The Kinase Chemogenomic Set (KCGS): An Open Science Resource for Kinase Vulnerability Identification
Abstract
1. Introduction
2. Results
2.1. Compound Selection
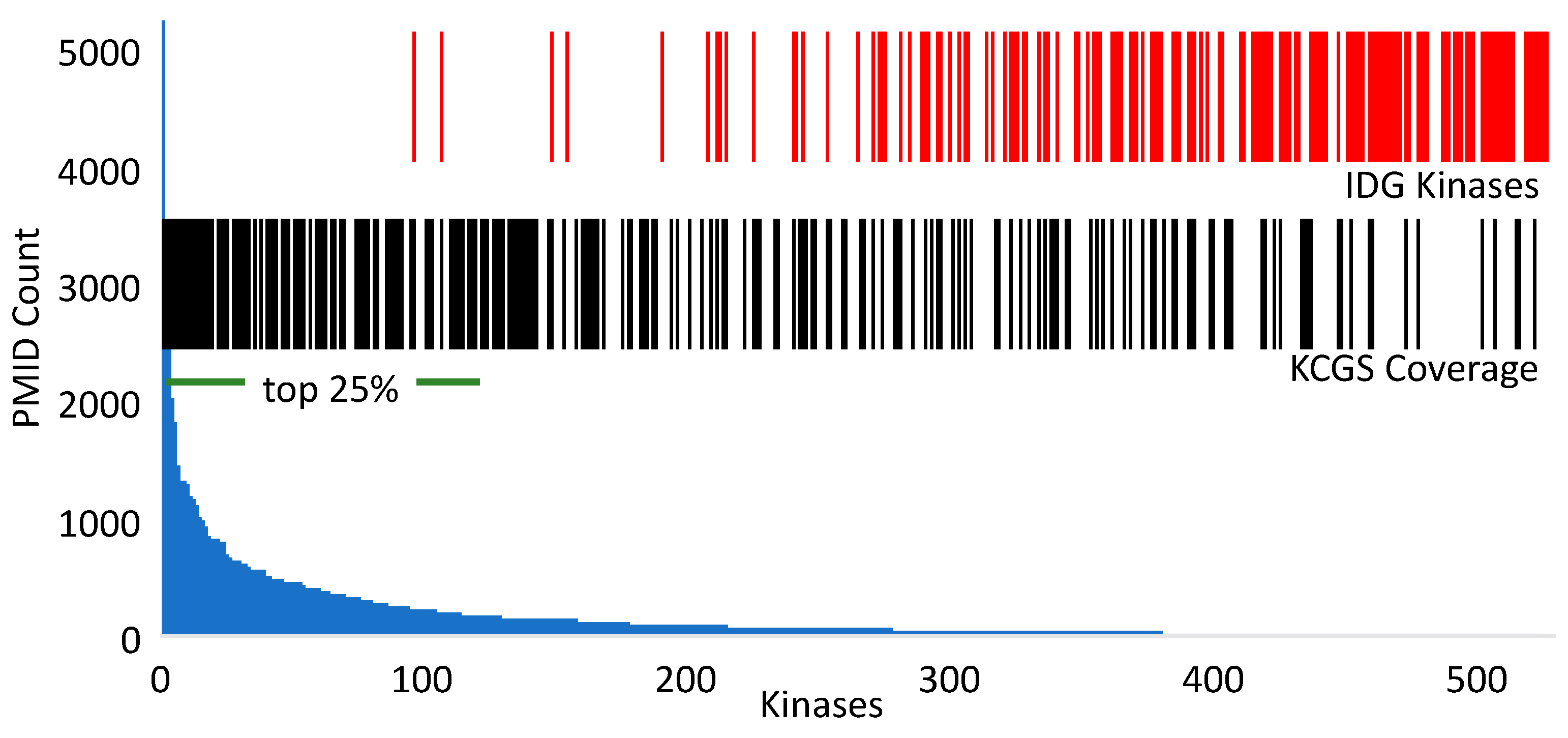
2.2. Kinase Coverage
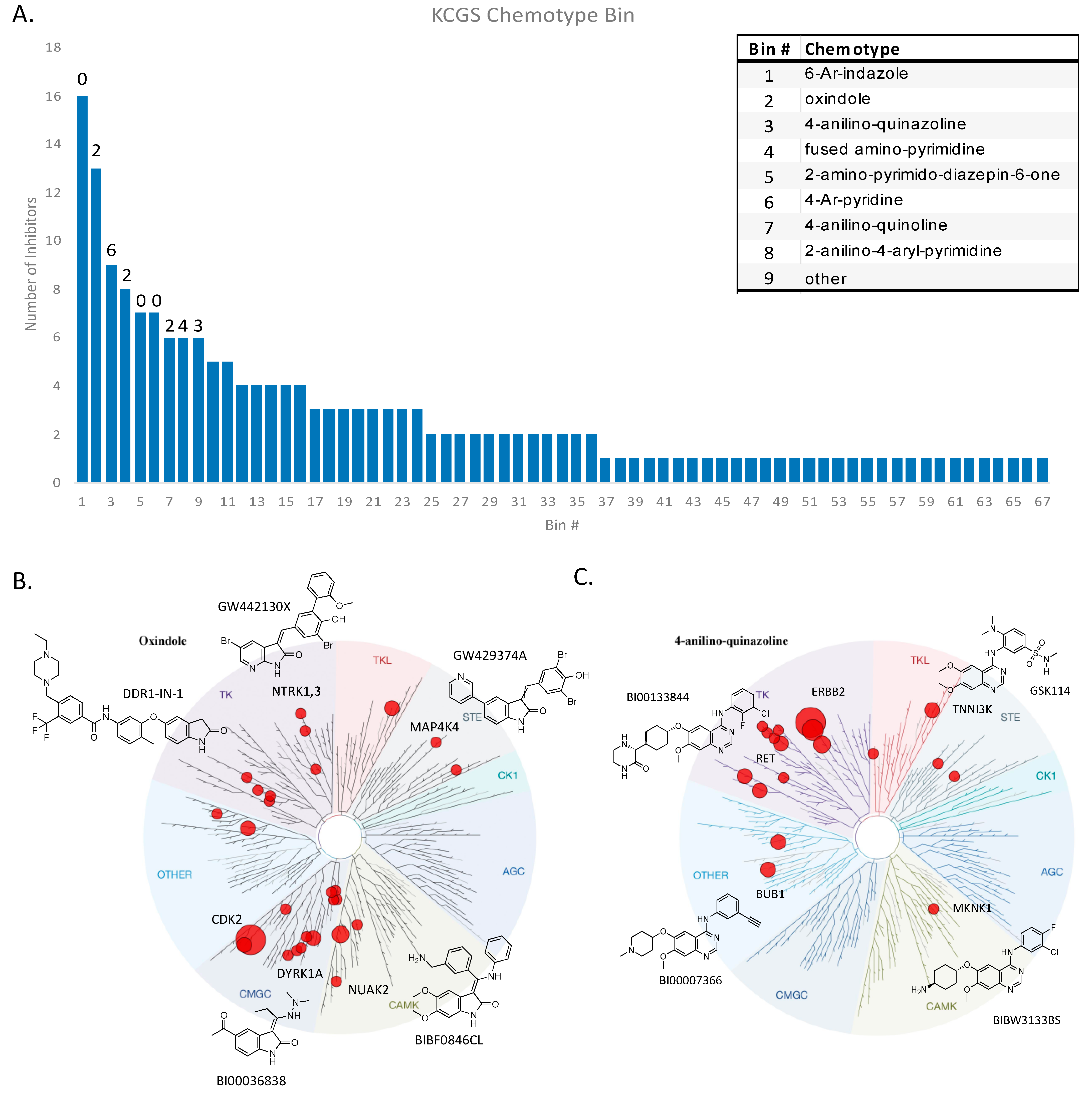
2.3. Chemotype Analysis
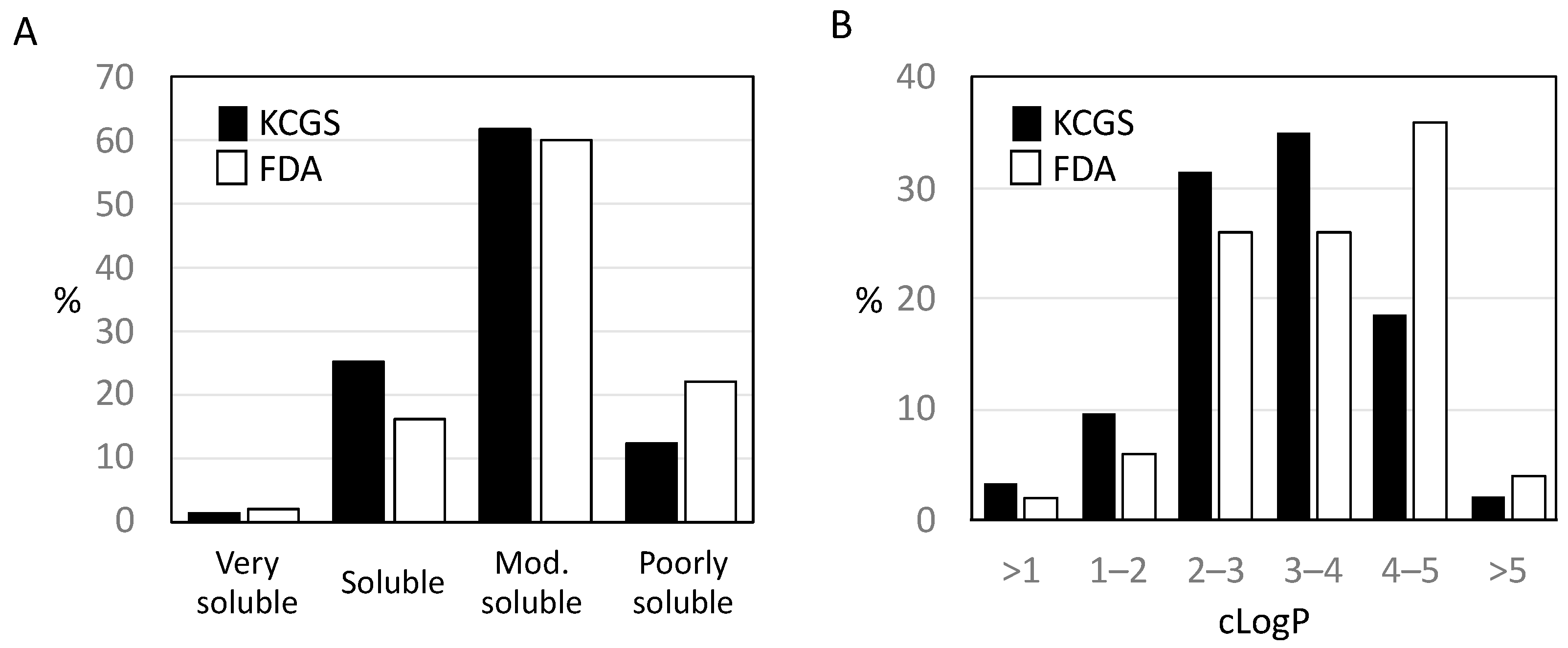
2.4. Calculated Properties
2.5. Chemogenomic Screening
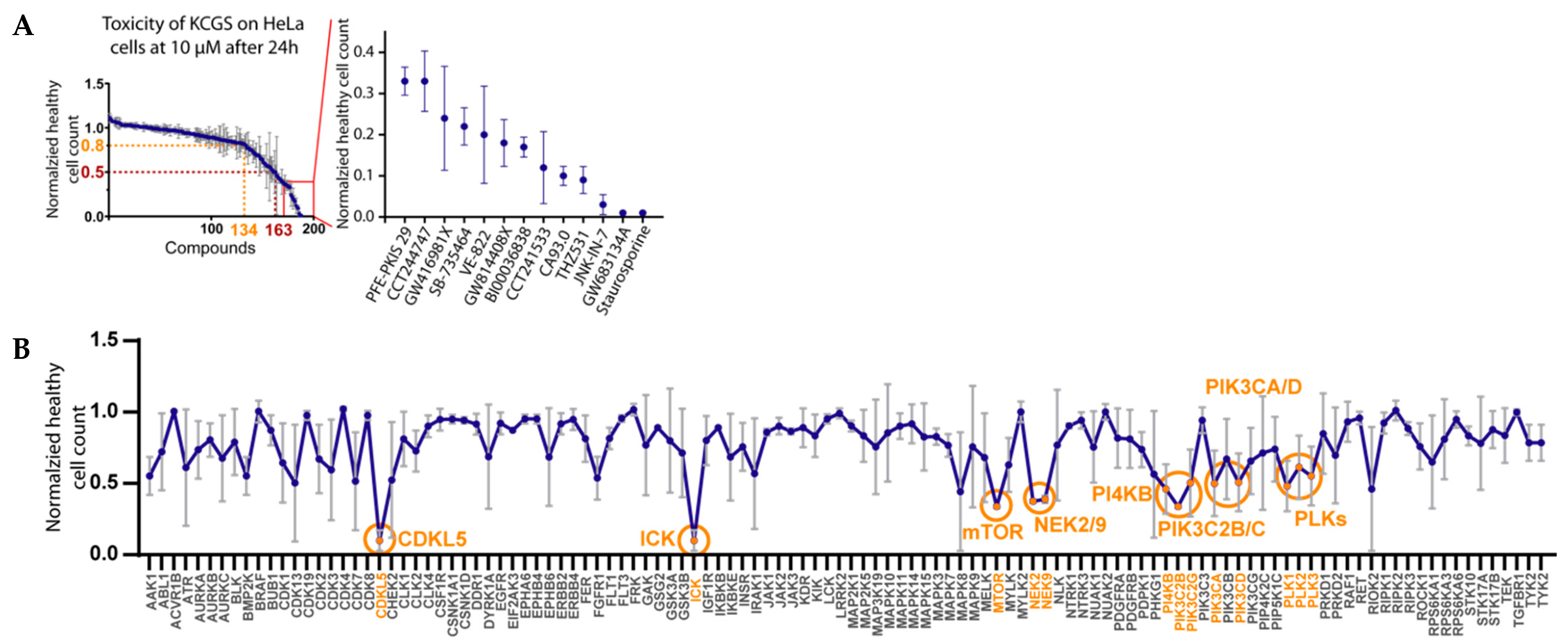
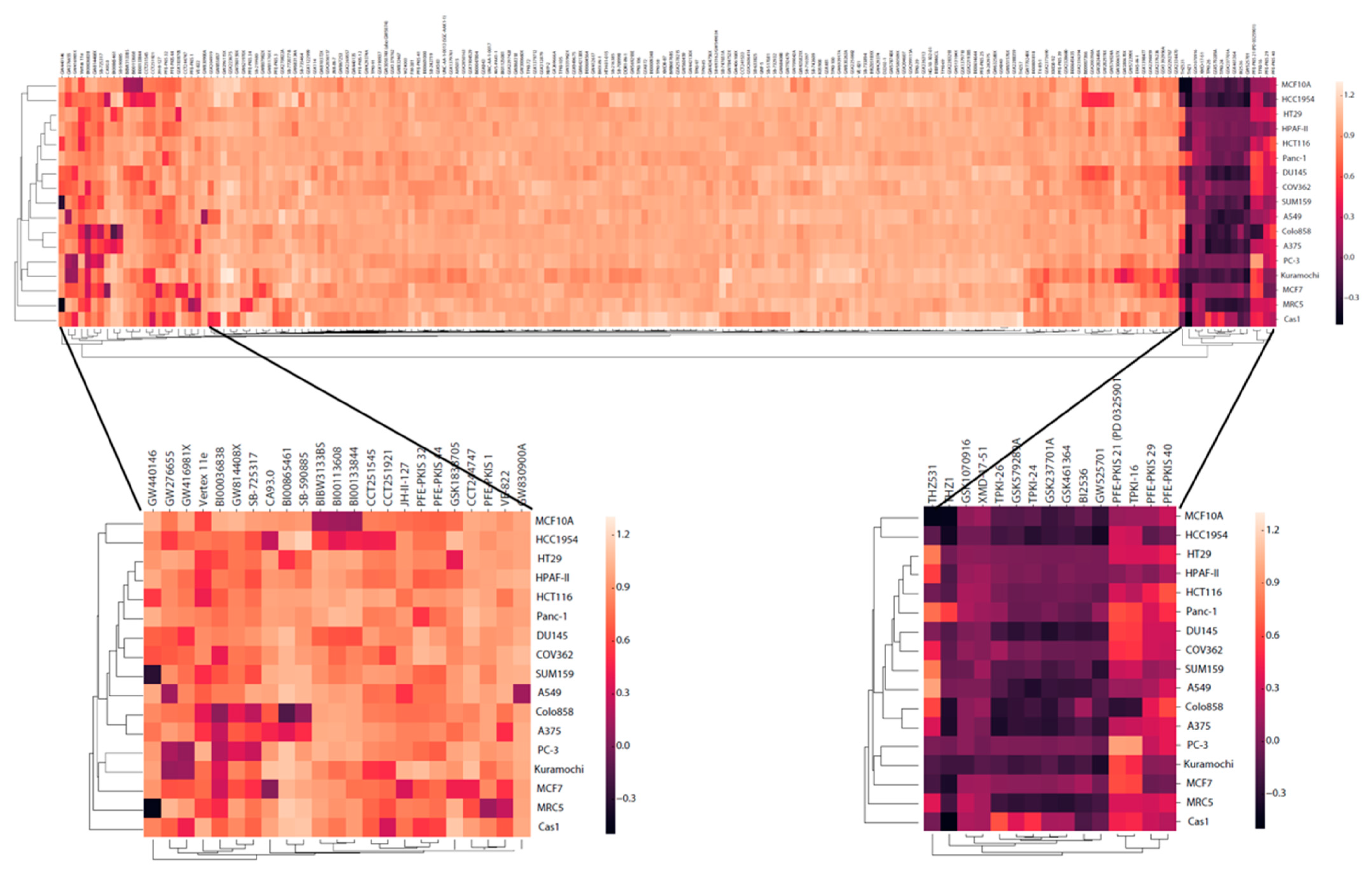
2.6. Cell Toxicity
2.7. Cell Growth
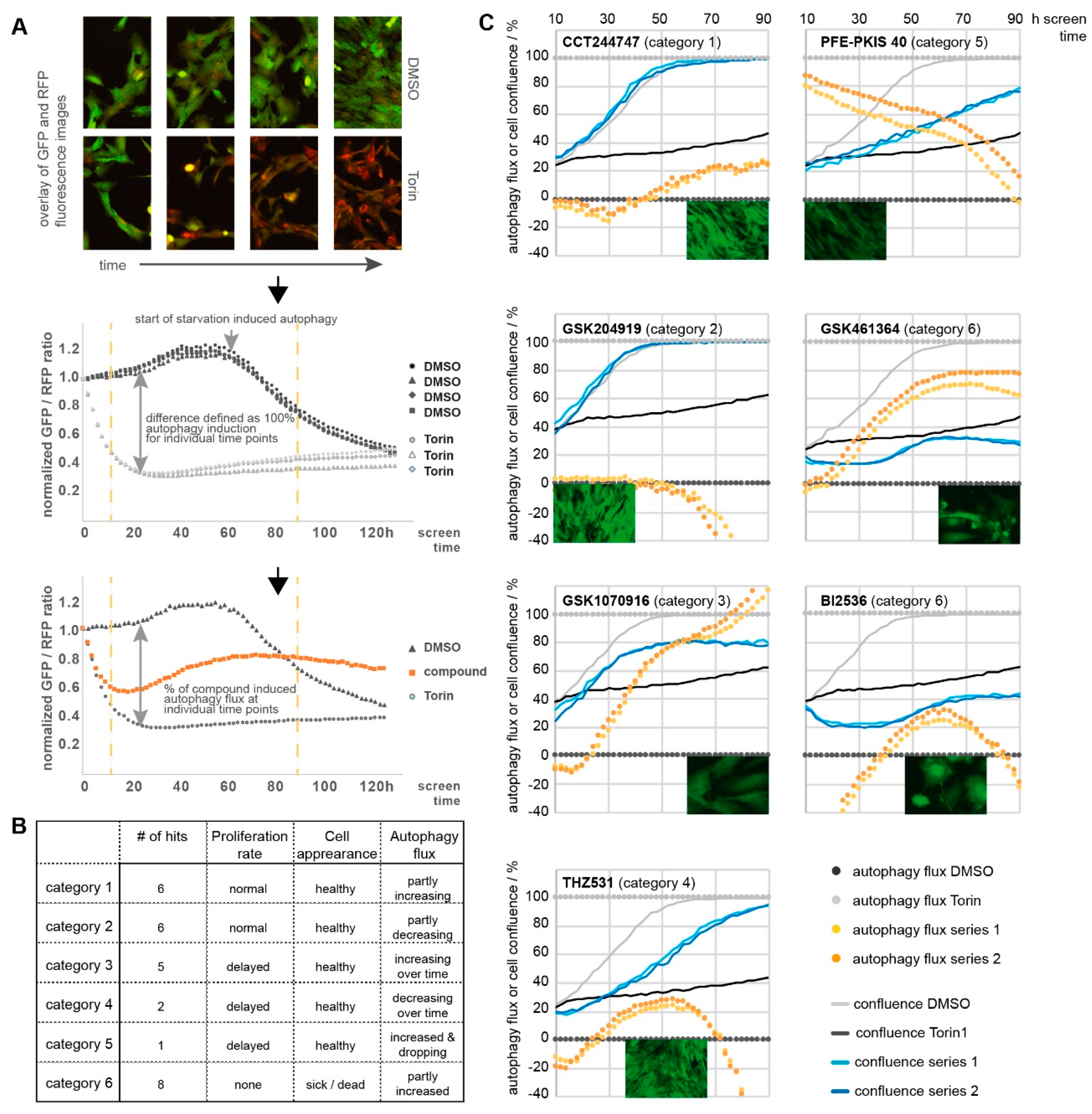
2.8. Kinases Linked to Autophagy
3. Discussion
4. Materials and Methods
4.1. KCGS
4.2. Kinase Assays
4.3. Chemotype Binning
4.4. Cytotoxicity Assays
4.5. Cell Growth Assays
4.6. Autophagy Assays
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roskoski, R., Jr. FDA-Approved Small Molecule Protein Kinase Inhibitors. Available online: http://www.brimr.org/PKI/PKIs.htm (accessed on 1 December 2020).
- Morphy, R. Selectively nonselective kinase inhibition: Striking the right balance. J. Med. Chem. 2010, 53, 1413–1437. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, F.M.; Gray, N.S. Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov. 2018, 17, 353–377. [Google Scholar] [CrossRef] [PubMed]
- Knapp, S.; Arruda, P.; Blagg, J.; Burley, S.; Drewry, D.H.; Edwards, A.; Fabbro, D.; Gillespie, P.; Gray, N.S.; Kuster, B.; et al. A public-private partnership to unlock the untargeted kinome. Nat. Chem. Biol. 2013, 9, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, O.; Muller, S.; Knapp, S. The (un)targeted cancer kinome. Nat. Chem. Biol. 2010, 6, 166–169. [Google Scholar] [CrossRef]
- Edwards, A.M.; Isserlin, R.; Bader, G.D.; Frye, S.V.; Willson, T.M.; Yu, F.H. Too many roads not taken. Nature 2011, 470, 163–165. [Google Scholar] [CrossRef]
- Oprea, T.I.; Bologa, C.G.; Brunak, S.; Campbell, A.; Gan, G.N.; Gaulton, A.; Gomez, S.M.; Guha, R.; Hersey, A.; Holmes, J.; et al. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 2018, 17, 377. [Google Scholar] [CrossRef]
- Klaeger, S.; Heinzlmeir, S.; Wilhelm, M.; Polzer, H.; Vick, B.; Koenig, P.A.; Reinecke, M.; Ruprecht, B.; Petzoldt, S.; Meng, C.; et al. The target landscape of clinical kinase drugs. Science 2017, 358. [Google Scholar] [CrossRef]
- Christmann-Franck, S.; van Westen, G.J.; Papadatos, G.; Beltran Escudie, F.; Roberts, A.; Overington, J.P.; Domine, D. Unprecedently Large-Scale Kinase Inhibitor Set Enabling the Accurate Prediction of Compound-Kinase Activities: A Way toward Selective Promiscuity by Design? J. Chem. Inf. Model. 2016, 56, 1654–1675. [Google Scholar] [CrossRef]
- Arrowsmith, C.H.; Audia, J.E.; Austin, C.; Baell, J.; Bennett, J.; Blagg, J.; Bountra, C.; Brennan, P.E.; Brown, P.J.; Bunnage, M.E.; et al. The promise and peril of chemical probes. Nat. Chem. Biol. 2015, 11, 536–541. [Google Scholar] [CrossRef]
- Jones, L.H.; Bunnage, M.E. Applications of chemogenomic library screening in drug discovery. Nat. Rev. Drug Discov. 2017, 16, 285–296. [Google Scholar] [CrossRef]
- Gautam, P.; Jaiswal, A.; Aittokallio, T.; Al-Ali, H.; Wennerberg, K. Phenotypic Screening Combined with Machine Learning for Efficient Identification of Breast Cancer-Selective Therapeutic Targets. Cell Chem. Biol. 2019, 26, 970–979.e974. [Google Scholar] [CrossRef] [PubMed]
- Bamborough, P.; Drewry, D.; Harper, G.; Smith, G.K.; Schneider, K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J. Med. Chem. 2008, 51, 7898–7914. [Google Scholar] [CrossRef] [PubMed]
- Posy, S.L.; Hermsmeier, M.A.; Vaccaro, W.; Ott, K.H.; Todderud, G.; Lippy, J.S.; Trainor, G.L.; Loughney, D.A.; Johnson, S.R. Trends in Kinase Selectivity: Insights for Target Class-Focused Library Screening. J. Med. Chem. 2011, 54, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Elkins, J.M.; Fedele, V.; Szklarz, M.; Abdul Azeez, K.R.; Salah, E.; Mikolajczyk, J.; Romanov, S.; Sepetov, N.; Huang, X.-P.; Roth, B.L.; et al. Comprehensive characterization of the Published Kinase Inhibitor Set. Nat. Biotech. 2016, 34, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Drewry, D.H.; Wells, C.I.; Andrews, D.M.; Angell, R.; Al-Ali, H.; Axtman, A.D.; Capuzzi, S.J.; Elkins, J.M.; Ettmayer, P.; Frederiksen, M.; et al. Progress towards a public chemogenomic set for protein kinases and a call for contributions. PLoS ONE 2017, 12, e0181585. [Google Scholar] [CrossRef]
- Drewry, D.H.; Wells, C.I.; Zuercher, W.J.; Willson, T.M. A Perspective on Extreme Open Science: Companies Sharing Compounds without Restriction. SLAS Discov. 2019, 24, 505–514. [Google Scholar] [CrossRef]
- Moret, N.; Clark, N.A.; Hafner, M.; Wang, Y.; Lounkine, E.; Medvedovic, M.; Wang, J.; Gray, N.; Jenkins, J.; Sorger, P.K. Cheminformatics Tools for Analyzing and Designing Optimized Small-Molecule Collections and Libraries. Cell Chem. Biol. 2019, 26, 765–777.e763. [Google Scholar] [CrossRef]
- Edwards, A.; Morgan, M.; Al Chawaf, A.; Andrusiak, K.; Charney, R.; Cynader, Z.; ElDessouki, A.; Lee, Y.; Moeser, A.; Stern, S.; et al. A trust approach for sharing research reagents. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- The Structural Genomics Consortium. Available online: www.thesgc.org (accessed on 1 December 2019).
- Fabian, M.A.; Biggs, W.H., 3rd; Treiber, D.K.; Atteridge, C.E.; Azimioara, M.D.; Benedetti, M.G.; Carter, T.A.; Ciceri, P.; Edeen, P.T.; Floyd, M.; et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [Google Scholar] [CrossRef]
- DiscoverX. scanMAX. Available online: https://www.discoverx.com/services/drug-discovery-development-services/kinase-profiling/kinomescan/scanmax (accessed on 1 December 2019).
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Zwick, M.; Kraemer, O.; Carter, A.J. Dataset of the frequency patterns of publications annotated to human protein-coding genes, their protein products and genetic relevance. Data Brief 2019, 25, 104284. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.J.; Kraemer, O.; Zwick, M.; Mueller-Fahrnow, A.; Arrowsmith, C.H.; Edwards, A.M. Target 2035: Probing the human proteome. Drug Discov. Today 2019, 24, 2111–2115. [Google Scholar] [CrossRef] [PubMed]
- DAYLIGHT Chemical Information Systems. Available online: www.daylight.com/dayhtml/doc/theory/theory.smarts.html (accessed on 1 December 2019).
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2019, 144, 19–50. [Google Scholar] [CrossRef]
- Varma, M.V.; Obach, R.S.; Rotter, C.; Miller, H.R.; Chang, G.; Steyn, S.J.; El-Kattan, A.; Troutman, M.D. Physicochemical space for optimum oral bioavailability: Contribution of human intestinal absorption and first-pass elimination. J. Med. Chem. 2010, 53, 1098–1108. [Google Scholar] [CrossRef]
- Montenegro, R.C.; Clark, P.G.; Howarth, A.; Wan, X.; Ceroni, A.; Siejka, P.; Nunez-Alonso, G.A.; Monteiro, O.; Rogers, C.; Gamble, V.; et al. BET inhibition as a new strategy for the treatment of gastric cancer. Oncotarget 2016, 7, 43997–44012. [Google Scholar] [CrossRef]
- Howarth, A.; Schroder, M.; Montenegro, R.C.; Drewry, D.H.; Sailem, H.; Millar, V.; Muller, S.; Ebner, D.V. HighVia—A Flexible Live-Cell High-Content Screening Pipeline to Assess Cellular Toxicity. SLAS Discov. 2020, 25, 801–811. [Google Scholar] [CrossRef]
- Hafner, M.; Niepel, M.; Chung, M.; Sorger, P.K. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat. Methods 2016, 13, 521–527. [Google Scholar] [CrossRef]
- Mills, C.; Gerosa, L. Optimized Experimental and Analytical Tools for Reproducible Drug-Response Studies. Available online: http://lincs.hms.harvard.edu/wordpress/wp-content/uploads/2018/06/DoseResponseNanocourse_2018_Final.pdf (accessed on 1 December 2019).
- Hafner, M.; Niepel, M.; Subramanian, K.; Sorger, P.K. Designing Drug-Response Experiments and Quantifying their Results. Curr. Protoc. Chem. Biol. 2017, 9, 96–116. [Google Scholar] [CrossRef]
- Stolz, A.; Putyrski, M.; Kutle, I.; Huber, J.; Wang, C.; Major, V.; Sidhu, S.S.; Youle, R.J.; Rogov, V.V.; Dotsch, V.; et al. Fluorescence-based ATG8 sensors monitor localization and function of LC3/GABARAP proteins. EMBO J. 2017, 36, 549–564. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Kaizuka, T.; Morishita, H.; Hama, Y.; Tsukamoto, S.; Matsui, T.; Toyota, Y.; Kodama, A.; Ishihara, T.; Mizushima, T.; Mizushima, N. An Autophagic Flux Probe That Releases an Internal Control. Mol. Cell 2016, 64, 835–849. [Google Scholar] [CrossRef] [PubMed]
- Ozfiliz-Kilbas, P.; Sarikaya, B.; Obakan-Yerlikaya, P.; Coker-Gurkan, A.; Arisan, E.D.; Temizci, B.; Palavan-Unsal, N. Cyclin-dependent kinase inhibitors, roscovitine and purvalanol, induce apoptosis and autophagy related to unfolded protein response in HeLa cervical cancer cells. Mol. Biol. Rep. 2018, 45, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.R.; Yang, Z.Z.; Wang, S.J.; Zhang, L.; Luo, J.R.; Feng, Y.; Yu, X.L.; Chen, X.X.; Guo, X.M. The Chk1 inhibitor MK-8776 increases the radiosensitivity of human triple-negative breast cancer by inhibiting autophagy. Acta Pharmacol. Sin. 2017, 38, 513–523. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, F.; Zhou, Z.W.; Xia, H.C.; Wang, X.Y.; Yang, Y.X.; He, Z.X.; Sun, T.; Zhou, S.F. Alisertib induces G2/M arrest, apoptosis, and autophagy via PI3K/Akt/mTOR- and p38 MAPK-mediated pathways in human glioblastoma cells. Am. J. Transl. Res. 2017, 9, 845–873. [Google Scholar] [PubMed]
- Wang, Y.; Zhang, H. Regulation of Autophagy by mTOR Signaling Pathway. Adv. Exp. Med. Biol. 2019, 1206, 67–83. [Google Scholar] [CrossRef]
- Li, Z.Y.; Zhang, X. Kinases Involved in Both Autophagy and Mitosis. Int. J. Mol. Sci. 2017, 18, 1884. [Google Scholar] [CrossRef]
- Vasta, J.D.; Corona, C.R.; Wilkinson, J.; Zimprich, C.A.; Hartnett, J.R.; Ingold, M.R.; Zimmerman, K.; Machleidt, T.; Kirkland, T.A.; Huwiler, K.G.; et al. Quantitative, Wide-Spectrum Kinase Profiling in Live Cells for Assessing the Effect of Cellular ATP on Target Engagement. Cell Chem. Biol. 2018, 25, 206–214.e211. [Google Scholar] [CrossRef]
- Open Babel: The Open Source Chemistry Toolbox. Available online: http://openbabel.org/ (accessed on 1 December 2019).
- MATLAB. Available online: https://www.mathworks.com/products/matlab.html (accessed on 1 December 2019).
- Bhullar, K.S.; Lagaron, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-Targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef]
- Kannaiyan, R.; Mahadevan, D. A comprehensive review of protein kinase inhibitors for cancer therapy. Expert. Rev. Anticancer Ther. 2018, 18, 1249–1270. [Google Scholar] [CrossRef]
- Burdova, K.; Yang, H.B.; Faedda, R.; Hume, S.; Chauhan, J.; Ebner, D.; Kessler, B.M.; Vendrell, I.; Drewry, D.H.; Wells, C.I.; et al. E2F1 proteolysis via SCF-cyclin F underlies synthetic lethality between cyclin F loss and Chk1 inhibition. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Vandoorne, T.; Steyaert, J.; Staats, K.A.; Van Den Bosch, L. The multifaceted role of kinases in amyotrophic lateral sclerosis: Genetic, pathological and therapeutic implications. Brain 2020, 143, 1651–1673. [Google Scholar] [CrossRef] [PubMed]
- Guttuso, T., Jr.; Andrzejewski, K.L.; Lichter, D.G.; Andersen, J.K. Targeting kinases in Parkinson’s disease: A mechanism shared by LRRK2, neurotrophins, exenatide, urate, nilotinib and lithium. J. Neurol. Sci. 2019, 402, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.M.; Cummings, E.; Pantos, C.; Singh, J. Protein kinase C and cardiac dysfunction: A review. Heart Fail. Rev. 2017, 22, 843–859. [Google Scholar] [CrossRef]
| Kinases | Assays | KCGS | % | |
|---|---|---|---|---|
| AGC | 63 | 46 | 20 | 43 |
| Atypical | 34 | 7 | 5 | 71 |
| CAMK | 74 | 58 | 28 | 48 |
| CK1 | 12 | 8 | 3 | 38 |
| CMGC | 64 | 60 | 37 | 62 |
| Lipid | 20 | 13 | 10 | 77 |
| Other | 81 | 51 | 26 | 51 |
| STE | 47 | 42 | 13 | 31 |
| TK | 90 | 81 | 54 | 67 |
| TKL | 43 | 35 | 19 | 54 |
| Total | 215 | 401 | 528 | 54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wells, C.I.; Al-Ali, H.; Andrews, D.M.; Asquith, C.R.M.; Axtman, A.D.; Dikic, I.; Ebner, D.; Ettmayer, P.; Fischer, C.; Frederiksen, M.; et al. The Kinase Chemogenomic Set (KCGS): An Open Science Resource for Kinase Vulnerability Identification. Int. J. Mol. Sci. 2021, 22, 566. https://doi.org/10.3390/ijms22020566
Wells CI, Al-Ali H, Andrews DM, Asquith CRM, Axtman AD, Dikic I, Ebner D, Ettmayer P, Fischer C, Frederiksen M, et al. The Kinase Chemogenomic Set (KCGS): An Open Science Resource for Kinase Vulnerability Identification. International Journal of Molecular Sciences. 2021; 22(2):566. https://doi.org/10.3390/ijms22020566
Chicago/Turabian StyleWells, Carrow I., Hassan Al-Ali, David M. Andrews, Christopher R. M. Asquith, Alison D. Axtman, Ivan Dikic, Daniel Ebner, Peter Ettmayer, Christian Fischer, Mathias Frederiksen, and et al. 2021. "The Kinase Chemogenomic Set (KCGS): An Open Science Resource for Kinase Vulnerability Identification" International Journal of Molecular Sciences 22, no. 2: 566. https://doi.org/10.3390/ijms22020566
APA StyleWells, C. I., Al-Ali, H., Andrews, D. M., Asquith, C. R. M., Axtman, A. D., Dikic, I., Ebner, D., Ettmayer, P., Fischer, C., Frederiksen, M., Futrell, R. E., Gray, N. S., Hatch, S. B., Knapp, S., Lücking, U., Michaelides, M., Mills, C. E., Müller, S., Owen, D., ... Drewry, D. H. (2021). The Kinase Chemogenomic Set (KCGS): An Open Science Resource for Kinase Vulnerability Identification. International Journal of Molecular Sciences, 22(2), 566. https://doi.org/10.3390/ijms22020566







