Differential Marker Expression between Keratinocyte Stem Cells and Their Progeny Generated from a Single Colony
Abstract
1. Introduction
2. Results
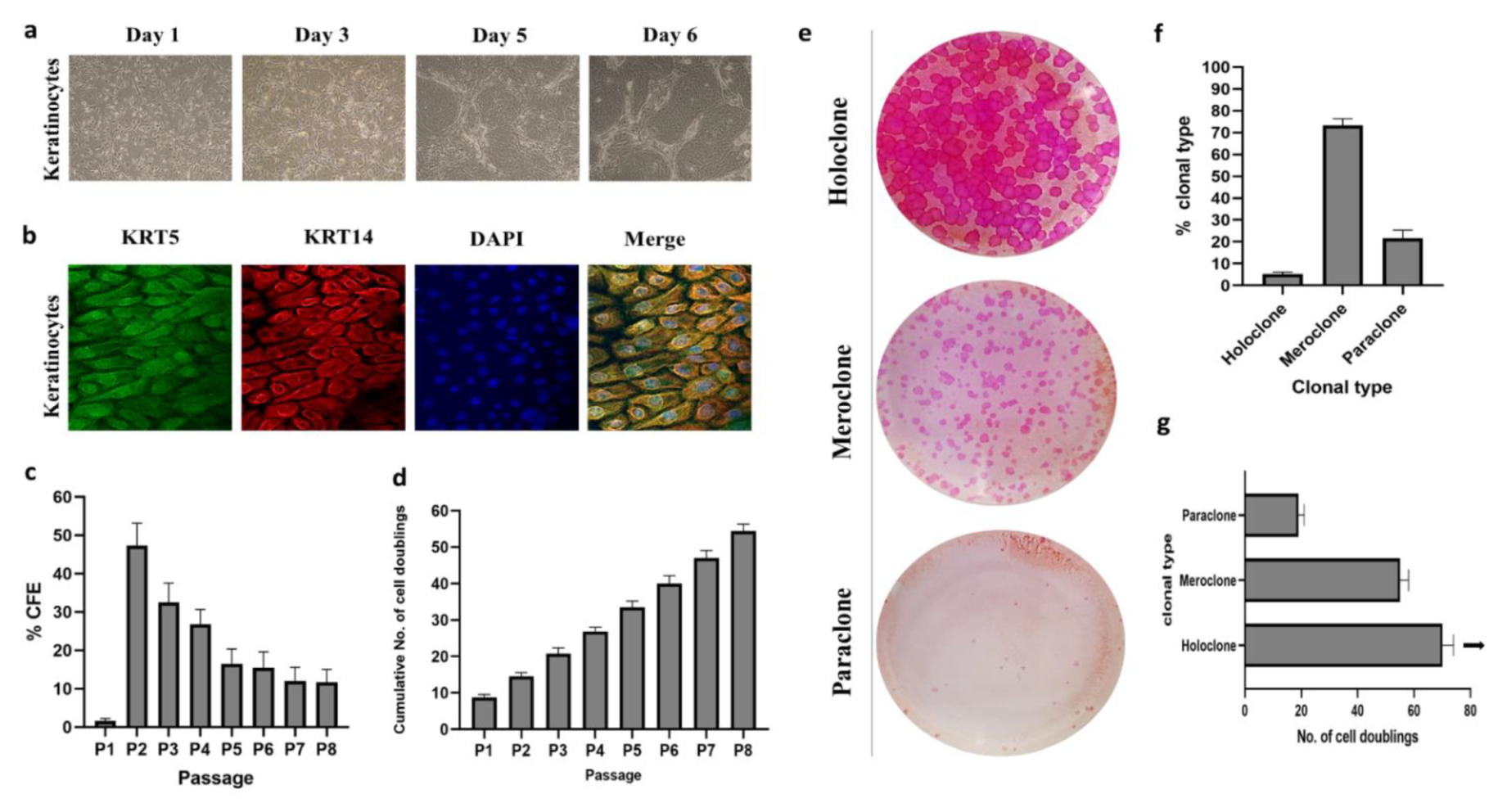
2.1. Isolation and Characterization of Cultured Human Epidermal Keratinocytes
2.2. Clonal Analysis
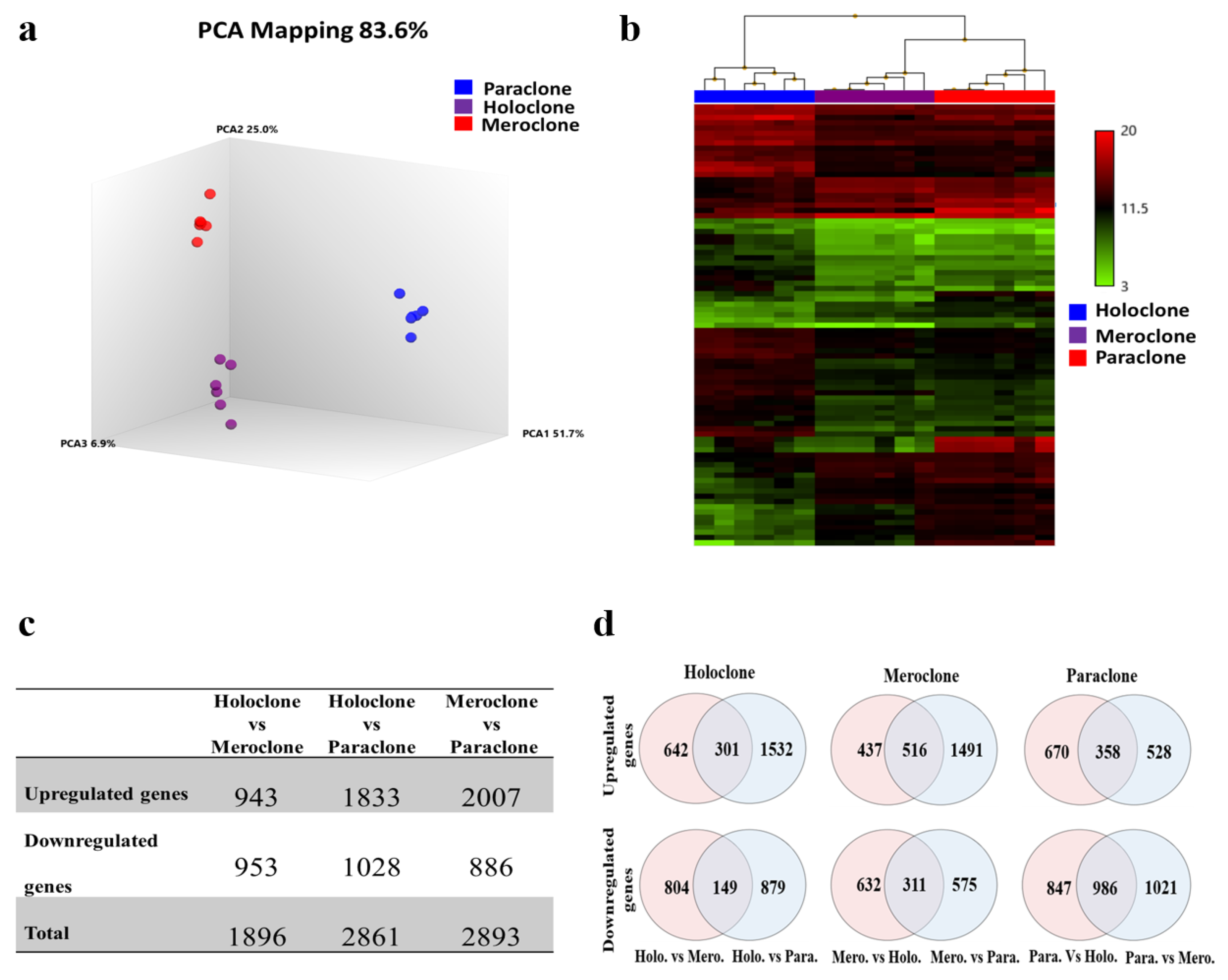
2.3. Identification of Differentially Expressed Genes (DEGs)
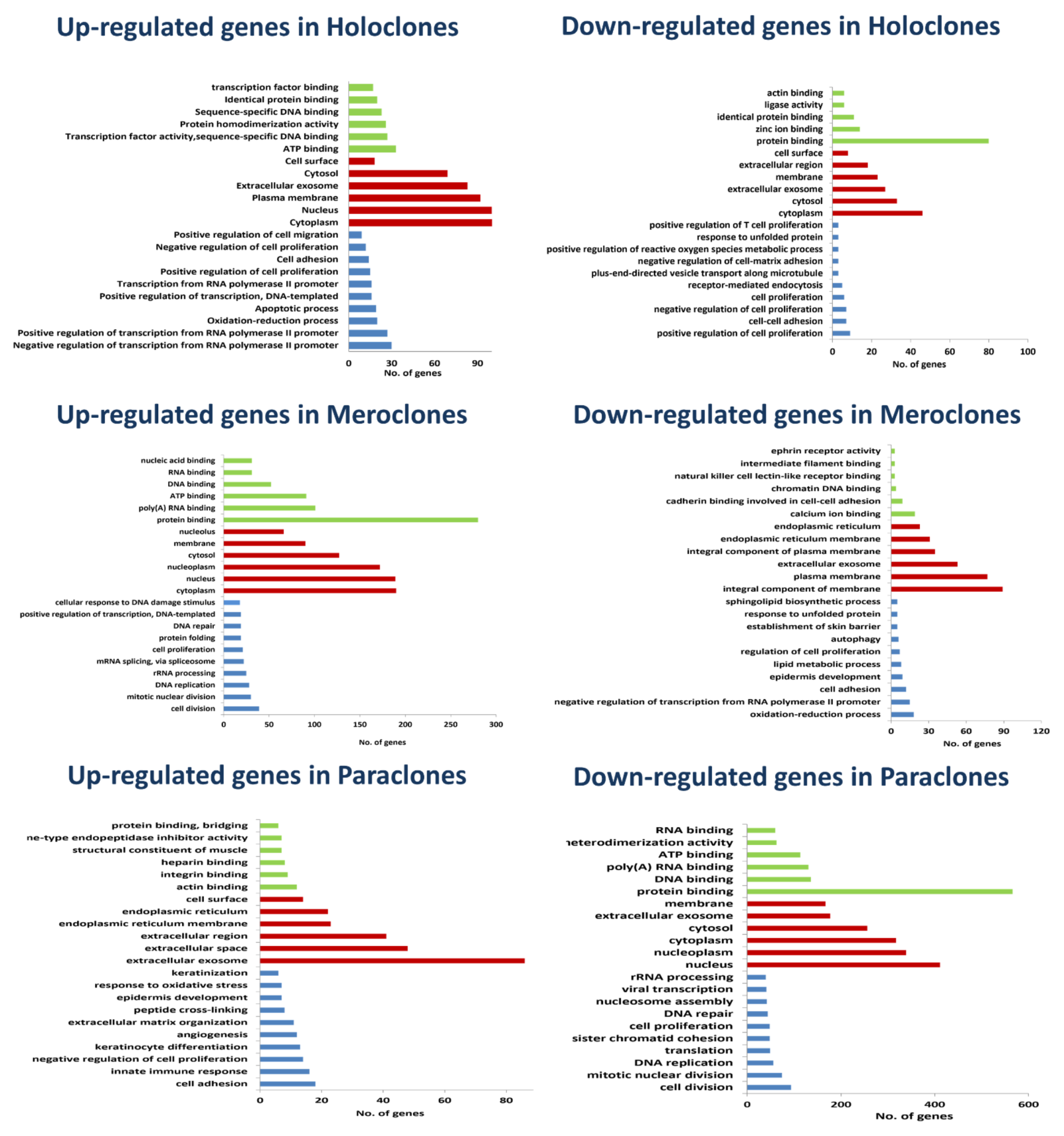
2.4. Signature Genes and Gene Ontology
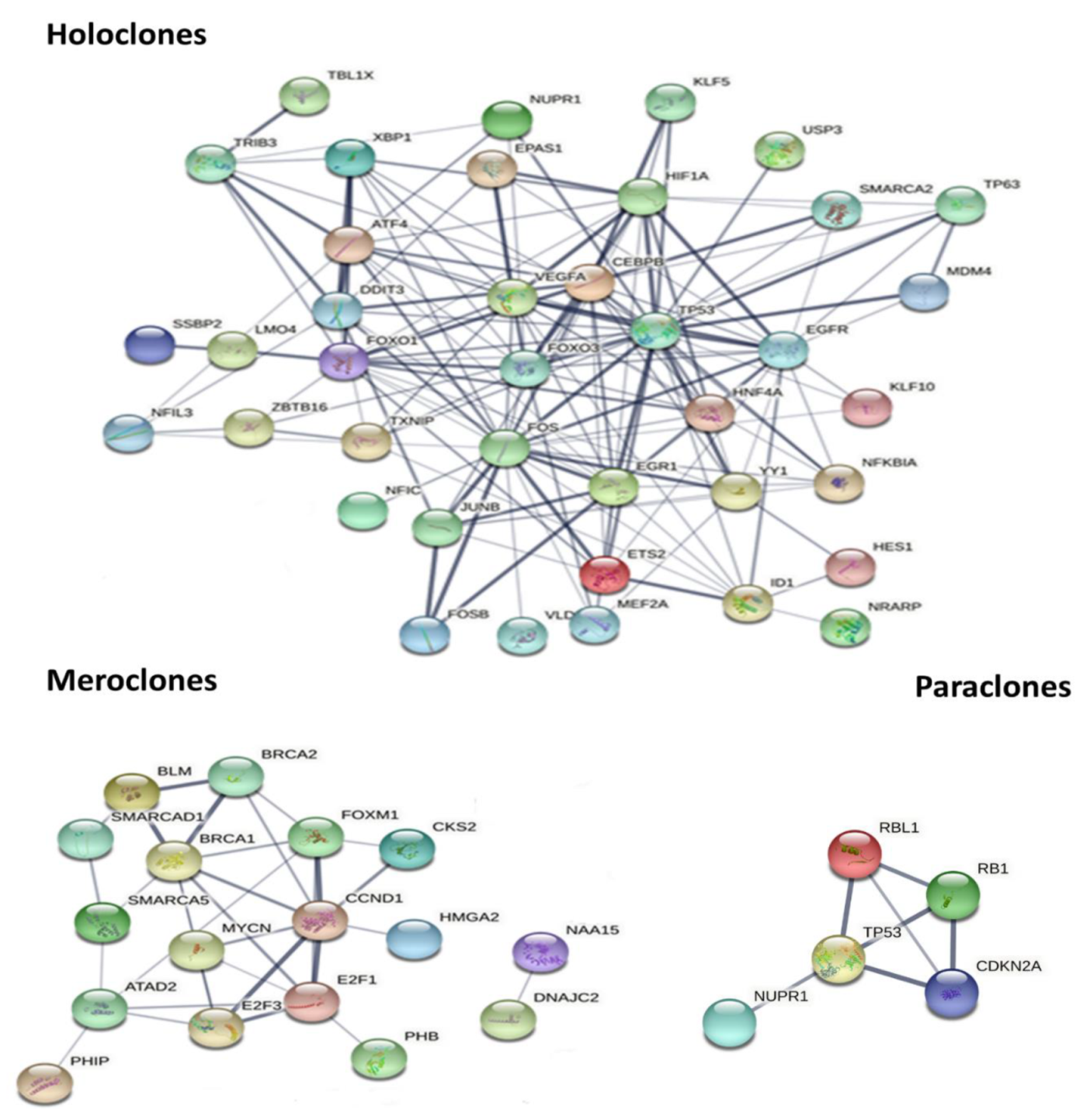
2.5. Signaling Pathway and Upstream Regulator Analysis
3. Discussion
4. Materials and Methods
4.1. Ethical Statement and Informed Consent
4.2. Isolation and Culture of Human Epidermal Keratinocytes (HEKs)
4.3. Characterization of Keratinocyte Cultures
4.3.1. Immunofluorescence Staining
4.3.2. Colony-Forming Efficiency
4.3.3. Proliferation Potential
4.4. Clonal Analysis
4.5. Whole Transcriptome Analysis
4.5.1. RNA Extraction
4.5.2. Global Gene Expression Profiling
4.5.3. Data Analysis
4.6. Validation of the Microarray Results
4.6.1. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.6.2. Flow Cytometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chu, D.H. Overview of biology, development, and structure of skin. In Fitzpatrick’s Dermatology in General Medicine, 7th ed.; Wolff, K., Goldsmith, L.A., Katz, S.I., Gilchrest, B.A., Paller, A.S., Leffell, D.J., Eds.; McGraw-Hill: New York, NY, USA, 2008; pp. 57–73. [Google Scholar]
- Murphy, G.F. Histology of the skin. In Lever’s Histopathology of the Skin, 8th ed.; Elder, R., Elenitsas, C., Jaworsky, B., Johnson, B., Jr., Eds.; Lippincott Williams & Wilkin: Philadelphia, PA, USA, 1997; pp. 5–45. [Google Scholar]
- Cotsarelis, G.; Sun, T.-T.; Lavker, R.M. Label-retaining cells reside in the bulge area of pilosebaceous unit: Implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 1990, 61, 1329–1337. [Google Scholar] [CrossRef]
- Rheinwald, J.G.; Green, H. Serial cultivation of strains of human epidermal keratinocytes: The formation of keratinizing colonies from single cells. Cell 1975, 6, 331–343. [Google Scholar] [CrossRef]
- Barrandon, Y. The epidermal stem cell: An overview. Semin. Dev. Biol. 1993, 4, 209–215. [Google Scholar] [CrossRef]
- Barrandon, Y.; Green, H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 1987, 84, 2302–2306. [Google Scholar] [CrossRef]
- Mathor, M.B.; Ferrari, G.; Dellambra, E.; Cilli, M.; Mavilio, F.; Cancedda, R.; De Luca, M. Clonal analysis of stably transduced human epidermal stem cells in culture. Proc. Natl. Acad. Sci. USA 1996, 93, 10371–10376. [Google Scholar] [CrossRef]
- Nanba, D.; Toki, F.; Matsushita, N.; Matsushita, S.; Higashiyama, S.; Barrandon, Y. Actin filament dynamics impacts keratinocyte stem cell maintenance. EMBO Mol. Med. 2013, 5, 640–653. [Google Scholar] [CrossRef]
- Ojeh, N.; Pastar, I.; Tomic-Canic, M.; Stojadinovic, O. Stem Cells in Skin Regeneration, Wound Healing, and Their Clinical Applications. Int. J. Mol. Sci. 2015, 16, 25476–25501. [Google Scholar] [CrossRef] [PubMed]
- Schreder, A.; Pierard, G.E.; Paquet, P.; Reginster, M.-A.; Pierard-Franchimont, C.; Quatresooz, P. Facing towards Epidermal Stem Cells (Review). Int. J. Mol. Med. 2010, 26, 171–174. [Google Scholar] [CrossRef][Green Version]
- Schlüter, H.; Paquet-Fifield, S.; Gangatirkar, P.; Li, J.; Kaur, P. Functional Characterization of Quiescent Keratinocyte Stem Cells and Their Progeny Reveals a Hierarchical Organization in Human Skin Epidermis. Stem Cells 2011, 29, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Matic, M. A Subpopulation of Human Basal Keratinocytes Has a Low/Negative MHC Class I Expression. Hum. Immunol. 2005, 66, 962–968. [Google Scholar] [CrossRef]
- Larderet, G.; Fortunel, N.O.; Vaigot, P.; Cegalerba, M.; Maltère, P.; Zobiri, O.; Gidrol, X.; Waksman, G.; Martin, M.T. Human Side Population Keratinocytes Exhibit Long-Term Proliferative Potential and a Specific Gene Expression Profile and Can Form a Pluristratified Epidermis. Stem Cells 2006, 24, 965–974. [Google Scholar] [CrossRef]
- Bisson, F.; Rochefort, É.; Lavoie, A.; Larouche, D.; Zaniolo, K.; Simard-Bisson, C.; Damour, O.; Auger, F.A.; Guérin, S.L.; Germain, L. Irradiated Human Dermal Fibroblasts Are as Efficient as Mouse Fibroblasts as a Feeder Layer to Improve Human Epidermal Cell Culture Lifespan. Int. J. Mol. Sci. 2013, 14, 4684–4704. [Google Scholar] [CrossRef] [PubMed]
- Coolen, N.A.; Verkerk, M.; Reijnen, L.; Vlig, M.; Van Den Bogaerdt, A.J.; Breetveld, M.; Gibbs, S.; Middelkoop, E.; Ulrich, M.M. Culture of Keratinocytes for Transplantation without the Need of Feeder Layer Cells. Cell Transplant. 2007, 16, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Kaviani, M.; Geramizadeh, B.; Rahsaz, M.; Marzban, S. Considerations in the Improvement of Human Epidermal Keratinocyte Culture in Vitro. Exp. Clin. Transplant. 2015, 13, 366–370. [Google Scholar]
- Lamb, R.; Ambler, C.A. Keratinocytes Propagated in Serum-Free, Feeder-Free Culture Conditions Fail to Form Stratified Epidermis in a Reconstituted Skin Model. PLoS ONE 2013, 8, e52494. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, G.; Ranno, R.; Stracuzzi, G.; Bondanza, S.; Guerra, L.; Zambruno, G.; Micali, G.; De Luca, M. The Control of Epidermal Stem Cells (Holoclones) in the Treatment of Massive Full-Thickness Burns with Autologous Keratinocytes Cultured on Fibrin1. Transplantation 1999, 68, 868–879. [Google Scholar] [CrossRef]
- Grande, E.; Aparicio, L.A. Stem Cells in Cancer: Should We Believe Or Not? Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Panacchia, L.; Dellambra, E.; Bondanza, S.; Paterna, P.; Maurelli, R.; Paionni, E.; Guerra, L. Nonirradiated Human Fibroblasts and Irradiated 3T3-J2 Murine Fibroblasts as a Feeder Layer for Keratinocyte Growth and Differentiation in Vitro on a Fibrin Substrate. Cells Tissues Organs 2010, 191, 21–35. [Google Scholar] [CrossRef]
- Papini, S.; Cecchetti, D.; Campani, D.; Fitzgerald, W.; Grivel, J.C.; Chen, S.; Margolis, L.; Revoltella, R.P. Isolation and Clonal Analysis of Human Epidermal Keratinocyte Stem Cells in Long-Term Culture. Stem Cells 2003, 21, 481–494. [Google Scholar] [CrossRef]
- Pellegrini, G.; Dellambra, E.; Golisano, O.; Martinelli, E.; Fantozzi, I.; Bondanza, S.; Ponzin, D.; McKeon, F.; De Luca, M. P63 Identifies Keratinocyte Stem Cells. Proc. Natl. Acad. Sci. USA 2001, 98, 3156–3161. [Google Scholar] [CrossRef]
- Legg, J.; Jensen, U.B.; Broad, S.; Leigh, I.; Watt, F.M. Role of Melanoma Chondroitin Sulphate Proteoglycan in Patterning Stem Cells in Human Interfollicular Epidermis. Development 2003, 130, 6049–6063. [Google Scholar] [CrossRef]
- Yang, A.; Schweitzer, R.; Sun, D.; Kaghad, M.; Walker, N.; Bronson, R.T.; Tabin, C.; Sharpe, A.; Caput, D.; Crum, C.; et al. P63 Is Essential for Regenerative Proliferation in Limb, Craniofacial and Epithelial Development. Nature 1999, 398, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Lowell, S.; Jones, P.; Le Roux, I.; Dunne, J.; Watt, F.M. Stimulation of Human Epidermal Differentiation by Delta–Notch Signalling at the Boundaries of Stem-Cell Clusters. Curr. Biol. 2000, 10, 491–500. [Google Scholar] [CrossRef]
- Kretzschmar, K.; Watt, F.M. Markers of Epidermal Stem Cell Subpopulations in Adult Mammalian Skin. Cold Spring Harb. Perspect. Med. 2014, 4, a013631. [Google Scholar] [CrossRef]
- Tan, D.W.M.; Jensen, K.B.; Trotter, M.W.B.; Connelly, J.T.; Broad, S.; Watt, F.M. Single-Cell Gene Expression Profiling Reveals Functional Heterogeneity of Undifferentiated Human Epidermal Cells. Development 2013, 140, 1433–1444. [Google Scholar] [CrossRef]
- Bose, A.; Teh, M.-T.; Mackenzie, I.C.; Waseem, A. Keratin K15 as a Biomarker of Epidermal Stem Cells. Int. J. Mol. Sci. 2013, 14, 19385–19398. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.Z.; Fong, S.; Yue, L.; Zhang, K.; Strachan, L.R.; Scalapino, K.; Mancianti, M.L.; Ghadially, R. The CD44+ ALDH+ Population of Human Keratinocytes Is Enriched for Epidermal Stem Cells with Long-Term Repopulating Ability. Stem Cells 2013, 31, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Drummond, M.L.; Guerrero-Juarez, C.F.; Tarapore, E.; MacLean, A.L.; Stabell, A.R.; Wu, S.C.; Gutierrez, G.; That, B.T.; Benavente, C.A.; et al. Single Cell Transcriptomics of Human Epidermis Identifies Basal Stem Cell Transition States. Nat. Commun. 2020, 11, 4239. [Google Scholar] [CrossRef] [PubMed]
- Rheinwald, J.G.; Green, H. Epidermal Growth Factor and the Multiplication of Cultured Human Epidermal Keratinocytes. Nature 1977, 265, 421–424. [Google Scholar] [CrossRef]
- Chen, J.; Chung, F.; Yang, G.; Pu, M.; Gao, H.; Jiang, W.; Yin, H.; Capka, V.; Kasibhatla, S.; Laffitte, B.; et al. Phosphoglycerate Dehydrogenase Is Dispensable for Breast Tumor Maintenance and Growth. Oncotarget 2013, 4, 2502. [Google Scholar] [CrossRef]
- Ichihara, A.; Greenberg, D.M. Further Studies on the Pathway of Serine Formation from Carbohydrate. J. Biol. Chem. 1957, 224, 331–340. [Google Scholar] [CrossRef]
- Gheller, B.J.; Blum, J.E.; Bender, E.L.; Gheller, M.E.; Lim, E.W.; Handzlik, M.K.; Stover, P.J.; Field, M.S.; Cosgrove, B.D.; Metallo, C.M.; et al. Serine and Glycine Are Essential for Human Muscle Progenitor Cell Population Expansion. bioRxiv 2019, 833798. [Google Scholar] [CrossRef]
- Hwang, I.-Y.; Kwak, S.; Lee, S.; Kim, H.; Lee, S.E.; Kim, J.-H.; Kim, Y.A.; Jeon, Y.K.; Chung, D.H.; Jin, X.; et al. Psat1-Dependent Fluctuations in α-Ketoglutarate Affect the Timing of ESC Differentiation. Cell Metab. 2016, 24, 494–501. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, C.; Gao, K.; Wang, D.; Mao, J.; An, J.; Xu, C.; Wu, D.; Yu, H.; Liu, J.O.; et al. The Ubiquitin Ligase Itch Regulates Apoptosis by Targeting Thioredoxin-Interacting Protein for Ubiquitin-Dependent Degradation. J. Biol. Chem. 2010, 285, 8869–8879. [Google Scholar] [CrossRef]
- Jung, H.; Kim, M.J.; Kim, D.O.; Kim, W.S.; Yoon, S.-J.; Park, Y.-J.; Yoon, S.R.; Kim, T.-D.; Suh, H.-W.; Yun, S.; et al. TXNIP Maintains the Hematopoietic Cell Pool by Switching the Function of P53 under Oxidative Stress. Cell Metab. 2013, 18, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, N.; Wang, J.; Wray, B.; Patel, P.; Yang, W.; Peng, H.; Lavker, R.M. Single-Cell RNA Transcriptome Helps Define the Limbal/Corneal Epithelial Stem/Early Transit Amplifying Cells and How Autophagy Affects This Population. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3570–3583. [Google Scholar] [CrossRef]
- Lou, F.; Sun, Y.; Xu, Z.; Niu, L.; Wang, Z.; Deng, S.; Liu, Z.; Zhou, H.; Bai, J.; Yin, Q.; et al. Excessive Polyamine Generation in Keratinocytes Promotes Self-RNA Sensing by Dendritic Cells in Psoriasis. Immunity 2020, 53, 204–216.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, R.; Zou, K.; Yu, W.; Guo, W.; Gao, Y.; Li, J.; Li, M.; Tai, Y.; Huang, W.; et al. Keratin 23 Promotes Telomerase Reverse Transcriptase Expression and Human Colorectal Cancer Growth. Cell Death Dis. 2017, 8, e2961. [Google Scholar] [CrossRef] [PubMed]
- Denecker, G.; Ovaere, P.; Vandenabeele, P.; Declercq, W. Caspase-14 Reveals Its Secrets. J. Cell Biol. 2008, 180, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Gkegkes, I.D.; Aroni, K.; Agrogiannis, G.; Patsouris, E.S.; Konstantinidou, A.E. Expression of Caspase-14 and Keratin-19 in the Human Epidermis and Appendages during Fetal Skin Development. Arch. Dermatol. Res. 2013, 305, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Handa, T.; Katayama, A.; Yokobori, T.; Yamane, A.; Horiguchi, J.; Kawabata-Iwakawa, R.; Rokudai, S.; Bao, P.; Gombodorj, N.; Altan, B.; et al. Caspase14 Expression Is Associated with Triple Negative Phenotypes and Cancer Stem Cell Marker Expression in Breast Cancer Patients. J. Surg. Oncol. 2017, 116, 706–715. [Google Scholar] [CrossRef]
- Jamora, C.; Fuchs, E. Intercellular Adhesion, Signalling and the Cytoskeleton. Nat. Cell Biol. 2002, 4, e101–e108. [Google Scholar] [CrossRef]
- Vespa, A.; Darmon, A.J.; Turner, C.E.; D’Souza, S.J.A.; Dagnino, L. Ca2+-Dependent Localization of Integrin-Linked Kinase to Cell Junctions in Differentiating Keratinocytes. J. Biol. Chem. 2003, 278, 11528–11535. [Google Scholar] [CrossRef]
- Song, X.; Zhu, C.-H.; Doan, C.; Xie, T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 2002, 296, 1855–1857. [Google Scholar] [CrossRef]
- Pan, L.; Chen, S.; Weng, C.; Call, G.; Zhu, D.; Tang, H.; Zhang, N.; Xie, T. Stem Cell Aging Is Controlled Both Intrinsically and Extrinsically in the Drosophila Ovary. Cell Stem Cell 2007, 1, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Rødningen, O.K.; Børresen-Dale, A.-L.; Alsner, J.; Hastie, T.; Overgaard, J. Radiation-Induced Gene Expression in Human Subcutaneous Fibroblasts Is Predictive of Radiation-Induced Fibrosis. Radiother. Oncol. 2008, 86, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, I.; Ojima, M.; Suzuki, S.; Yamaga, M.; Horie, M.; Koga, H.; Tsuji, K.; Miyaguchi, K.; Ogishima, S.; Tanaka, H.; et al. Human Mesenchymal Stem Cells in Synovial Fluid Increase in the Knee with Degenerated Cartilage and Osteoarthritis. J. Orthop. Res. 2012, 30, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Cu, N.H.; Fierro, F.A.; Forward, K.I.; Nolta, J.; Gordon, L.K.; Telander, D.G. Expression of Epithelial Membrane Protein 2 (EMP2) in Stem Cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5992. [Google Scholar]
- Cao, Z.; Said, N.; Wu, H.K.; Kuwabara, I.; Liu, F.-T.; Panjwani, N. Galectin-7 as a Potential Mediator of Corneal Epithelial Cell Migration. Arch. Ophthalmol. 2003, 121, 82–86. [Google Scholar] [CrossRef]
- Alhattab, D.; Jamali, F.; Ali, D.; Hammad, H.; Adwan, S.; Rahmeh, R.; Samarah, O.; Salah, B.; Hamdan, M.; Awidi, A. An Insight into the Whole Transcriptome Profile of Four Tissue-Specific Human Mesenchymal Stem Cells. Regen. Med. 2019, 14, 841–865. [Google Scholar] [CrossRef]
- La Rocca, G.; Anzalone, R.; Farina, F. The Expression of CD68 in Human Umbilical Cord Mesenchymal Stem Cells: New Evidences of Presence in Non-Myeloid Cell Types. Scand. J. Immunol. 2009, 70, 161–162. [Google Scholar] [CrossRef]
- Mii, S.; Murakumo, Y.; Asai, N.; Jijiwa, M.; Hagiwara, S.; Kato, T.; Asai, M.; Enomoto, A.; Ushida, K.; Sobue, S.; et al. Epidermal Hyperplasia and Appendage Abnormalities in Mice Lacking CD109. Am. J. Pathol. 2012, 181, 1180–1189. [Google Scholar] [CrossRef]
- Liu, T.; Jiang, L.; Tavana, O.; Gu, W. The Deubiquitylase OTUB1 Mediates Ferroptosis via Stabilization of SLC7A11. Cancer Res. 2019, 79, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, X.; Wei, C.; Zheng, D.; Lu, X.; Yang, Y.; Luo, A.; Zhang, K.; Duan, X.; Wang, Y. Targeting SLC7A11 Specifically Suppresses the Progression of Colorectal Cancer Stem Cells via Inducing Ferroptosis. Eur. J. Pharm. Sci. 2020, 152, 105450. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Morris, R.J.; Kaur, P. Enrichment for Murine Keratinocyte Stem Cells Based on Cell Surface Phenotype. Proc. Natl. Acad. Sci. USA 2000, 97, 10960–10965. [Google Scholar] [CrossRef]
- Freije, A.; Molinuevo, R.; Ceballos, L.; Cagigas, M.; Alonso-Lecue, P.; Rodriguez, R.; Menendez, P.; Aberdam, D.; De Diego, E.; Gandarillas, A. Inactivation of P53 in Human Keratinocytes Leads to Squamous Differentiation and Shedding via Replication Stress and Mitotic Slippage. Cell Rep. 2014, 9, 1349–1360. [Google Scholar] [CrossRef]
- Mills, A.A.; Zheng, B.; Wang, X.-J.; Vogel, O.H.; Roop, D.R.; Bradley, A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999, 398, 708–713. [Google Scholar] [CrossRef]
- Nair, R.P.; Krishnan, L.K. Identification of P63+ Keratinocyte Progenitor Cells in Circulation and Their Matrix-Directed Differentiation to Epithelial Cells. Stem Cell Res. Ther. 2013, 4, 38. [Google Scholar] [CrossRef]
- Senoo, M.; Pinto, F.; Crum, C.P.; McKeon, F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 2007, 129, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.L.; Adhikary, G.; Young, C.A.; Jans, R.; Crish, J.F.; Xu, W.; Rorke, E.A. AP1 Transcription Factors in Epidermal Differentiation and Skin Cancer. J. Ski. Cancer 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 in Cell Proliferation and Survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef]
- Paik, J.-H.; Ding, Z.; Narurkar, R.; Ramkissoon, S.; Muller, F.; Kamoun, W.S.; Chae, S.-S.; Zheng, H.; Ying, H.; Mahoney, J.; et al. FoxOs Cooperatively Regulate Diverse Pathways Governing Neural Stem Cell Homeostasis. Cell Stem Cell 2009, 5, 540–553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yalcin, S.; Lee, D.-F.; Yeh, T.-Y.J.; Lee, S.-M.; Su, J.; Mungamuisk, S.K.; Rimmelé, P.; Kennedy, M.; Sellers, R.; et al. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat. Cell Biol. 2011, 13, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Araki, K.Y.; Naka, K.; Arai, F.; Takubo, K.; Yamazaki, S.; Matsuoka, S.; Miyamoto, T.; Ito, K.; Ohmura, M.; et al. Foxo3a Is Essential for Maintenance of the Hematopoietic Stem Cell Pool. Cell Stem Cell 2007, 1, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Nakae, J.; Kitamura, T.; Kitamura, Y.; Biggs, W.H.; Arden, K.C.; Accili, D. The Forkhead Transcription Factor Foxo1 Regulates Adipocyte Differentiation. Dev. Cell 2003, 4, 119–129. [Google Scholar] [CrossRef]
- Kitamura, T.; Kitamura, Y.I.; Funahashi, Y.; Shawber, C.J.; Castrillon, D.H.; Kollipara, R.; DePinho, R.; Kitajewski, J.; Accili, D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Investig. 2007, 117, 2477–2485. [Google Scholar] [CrossRef] [PubMed]
- Ponugoti, B.; Xu, F.; Zhang, C.; Tian, C.; Pacios, S.; Graves, D.T. FOXO1 Promotes Wound Healing through the Up-Regulation of TGF-Β1 and Prevention of Oxidative Stress. J. Cell Biol. 2013, 203, 327–343. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Trinh, T.; Aljoufi, A.; Broxmeyer, H.E. Hypoxia Signaling Pathway in Stem Cell Regulation: Good and Evil. Curr. Stem Cell Rep. 2018, 4, 149–157. [Google Scholar] [CrossRef] [PubMed]
| Gene Symbol | Description | Holo vs. Mero FC | Holo vs. Para FC |
|---|---|---|---|
| Holoclone Upregulated Genes | |||
| CASP14 | caspase 14 | 50.69 | 3.85 |
| ASS1 | argininosuccinate synthase 1 | 25.53 | 6.51 |
| PSAT1 | phosphoserine aminotransferase 1 | 21.47 | 14.64 |
| H19; MIR675 | H19, imprinted maternally expressed transcript (non-protein coding); microRNA 675 | 21.11 | 32.67 |
| SLC7A11 | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 | 16.75 | 19.82 |
| ASNS | asparagine synthetase (glutamine-hydrolyzing) | 10.58 | 6.26 |
| LGALS7; LGALS7B | lectin, galactoside-binding, soluble, 7; lectin, galactoside-binding, soluble, 7B | 9.64 | 2.11 |
| IMPA2 | inositol(myo)-1(or 4)-monophosphatase 2 | 9.57 | 5.86 |
| TXNIP | thioredoxin interacting protein | 9.32 | 1.89 |
| CHAC1 | ChaC glutathione-specific gamma-glutamylcyclotransferase 1 | 9.19 | 3.03 |
| Holoclone Downregulated Genes | |||
| PTHLH | parathyroid hormone-like hormone | −9.81 | −4.33 |
| DCBLD2 | discoidin, CUB and LCCL domain containing 2 | −9.18 | −5.71 |
| THBS1 | thrombospondin 1 | −8.71 | −6.22 |
| DKK1 | dickkopf WNT signaling pathway inhibitor 1 | −8.52 | −4.72 |
| KRTAP2-3 | keratin associated protein 2–3 | −6.28 | −50.55 |
| TSPAN1 | tetraspanin 1 | −5.6 | −6.74 |
| PLAT | plasminogen activator, tissue | −5.41 | −9.5 |
| FAP | fibroblast activation protein alpha | −5.32 | −2.45 |
| ENC1 | ectodermal-neural cortex 1 (with BTB domain) | −4.87 | −7.82 |
| ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 | −4.8 | −12.59 |
| Meroclone Upregulated Genes | |||
| FST | follistatin | 6.81 | 5.42 |
| RNU6-26P | RNA, U6 small nuclear 26, pseudogene | 3.95 | 1.89 |
| TFRC | transferrin receptor | 3.88 | 2.8 |
| SMURF2 | SMAD specific E3 ubiquitin protein ligase 2 | 3.73 | 2.14 |
| ADAMTS6 | ADAM metallopeptidase with thrombospondin type 1 motif 6 | 3.64 | 2.24 |
| HMGA2 | high mobility group AT-hook 2 | 3.48 | 3.28 |
| CENPE | centromere protein E | 3.09 | 6.5 |
| C20orf197 | chromosome 20 open reading frame 197 | 3.08 | 2.8 |
| CLSPN | claspin | 3.02 | 4.74 |
| UTP20 | UTP20 small subunit (SSU) processome component | 2.98 | 2.53 |
| Meroclone Downregulated Genes | |||
| CASP14 | caspase 14 | −50.69 | −13.15 |
| ALOX12B | arachidonate 12-lipoxygenase, 12R type | −13.52 | −26.39 |
| ATP12A | ATPase, H+/K+ transporting, nongastric, alpha polypeptide | −10.69 | −18.9 |
| C10orf99 | chromosome 10 open reading frame 99 | −10.26 | −6.47 |
| LGALS7; LGALS7B | lectin, galactoside-binding, soluble, 7; lectin, galactoside-binding, soluble, 7B | −9.64 | −4.58 |
| TXNIP | thioredoxin interacting protein | −9.32 | −4.93 |
| CHAC1 | ChaC glutathione-specific gamma-glutamylcyclotransferase 1 | −9.19 | −3.04 |
| LGALS7B | lectin, galactoside-binding, soluble, 7B | −8.7 | −4.14 |
| SCNN1B | sodium channel, non voltage gated 1 beta subunit | −7.77 | −8.19 |
| CDHR1 | cadherin-related family member 1 | −7.51 | −4.02 |
| Paraclone Upregulated Genes | |||
| FLG | filaggrin | 269.82 | 86.31 |
| KRT1 | keratin 1, type II | 156.48 | 36.68 |
| LY6G6C | lymphocyte antigen 6 complex, locus G6C | 41.26 | 15.44 |
| CRNN | cornulin | 24.83 | 23.29 |
| SERPINB12 | serpin peptidase inhibitor, clade B (ovalbumin), member 12 | 22.69 | 6.67 |
| THEM5 | thioesterase superfamily member 5 | 19.34 | 9.05 |
| KRT10 | keratin 10, type I | 18.75 | 9.74 |
| CEACAM5 | carcinoembryonic antigen-related cell adhesion molecule 5 | 13.64 | 3.58 |
| PTPRM | protein tyrosine phosphatase, receptor type, M | 10.45 | 4.36 |
| MIR3687-1 | microRNA 3687-1 | 9.93 | 6.15 |
| Paraclone Downregulated Genes | |||
| CCNA2 | cyclin A2 | −20.09 | −13.53 |
| MKI67 | marker of proliferation Ki-67 | −17.62 | −13.92 |
| KIF11 | kinesin family member 11 | −15.14 | −8.72 |
| DLGAP5 | discs, large (Drosophila) homolog-associated protein 5 | −14.97 | −8.49 |
| HIST1H3B | histone cluster 1, H3b | −14.48 | −12.24 |
| NUSAP1 | nucleolar and spindle associated protein 1 | −14.14 | −11.9 |
| TOP2A | topoisomerase (DNA) II alpha | −13.94 | −10.46 |
| CENPF | centromere protein F | −12.41 | −9.44 |
| NCAPG | non-SMC condensin I complex subunit G | −10.75 | −5.57 |
| HIST1H3I | histone cluster 1, H3i | −10.32 | −9.18 |
| Gene Symbol | Description | Holo vs. Mero FC | Holo vs. Para FC | Colony F-Test |
|---|---|---|---|---|
| Holoclones | ||||
| SLC7A11 | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 | 16.7 | 19.8 | 3.55 × 10−7 |
| CEL | carboxyl ester lipase | 2.5 | 6 | 0.0052 |
| VEGFA | vascular endothelial growth factor A | 4 | 4 | 5.64 × 10−6 |
| LGALS1 | lectin, galactoside-binding, soluble, 1 | 3.3 | 10 | 1.39 × 10−5 |
| CD68 | CD68 molecule | 3 | 3.8 | 4.10 × 10−3 |
| THBD | thrombomodulin | 3.2 | 1.5 | 0.001 |
| SLC3A2 | solute carrier family 3 (amino acid transporter heavy chain), member 2 | 2.9 | 2.2 | 0.0004 |
| CD109 | CD109 molecule | 2.11 | 2.3 | 0.048 |
| MSLN | mesothelin | 2.5 | 4 | 0.0234 |
| PLA2R1 | phospholipase A2 receptor 1 | 2 | 1.5 | 0.0112 |
| EMP2 | epithelial membrane protein 2 | 2.2 | 2.4 | 1.39 × 10−5 |
| WNT4 | wingless-type MMTV integration site family, member 4 | 1.8 | 1.9 | 2.99 × 10−5 |
| ADAM17 | ADAM metallopeptidase domain 17 | 2 | 1.7 | 0.0003 |
| SFRP1 | secreted frizzled-related protein 1 | 1.69 | 2.1 | 7.75 × 10−5 |
| FGFR3 | fibroblast growth factor receptor 3 | 2.2 | 2.6 | 0.0011 |
| EGFR | epidermal growth factor receptor | 1.6 | 1.7 | 0.0052 |
| SLC1A4 | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 2.4 | 1.73 | 0.0102 |
| BACE1 | beta-site APP-cleaving enzyme 1 | 2.8 | 1.5 | 3.00 × 10−5 |
| CD59; C11orf91 | CD59 molecule, complement regulatory protein; chromosome 11 open reading frame 91 | −1.5 | −1.7 | 0.0078 |
| PLAT | plasminogen activator, tissue | −3.8 | −6 | 0.0004 |
| AREG | amphiregulin | −2.2 | −2 | 0.001 |
| TFRC | transferrin receptor | −3.9 | −1.77 | 0.0048 |
| SLIT2 | slit guidance ligand 2 | −2.8 | −1.6 | 0.0018 |
| FAP | fibroblast activation protein alpha | −5.3 | −2.5 | 0.0039 |
| THBS1 | thrombospondin 1 | −8.7 | −6.2 | 0.0002 |
| DCBLD2 | discoidin, CUB and LCCL domain containing 2 | −9.2 | −5.7 | 8.94 × 10−5 |
| Paraclones | ||||
| TNFSF18 | tumor necrosis factor (ligand) superfamily, member 18 | 1.57 | 1.55 | 0.0005 |
| PROM2 | prominin 2 | 3.62 | 3.13 | 0.0154 |
| ICAM1 | intercellular adhesion molecule 1 | 1.68 | 1.52 | 0.0394 |
| HLA-H | major histocompatibility complex, class I, H (pseudogene) | 1.67 | 2.14 | 0.0059 |
| HLA-C | major histocompatibility complex, class I, C | 2.2 | 2.63 | 0.0049 |
| HLA-B | major histocompatibility complex, class I, B | 1.88 | 2.28 | 0.0185 |
| HLA-A | major histocompatibility complex, class I, A | 2.41 | 3.8 | 0.0005 |
| CPM | carboxypeptidase M | 1.97 | 4.58 | 0.0035 |
| CLU | clusterin | 1.62 | 1.52 | 0.013 |
| CEP295NL; TIMP2 | CEP295 N-terminal like; TIMP metallopeptidase inhibitor 2 | 1.65 | 1.64 | 0.0333 |
| CD36 | CD36 molecule (thrombospondin receptor) | 1.85 | 1.93 | 0.0237 |
| ANXA9 | annexin A9 | 1.77 | 3.2 | 0.031 |
| AMOT; MIR4329 | angiomotin; microRNA 4329 | 3.31 | 2.81 | 0.0006 |
| ACE2 | angiotensin I converting enzyme 2 | 1.5 | 1.55 | 0.0123 |
| Upstream Regulator | Predicted Activation State | Activation Z-Score |
|---|---|---|
| Holoclone vs. Meroclone | ||
| MYC | Inhibited | −2.5 |
| ATF4 | Activated | 3.9 |
| TP53 | Activated | 3.6 |
| TP63 | Activated | 1.7 |
| HNF4A | Activated | 2.7 |
| NUPR1 | Activated | 6 |
| HIF1A | Activated | 2.8 |
| EPAS1 | Activated | 2 |
| CCND1 | Inhibited | −2.7 |
| Meroclone vs. Paraclone | ||
| E2F1 | Activated | 4.8 |
| TP53 | Inhibited | −5.5 |
| MYC | Activated | 8.5 |
| CCND1 | Activated | 3.8 |
| E2F3 | Activated | 6.2 |
| TBX2 | Activated | 4.8 |
| NUPR1 | Inhibited | −9.5 |
| CDKN2A | Inhibited | −7.8 |
| RB1 | Inhibited | −3.8 |
| FOXM1 | Activated | 5.8 |
| E2F2 | Activated | 3 |
| RBL1 | Inhibited | −4.5 |
| MAX | Activated | 2.4 |
| Holoclone vs. Paraclone | ||
| TP53 | Inhibited | −4.7 |
| MYC | Activated | 7.6 |
| E2F1 | Activated | 4.8 |
| CDKN2A | Inhibited | −7.1 |
| RB1 | Inhibited | −4.1 |
| NUPR1 | Inhibited | −6.5 |
| CCND1 | Activated | 3.4 |
| TBX2 | Activated | 4.3 |
| E2F3 | Activated | 4.7 |
| FOXM1 | Activated | 5.5 |
| RBL1 | Inhibited | −4.9 |
| EP400 | Activated | 4.8 |
| MYCN | Activated | 6.7 |
| E2F2 | Activated | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, D.; Alhattab, D.; Jafar, H.; Alzubide, M.; Sharar, N.; Bdour, S.; Awidi, A. Differential Marker Expression between Keratinocyte Stem Cells and Their Progeny Generated from a Single Colony. Int. J. Mol. Sci. 2021, 22, 10810. https://doi.org/10.3390/ijms221910810
Ali D, Alhattab D, Jafar H, Alzubide M, Sharar N, Bdour S, Awidi A. Differential Marker Expression between Keratinocyte Stem Cells and Their Progeny Generated from a Single Colony. International Journal of Molecular Sciences. 2021; 22(19):10810. https://doi.org/10.3390/ijms221910810
Chicago/Turabian StyleAli, Dema, Dana Alhattab, Hanan Jafar, Malak Alzubide, Nour Sharar, Salwa Bdour, and Abdalla Awidi. 2021. "Differential Marker Expression between Keratinocyte Stem Cells and Their Progeny Generated from a Single Colony" International Journal of Molecular Sciences 22, no. 19: 10810. https://doi.org/10.3390/ijms221910810
APA StyleAli, D., Alhattab, D., Jafar, H., Alzubide, M., Sharar, N., Bdour, S., & Awidi, A. (2021). Differential Marker Expression between Keratinocyte Stem Cells and Their Progeny Generated from a Single Colony. International Journal of Molecular Sciences, 22(19), 10810. https://doi.org/10.3390/ijms221910810





