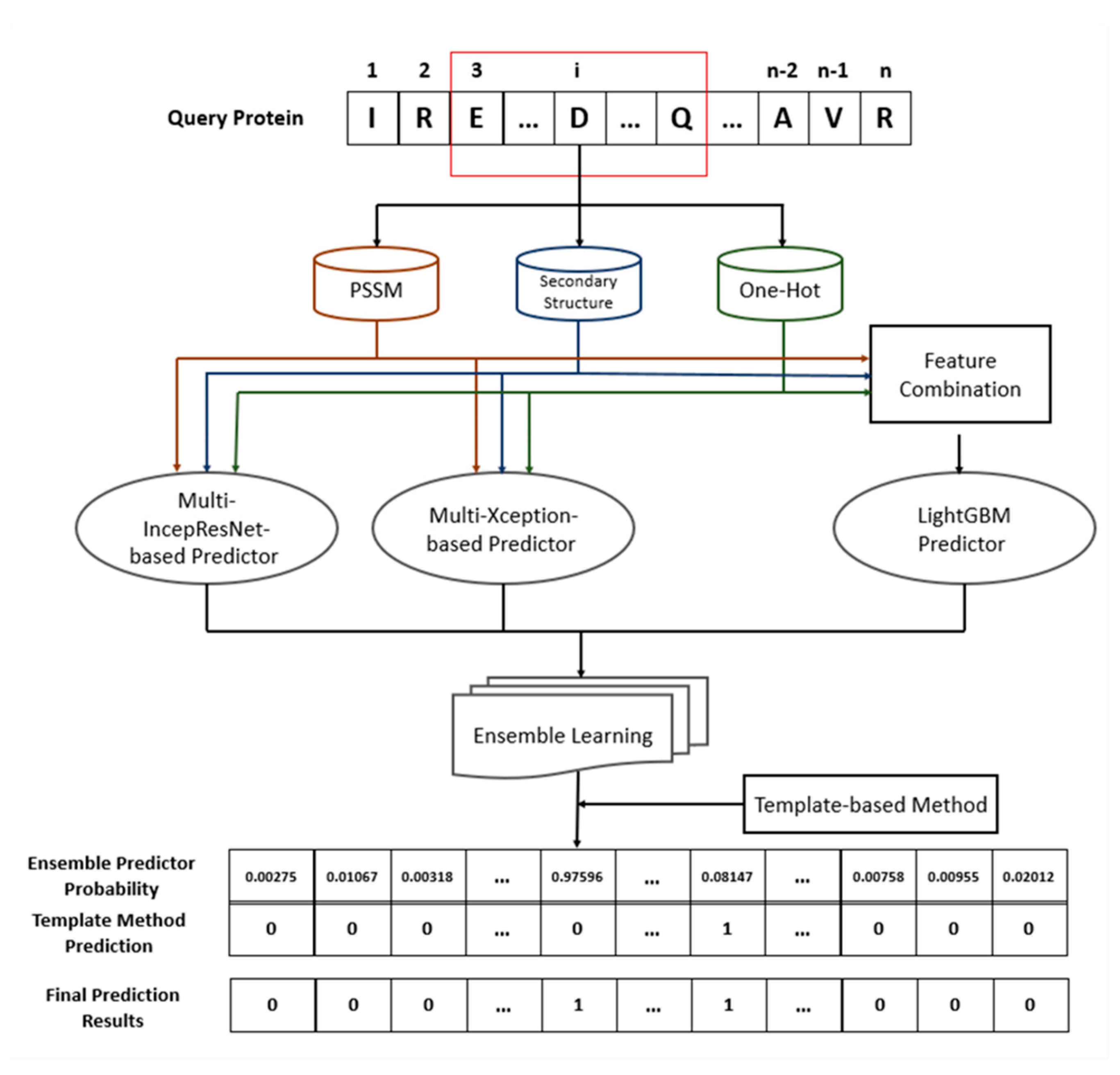
4.7. Architecture of Proposed Ensemble Prediction Method
4.8. Performance Evaluation
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Souza, P.C.T.; Thallmair, S.; Conflitti, P.; Ramírez-Palacios, C.; Alessandri, R.; Raniolo, S.; Limongelli, V.; Marrink, S.J. Protein–ligand binding with the coarse-grained Martini model. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Xu, L.; Chang, S.; Xu, X.; Meng, L. Multitask deep networks with grid featurization achieve improved scoring performance for protein–ligand binding. Chem. Biol. Drug Des. 2020, 96, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Verteramo, M.L.; Stenström, O.; Ignjatović, M.M.; Caldararu, O.; Olsson, M.A.; Manzoni, F.; Leffler, H.; Oksanen, E.; Logan, D.T.; Nilsson, U.J.; et al. Interplay between conformational entropy and solvation entropy in protein–ligand binding. J. Am. Chem. Soc. 2019, 141, 2012–2026. [Google Scholar] [CrossRef]
- Yuan, C.; Shui, I.M.; Wilson, K.M.; Stampfer, M.J.; Mucci, L.A.L.; Giovannucci, E.L. Circulating 25-hydroxyvitamin D, vitamin D binding proteinand risk of advanced and lethal prostate cancer. Int. J. Cancer 2019, 144, 2401–2407. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.P.; Sunilkumar, S.; Giordano, J.F.; Toro, A.L.; Barber, A.J.; Dennis, M.D. The stress response protein REDD1 promotes diabetes-induced oxidative stress in the retina by Keap1-independent Nrf2 degradation. J. Biol. Chem. 2020, 295, 7350–7361. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Qiao, Y.; Jiang, X.; Li, P.; Kuai, Z.; Gong, X.; Liu, D.; Fu, Q.; Sun, L.; Li, H.; et al. Multiple antigenic peptide system coupled with amyloid beta protein epitopes as an immunization approach to treat alzheimer’s disease. ACS Chem. Neurosci. 2019, 10, 2794–2800. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, A.; Lawson, D.M. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 2003, 3, 283–303. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Hu, J.; Tang, Z.; Shen, H.B.; Yang, J.; Yang, J.Y. Improving protein-ATP binding residues pre-diction by boosting SVMs with random under-sampling. Neurocomputing 2013, 104, 180–190. [Google Scholar] [CrossRef]
- Zhang, Y.-N.; Yu, D.-J.; Li, S.-S.; Fan, Y.-X.; Huang, Y.; Shen, H.-B. Predicting protein-ATP binding sites from primary sequence through fusing bi-profile sampling of multi-view features. BMC Bioinform. 2012, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Boutet, S.; Lomb, L.; Williams, G.J.; Barends, T.R.M.; Aquila, A.; Doak, R.B.; Weierstall, U.; DePonte, D.P.; Steinbrener, J.; Shoeman, R.L.; et al. High-resolution protein structure determination by serial femtosecond crystallography. Science 2012, 337, 362–364. [Google Scholar] [CrossRef]
- Cavalli, A.; Salvatella, X.; Dobson, C.M.; Vendruscolo, M. Protein structure determination from NMR chemical shifts. Proc. Natl. Acad. Sci. USA 2007, 104, 9615–9620. [Google Scholar] [CrossRef]
- Vangone, A.; Schaarschmidt, J.; Koukos, P.; Geng, C.; Citro, N.; Trellet, M.E.; Xue, L.C.; Bonvin, A.M.J.J. Large-scale prediction of binding affinity in protein–small ligand complexes: The PRODIGY-LIG web server. Bioinformatics 2019, 35, 1585–1587. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucl. Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R. The swiss-prot protein sequence data bank and its new supplement TREMBL. Nucl. Acids Res. 1996, 21, 21–25. [Google Scholar] [CrossRef]
- Chauhan, J.S.; Mishra, N.K.; Raghava, G.P. Identification of ATP binding residues of a protein from its primary sequence. BMC Bioinform. 2009, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Mizianty, M.; Kurgan, L. ATPsite: Sequence-based prediction of ATP-binding residues. Proteom. Sci. 2011, 9, S4. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Hu, J.; Huang, Y.; Shen, H.-B.; Qi, Y.; Tang, Z.-M.; Yang, J.-Y. TargetATPsite: A template-free method for ATP-binding sites prediction with residue evolution image sparse representation and classifier ensemble. J. Comput. Chem. 2013, 34, 974–985. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Noguchi, T.; Yamana, H. Simplified sequence-based method for ATP-binding prediction using contextual local evolutionary conservation. Algorithms Mol. Biol. 2014, 9, 7. [Google Scholar] [CrossRef]
- Hu, J.; Li, Y.; Zhang, Y.; Yu, D. ATPbind: Accurate protein-ATP binding site prediction by combining se-quence-profiling and structure-based comparisons. J. Chem. Inf. Model. 2018, 58, 501–510. [Google Scholar] [CrossRef]
- Cheng, F.; Zhang, H.; Fan, W.; Harris, B. Image recognition technology based on deep learning. Wirel. Pers. Commun. 2018, 102, 1917–1933. [Google Scholar] [CrossRef]
- Yu, Z.; Li, T.; Luo, G.; Fujita, H.; Yu, N.; Pan, Y. Convolutional networks with cross-layer neurons for image recognition. Inf. Sci. 2018, 433, 241–254. [Google Scholar] [CrossRef]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep Learning for Computer Vision: A Brief Review. Comput. Intell. Neurosci. 2018, 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Singhal, A.; Sinha, P.; Pant, R. Use of deep learning in modern recommendation system: A summary of recent works. Int. J. Comput. Appl. 2017, 180, 17–22. [Google Scholar] [CrossRef]
- Wang, W.; Gang, J. Application of convolutional neural network in natural language processing. In Proceedings of the 2018 International Conference on Information Systems and Computer Aided Education (ICISCAE), Changchun, China, 6–8 July 2018. [Google Scholar]
- Cheng, J.; Liu, Y.; Ma, Y. Protein secondary structure prediction based on integration of CNN and LSTM model. J. Vis. Commun. Image Represent 2020, 71, 102844. [Google Scholar] [CrossRef]
- Li, Y.; Shibuya, T. Malphite: A convolutional neural network and ensemble learning based protein secondary structure predictor. In Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 1260–1266. [Google Scholar]
- Cao, Z.; Zhang, S. Simple tricks of convolutional neural network architectures improve DNA-protein binding pre-diction. Bioinformatics 2019, 35, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Shen, H.-B. Predicting RNA–protein binding sites and motifs through combining local and global deep convolutional neural networks. Bioinformatics 2018, 34, 3427–3436. [Google Scholar] [CrossRef]
- Golkov, V.; Skwark, M.J.; Golkov, A.; Dosovitskiy, A.; Brox, T.; Meiler, J.; Cremers, D. Protein contact prediction from amino acid co-evolution using convolutional networks for graph-valued images. In Proceedings of the Conference on Neural Information Processing Systems (NeurlIPS), Barcelona, Spain, 5–10 December 2016. [Google Scholar]
- Zhou, J.; Lu, Q.; Xu, R.; Gui, L.; Wang, H. CNNsite: Prediction of DNA-binding residues in proteins using convolutional neural network with sequence features. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 78–85. [Google Scholar]
- Nguyen, T.-T.-D.; Le, N.-Q.-K.; Kusuma, R.M.I.; Ou, Y.-Y. Prediction of ATP-binding sites in membrane proteins using a two-dimensional convolutional neural network. J. Mol. Graph. Model. 2019, 92, 86–93. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. AIP Conf. Proc. 2016, 2818–2826. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chem, W.; Ma, W.; Ye, O.; Liu, T. LightGBM: A highly efficient gradient boosting decision tree. In Proceedings of the 31st Conference on Neural Information Processing System, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Do, D.T.; Hung, T.N.K.; Lam, L.H.T.; Lin, C.-M.; Nguyen, N.T.K. A computational framework based on ensemble deep neural networks for essential genes identification. Int. J. Mol. Sci. 2020, 21, 9070. [Google Scholar] [CrossRef]
- Zhou, J.; Lu, Q.; Xu, R.; He, Y.; Wang, H. EL_PSSM-RT: DNA-binding residue prediction by integrating ensemble learning with PSSM relation transformation. BMC Bioinform. 2017, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lam, L.H.T.; Le, N.H.; Van Tuan, L.; Ban, H.T.; Hung, T.N.K.; Nguyen, N.T.K.; Dang, L.H.; Le, N.-Q.-K. Machine Learning Model for Identifying Antioxidant Proteins Using Features Calculated from Primary Sequences. Biology 2020, 9, 325. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef] [PubMed]
- Wuthrich, K.; Billeter, M.; Braun, W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J. Mol. Biol. 1983, 169, 949–961. [Google Scholar] [CrossRef]
- Fang, C.; Shang, Y.; Xu, D. MUFOLD-SS: New deep inception-inside-inception networks for protein secondary structure prediction. Proteins Struct. Funct. Bioinform. 2018, 86, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Hong, Q.; Wang, B.; Wang, H. Efficient resnet model to predict protein-protein interactions with gpu computing. IEEE Access 2020, 8, 127834–127844. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Walia, R.R.; Xue, L.C.; Wilkins, K.; El-Manzalawy, Y.; Dobbs, D.; Honavar, V. RNABindRPlus: A predictor that combines machine learning and sequence homology-based methods to improve the reliability of predicted RNA-binding residues in proteins. PLoS ONE 2014, 9, e97725. [Google Scholar] [CrossRef]
- Chen, K.; Mizianty, M.J.; Kurgan, L. Prediction and analysis of nucleotide-binding residues using sequence and sequence-derived structural descriptors. Bioinformatics 2012, 28, 331–341. [Google Scholar] [CrossRef]
- Hu, J.; Li, Y.; Yan, W.-X.; Yang, J.-Y.; Shen, H.-B.; Yu, D.-J. KNN-based dynamic query-driven sample rescaling strategy for class imbalance learning. Neurocomputing 2016, 191, 363–373. [Google Scholar] [CrossRef]



| Hyperparameter | Value |
|---|---|
| Optimizer | Adamdelta, Adamgrad, Adam, RMSprop, SGD, Adamax |
| Number of epochs | 30 to 120 |
| Batch size | 32, 64, 128, 256 |
| Learning rate | 1 × 10−1, 1 × 10−2, 1 × 10−3, 1 × 10−4 |
| Dropout rate | 0.1, 0.2, 0.3, 0.4, 0.5 |
| Method | Accuracy (ACC) | Sensitivity (Sen) | Specificity (Spe) | Matthews Correlation Coefficient (MCC) | AUC |
|---|---|---|---|---|---|
| Fine-tuned 2D CNN model | 0.950 | 0.512 | 0.967 | 0.417 | 0.871 |
| Multi-IncepResNet-based predictor | 0.965 | 0.489 | 0.984 | 0.501 | 0.886 |
| Multi-Xception-based predictor | 0.967 | 0.491 | 0.986 | 0.519 | 0.892 |
| Hyperparameter | CNN Structure with Combined Feature | CNN Structure with Separate Features |
|---|---|---|
| Optimizer | Adam | Adam |
| Training epochs | 60 | 60 |
| Initial learning rate | 0.001 | 0.0001 |
| Batch size | 128 | 256 |
| Dropout rate | 0.4 | 0.4 |
| Strategy | ACC | Sen | Spe | MCC | AUC |
|---|---|---|---|---|---|
| ATP-388 | |||||
| maximum | 0.963 | 0.557 | 0.980 | 0.523 | 0.902 |
| minimum | 0.965 | 0.481 | 0.982 | 0.528 | 0.892 |
| mean | 0.966 | 0.568 | 0.981 | 0.544 | 0.906 |
| weighted | 0.968 | 0.599 | 0.981 | 0.549 | 0.910 |
| ATP-227 | |||||
| maximum | 0.965 | 0.537 | 0.983 | 0.536 | 0.898 |
| minimum | 0.967 | 0.529 | 0.984 | 0.544 | 0.893 |
| mean | 0.968 | 0.483 | 0.989 | 0.552 | 0.903 |
| weighted | 0.969 | 0.493 | 0.989 | 0.556 | 0.907 |
| Dataset | No. of ATP-Binding Residues | No. of Nonbinding Residues | Ratio a |
|---|---|---|---|
| ATP-227 | 3393 | 80,409 | 23.7 |
| ATP-17 | 248 | 6974 | 28.1 |
| ATP-388 | 5657 | 142,086 | 25.1 |
| ATP-41 | 681 | 14,152 | 20.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Liu, G.; Jiang, J.; Zhang, P.; Liang, Y. Prediction of Protein–ATP Binding Residues Based on Ensemble of Deep Convolutional Neural Networks and LightGBM Algorithm. Int. J. Mol. Sci. 2021, 22, 939. https://doi.org/10.3390/ijms22020939
Song J, Liu G, Jiang J, Zhang P, Liang Y. Prediction of Protein–ATP Binding Residues Based on Ensemble of Deep Convolutional Neural Networks and LightGBM Algorithm. International Journal of Molecular Sciences. 2021; 22(2):939. https://doi.org/10.3390/ijms22020939
Chicago/Turabian StyleSong, Jiazhi, Guixia Liu, Jingqing Jiang, Ping Zhang, and Yanchun Liang. 2021. "Prediction of Protein–ATP Binding Residues Based on Ensemble of Deep Convolutional Neural Networks and LightGBM Algorithm" International Journal of Molecular Sciences 22, no. 2: 939. https://doi.org/10.3390/ijms22020939
APA StyleSong, J., Liu, G., Jiang, J., Zhang, P., & Liang, Y. (2021). Prediction of Protein–ATP Binding Residues Based on Ensemble of Deep Convolutional Neural Networks and LightGBM Algorithm. International Journal of Molecular Sciences, 22(2), 939. https://doi.org/10.3390/ijms22020939





