Genomic Changes Driven by Radiation-Induced DNA Damage and Microgravity in Human Cells
Abstract
1. Introduction
2. Space Radiation Environment
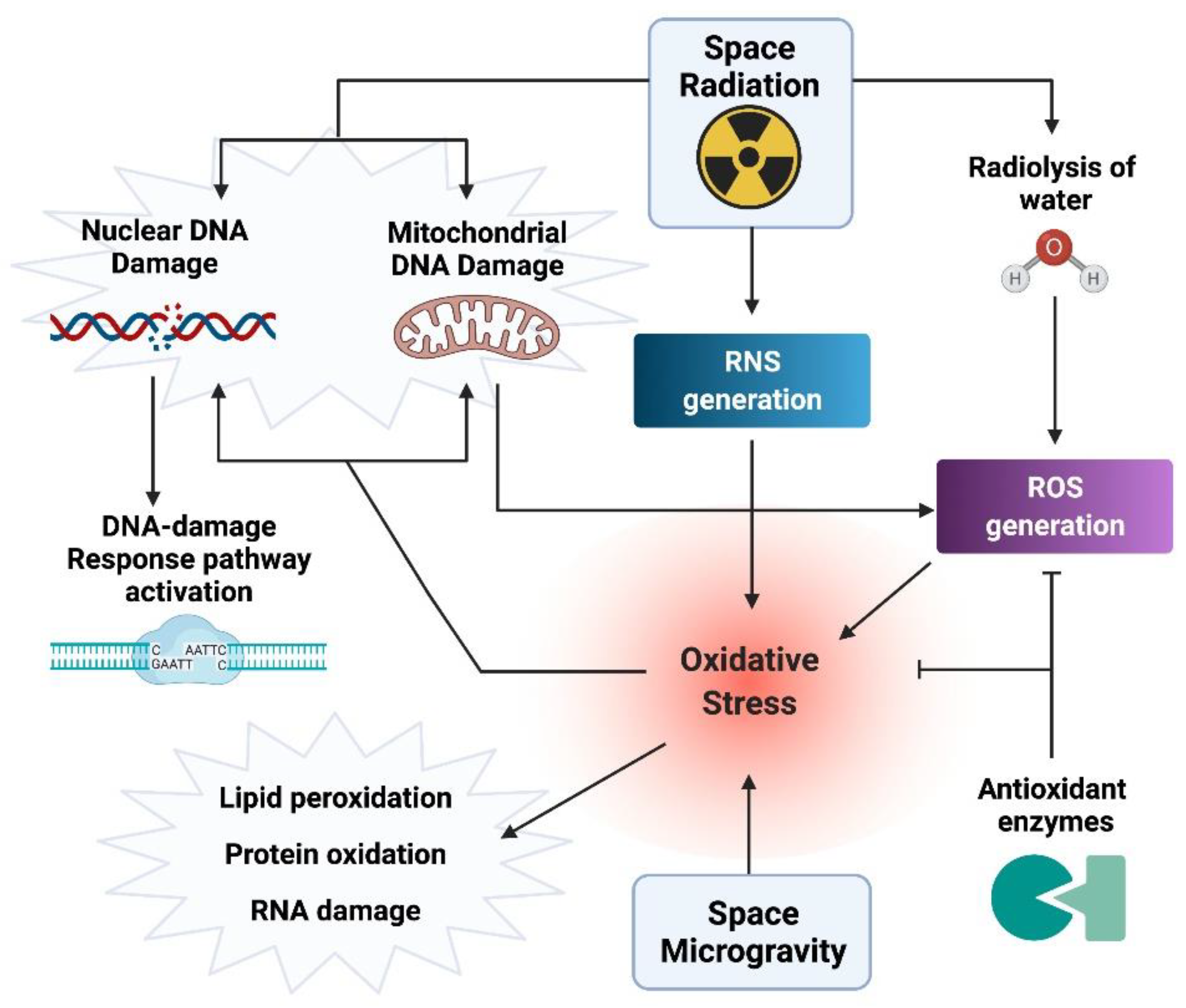
3. Oxidative Stress and Damage (OsaD) Generation in the Space Environment
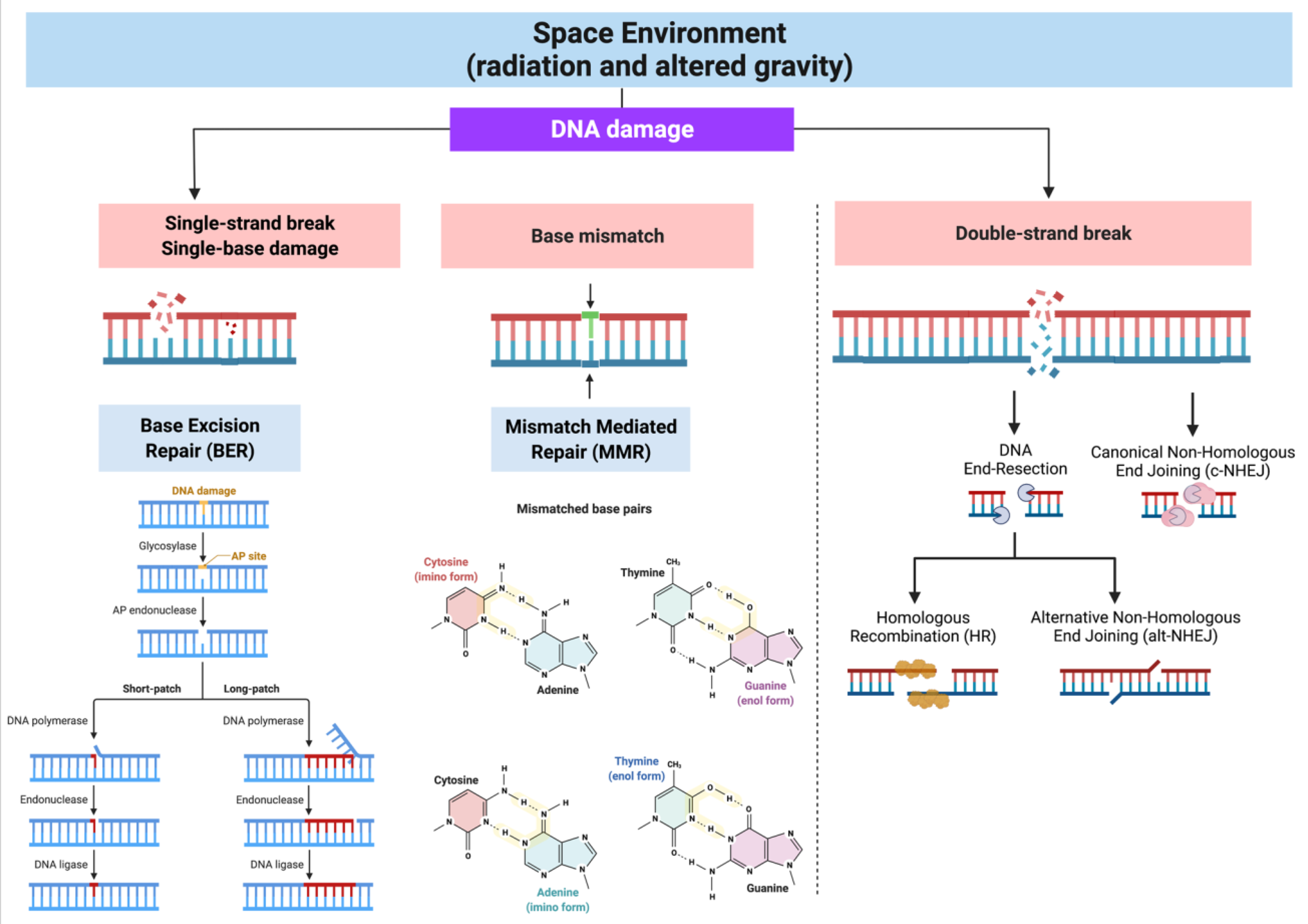
Repair of DNA Damage Originated in the Space Environment
4. Genetic and Epigenetic Changes in Space-Flown Human Cells
4.1. Genetic Polymorphism
4.2. Chromatin Structure
4.3. DNA Methylation and Histone PTMs
4.4. Non-Coding RNAs (ncRNAs)
4.5. General Transcriptomic Changes
5. Genomic Alterations in Human Cells Cultured in Simulated Space Conditions
5.1. Nucleotide Structure Variation
5.2. Chromatin and Histone PTMs Changes
5.3. DNA Methylation Alterations
5.4. Non-Coding RNAs and Gene Expression Changes
6. Genetic and Epigenetic Changes Associated with Radiation Carcinogenesis Risk
7. Countermeasures Development
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Longnecker, D.E.; Manning, F.J.; Worth, M.H., Jr. (Eds.) Review of NASA’s Longitudinal Study of Astronaut. Health; National Academies Press: Washington, DC, USA, 2004. [Google Scholar]
- Afshinnekoo, E.; Scott, R.T.; MacKay, M.J.; Pariset, E.; Cekanaviciute, E.; Barker, R.; Gilroy, S.; Hassane, D.; Smith, S.M.; Zwart, S.R.; et al. Fundamental Biological Features of Spaceflight: Advancing the Field to Enable Deep-Space Exploration. Cell 2020, 183, 1162–1184. [Google Scholar] [CrossRef]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science 2019, 364. [Google Scholar] [CrossRef]
- Luxton, J.J.; McKenna, M.J.; Lewis, A.; Taylor, L.E.; George, K.A.; Dixit, S.M.; Moniz, M.; Benegas, W.; Mackay, M.J.; Mozsary, C.; et al. Telomere Length Dynamics and DNA Damage Responses Associated with Long-Duration Spaceflight. Cell Rep. 2020, 33, 108457. [Google Scholar] [CrossRef]
- Durante, M.; Cucinotta, F.A. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer 2008, 8, 465–472. [Google Scholar] [CrossRef]
- Human Radiosensitivity; Public Health England: London, UK, 2013.
- Palumbo, E.; Piotto, C.; Calura, E.; Fasanaro, E.; Groff, E.; Busato, F.; El Khouzai, B.; Rigo, M.; Baggio, L.; Romualdi, C.; et al. Individual Radiosensitivity in Oncological Patients: Linking Adverse Normal Tissue Reactions and Genetic Features. Front. Oncol. 2019, 9, 987. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; Kim, M.H.; Willingham, V.; George, K.A. Physical and biological organ dosimetry analysis for international space station astronauts. Radiat. Res. 2008, 170, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Malkani, S.; Chin, C.R.; Cekanaviciute, E.; Mortreux, M.; Okinula, H.; Tarbier, M.; Schreurs, A.S.; Shirazi-Fard, Y.; Tahimic, C.G.T.; Rodriguez, D.N.; et al. Circulating miRNA Spaceflight Signature Reveals Targets for Countermeasure Development. Cell Rep. 2020, 33, 108448. [Google Scholar] [CrossRef] [PubMed]
- Benton, E.R.; Benton, E.V. Space radiation dosimetry in low-Earth orbit and beyond. Nucl. Instrum. Methods Phys. Res. B 2001, 184, 255–294. [Google Scholar] [CrossRef]
- Furukawa, S.; Nagamatsu, A.; Nenoi, M.; Fujimori, A.; Kakinuma, S.; Katsube, T.; Wang, B.; Tsuruoka, C.; Shirai, T.; Nakamura, A.J.; et al. Space Radiation Biology for “Living in Space”. Biomed. Res. Int. 2020, 2020, 4703286. [Google Scholar] [CrossRef]
- Sato, T.; Nagamatsu, A.; Ueno, H.; Kataoka, R.; Miyake, S.; Takeda, K.; Niita, K. Comparison of Cosmic-Ray Environments on Earth, Moon, Mars and in Spacecraft Using Phits. Radiat Prot. Dosim. 2018, 180, 146–149. [Google Scholar] [CrossRef]
- Simonsen, L.C.; Slaba, T.C.; Guida, P.; Rusek, A. NASA’s first ground-based Galactic Cosmic Ray Simulator: Enabling a new era in space radiobiology research. PLoS Biol. 2020, 18, e3000669. [Google Scholar] [CrossRef]
- Borak, T.B.; Heilbronn, L.H.; Krumland, N.; Weil, M.M. Design and dosimetry of a facility to study health effects following exposures to fission neutrons at low dose rates for long durations. Int. J. Radiat. Biol. 2021, 97, 1063–1076. [Google Scholar] [CrossRef]
- Nelson, G.A. Space Radiation and Human Exposures, A Primer. Radiat. Res. 2016, 185, 349–358. [Google Scholar] [CrossRef]
- Norbury, J.W.; Slaba, T.C. Space radiation accelerator experiments—The role of neutrons and light ions. Life Sci. Space Res. 2014, 3, 90–94. [Google Scholar] [CrossRef]
- Ikeda, H.; Souda, H.; Puspitasari, A.; Held, K.D.; Hidema, J.; Nikawa, T.; Yoshida, Y.; Kanai, T.; Takahashi, A. A New System for Three-dimensional Clinostat Synchronized X-irradiation with a High-speed Shutter for Space Radiation Research. Biol. Sci. Space 2016, 30, 8–16. [Google Scholar] [CrossRef]
- Ikeda, H.; Souda, H.; Puspitasari, A.; Held, K.D.; Hidema, J.; Nikawa, T.; Yoshida, Y.; Kanai, T.; Takahashi, A. Development and performance evaluation of a three-dimensional clinostat synchronized heavy-ion irradiation system. Life Sci. Space Res. 2017, 12, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Yamanouchi, S.; Takeuchi, K.; Takahashi, S.; Tashiro, M.; Hidema, J.; Higashitani, A.; Adachi, T.; Zhang, S.; Guirguis, F.N.L.; et al. Combined Environment Simulator for Low-Dose-Rate Radiation and Partial Gravity of Moon and Mars. Life 2020, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Rusin, M.; Ghobrial, N.; Takacs, E.; Willey, J.S.; Dean, D. Changes in ionizing radiation dose rate affect cell cycle progression in adipose derived stem cells. PLoS ONE 2021, 16, e0250160. [Google Scholar] [CrossRef] [PubMed]
- McBride, W.H.; Schaue, D. Radiation-induced tissue damage and response. J. Pathol. 2020, 250, 647–655. [Google Scholar] [CrossRef]
- Corcoran, N.M.; Clarkson, M.J.; Stuchbery, R.; Hovens, C.M. Molecular Pathways: Targeting DNA Repair Pathway Defects Enriched in Metastasis. Clin. Cancer Res. 2016, 22, 3132–3137. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Shiloh, Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef]
- Marteijn, J.A.; Vermeulen, W.; Tresini, M. Noncanonical ATM Activation and Signaling in Response to Transcription-Blocking DNA Damage. Methods Mol. Biol. 2017, 1599, 347–361. [Google Scholar]
- Zhang, X.; Wan, G.; Berger, F.G.; He, X.; Lu, X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol. Cell 2011, 41, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Azzam, E.I.; Jay-Gerin, J.P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef]
- Goodwin, T.J.; Christofidou-Solomidou, M. Oxidative Stress and Space Biology: An Organ-Based Approach. Int. J. Mol. Sci. 2018, 19, 959. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, R.B.; Wardman, P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene 2003, 22, 5734–5754. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Sorrell, M.; Berman, Z. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol. Life Sci. 2014, 71, 3951–3967. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kozlov, S.; Lavin, M.F.; Person, M.D.; Paull, T.T. ATM activation by oxidative stress. Science 2010, 330, 517–521. [Google Scholar] [CrossRef]
- da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Stear, B.; et al. Comprehensive Multi-omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 2020, 183, 1185–1201.e20. [Google Scholar] [CrossRef]
- Barnes, D.E.; Lindahl, T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004, 38, 445–476. [Google Scholar] [CrossRef]
- Izumi, T.; Wiederhold, L.R.; Roy, G.; Roy, R.; Jaiswal, A.; Bhakat, K.K.; Mitra, S.; Hazra, T.K. Mammalian DNA base excision repair proteins: Their interactions and role in repair of oxidative DNA damage. Toxicology 2003, 193, 43–65. [Google Scholar] [CrossRef]
- Weidinger, A.; Kozlov, A.V. Biological Activities of Reactive Oxygen and Nitrogen Species: Oxidative Stress versus Signal Transduction. Biomolecules 2015, 5, 472–484. [Google Scholar] [CrossRef]
- Grifalconi, M.; Celotti, L.; Mognato, M. Bystander response in human lymphoblastoid TK6 cells. Mutat. Res. 2007, 625, 102–111. [Google Scholar] [CrossRef]
- Shcherbik, N.; Pestov, D.G. The Impact of Oxidative Stress on Ribosomes: From Injury to Regulation. Cells 2019, 8, 1379. [Google Scholar] [CrossRef] [PubMed]
- Autsavapromporn, N.; de Toledo, S.M.; Little, J.B.; Jay-Gerin, J.P.; Harris, A.L.; Azzam, E.I. The role of gap junction communication and oxidative stress in the propagation of toxic effects among high-dose alpha-particle-irradiated human cells. Radiat. Res. 2011, 175, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Bezdan, D.; Grigorev, K.; Meydan, C.; Pelissier Vatter, F.A.; Cioffi, M.; Rao, V.; MacKay, M.; Nakahira, K.; Burnham, P.; Afshinnekoo, E.; et al. Cell-free DNA (cfDNA) and Exosome Profiling from a Year-Long Human Spaceflight Reveals Circulating Biomarkers. iScience 2020, 23, 101844. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.P. Space flight and oxidative stress. Nutrition 2002, 18, 867–871. [Google Scholar] [CrossRef]
- Markin, A.; Strogonova, L.; Balashov, O.; Polyakov, V.; Tigner, T. The dynamics of blood biochemical parameters in cosmonauts during long-term space flights. Acta Astronaut. 1998, 42, 247–253. [Google Scholar] [CrossRef]
- Luxton, J.J.; McKenna, M.J.; Taylor, L.E.; George, K.A.; Zwart, S.R.; Crucian, B.E.; Drel, V.R.; Garrett-Bakelman, F.E.; Mackay, M.J.; Butler, D.; et al. Temporal Telomere and DNA Damage Responses in the Space Radiation Environment. Cell Rep. 2020, 33, 108435. [Google Scholar] [CrossRef]
- Tauber, S.; Christoffel, S.; Thiel, C.S.; Ullrich, O. Transcriptional Homeostasis of Oxidative Stress-Related Pathways in Altered Gravity. Int. J. Mol. Sci. 2018, 19, 2814. [Google Scholar] [CrossRef]
- Overbey, E.G.; da Silveira, W.A.; Stanbouly, S.; Nishiyama, N.C.; Roque-Torres, G.D.; Pecaut, M.J.; Zawieja, D.C.; Wang, C.; Willey, J.S.; Delp, M.D.; et al. Spaceflight influences gene expression, photoreceptor integrity, and oxidative stress-related damage in the murine retina. Sci. Rep. 2019, 9, 13304. [Google Scholar] [CrossRef]
- Ohnishi, T.; Ohnishi, K.; Takahashi, A.; Taniguchi, Y.; Sato, M.; Nakano, T.; Nagaoka, S. Detection of DNA damage induced by space radiation in Mir and space shuttle. J. Radiat. Res. 2002, 43, S133–S136. [Google Scholar] [CrossRef]
- Lu, T.; Zhang, Y.; Wong, M.; Feiveson, A.; Gaza, R.; Stoffle, N.; Wang, H.; Wilson, B.; Rohde, L.; Stodieck, L.; et al. Detection of DNA damage by space radiation in human fibroblasts flown on the International Space Station. Life Sci. Space Res. 2017, 12, 24–31. [Google Scholar] [CrossRef]
- Ohnishi, T.; Takahashi, A.; Nagamatsu, A.; Omori, K.; Suzuki, H.; Shimazu, T.; Ishioka, N. Detection of space radiation-induced double strand breaks as a track in cell nucleus. Biochem. Biophys. Res. Commun. 2009, 390, 485–488. [Google Scholar] [CrossRef]
- Deriano, L.; Roth, D.B. Modernizing the nonhomologous end-joining repertoire: Alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013, 47, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Karanam, K.; Kafri, R.; Loewer, A.; Lahav, G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell 2012, 47, 320–329. [Google Scholar] [CrossRef] [PubMed]
- David, S.S.; O’Shea, V.L.; Kundu, S. Base-excision repair of oxidative DNA damage. Nature 2007, 447, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Maher, R.L.; Prasad, A.; Rizvanova, O.; Wallace, S.S.; Pederson, D.S. Contribution of DNA unwrapping from histone octamers to the repair of oxidatively damaged DNA in nucleosomes. DNA Repair 2013, 12, 964–971. [Google Scholar] [CrossRef]
- Mazurek, A.; Berardini, M.; Fishel, R. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J. Biol. Chem. 2002, 277, 8260–8266. [Google Scholar] [CrossRef] [PubMed]
- Neri, S.; Guidotti, S.; Bini, C.; Pelotti, S.; D’Adamo, S.; Minguzzi, M.; Platano, D.; Santi, S.; Mariani, E.; Cattini, L.; et al. Oxidative stress-induced DNA damage and repair in primary human osteoarthritis chondrocytes: Focus on IKKalpha and the DNA Mismatch Repair System. Free Radic. Biol. Med. 2021, 166, 212–225. [Google Scholar] [CrossRef]
- Roychoudhury, S.; Pramanik, S.; Harris, H.L.; Tarpley, M.; Sarkar, A.; Spagnol, G.; Sorgen, P.L.; Chowdhury, D.; Band, V.; Klinkebiel, D.; et al. Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome. Proc. Natl. Acad. Sci. USA 2020, 117, 11409–11420. [Google Scholar] [CrossRef]
- Bucker, H.; Horneck, G.; Reitz, G.; Graul, E.H.; Berger, H.; Hoffken, H.; Ruther, W.; Heinrich, W.; Beaujean, R. Embryogenesis and organogenesis of Carausius morosus under spaceflight conditions. Naturwissenschaften 1986, 73, 433–434. [Google Scholar] [CrossRef]
- Ikenaga, M.; Yoshikawa, I.; Kojo, M.; Ayaki, T.; Ryo, H.; Ishizaki, K.; Kato, T.; Yamamoto, H.; Hara, R. Mutations induced in Drosophila during space flight. Biol. Sci. Space 1997, 11, 346–350. [Google Scholar] [CrossRef]
- Horneck, G.; Rettberg, P.; Kozubek, S.; Baumstark-Khan, C.; Rink, H.; Schafer, M.; Schmitz, C. The influence of microgravity on repair of radiation-induced DNA damage in bacteria and human fibroblasts. Radiat. Res. 1997, 147, 376–384. [Google Scholar] [CrossRef]
- Pross, H.D.; Kiefer, J. Repair of cellular radiation damage in space under microgravity conditions. Radiat. Environ. Biophys. 1999, 38, 133–138. [Google Scholar] [CrossRef]
- Takahashi, A.; Ohnishi, K.; Takahashi, S.; Masukawa, M.; Sekikawa, K.; Amano, T.; Nakano, T.; Nagaoka, S.; Ohnishi, T. The effects of microgravity on ligase activity in the repair of DNA double-strand breaks. Int. J. Radiat. Biol. 2000, 76, 783–788. [Google Scholar] [CrossRef]
- Takahashi, A.; Ohnishi, K.; Takahashi, S.; Masukawa, M.; Sekikawa, K.; Amano, T.; Nakano, T.; Nagaoka, S.; Ohnishi, T. The effects of microgravity on induced mutation in Escherichia coli and Saccharomyces cerevisiae. Adv. Space Res. 2001, 28, 555–561. [Google Scholar] [CrossRef]
- Horneck, G. Impact of microgravity on radiobiological processes and efficiency of DNA repair. Mutat. Res. 1999, 430, 221–228. [Google Scholar] [CrossRef]
- Carvalho, G.; Repoles, B.M.; Mendes, I.; Wanrooij, P.H. Mitochondrial DNA Instability in Mammalian Cells. Antioxid. Redox Signal. 2021. [Google Scholar] [CrossRef] [PubMed]
- Fontana, G.A.; Gahlon, H.L. Mechanisms of replication and repair in mitochondrial DNA deletion formation. Nucleic Acids Res. 2020, 48, 11244–11258. [Google Scholar] [CrossRef]
- Guo, J.; Han, N.; Zhang, Y.; Wang, H.; Zhang, X.; Su, L.; Liu, C.; Li, J.; Chen, C.; Liu, C. Use of genome sequencing to assess nucleotide structure variation of Staphylococcus aureus strains cultured in spaceflight on Shenzhou-X, under simulated microgravity and on the ground. Microbiol. Res. 2015, 170, 61–68. [Google Scholar] [CrossRef]
- Urbaniak, C.; Lorenzi, H.; Thissen, J.; Jaing, C.; Crucian, B.; Sams, C.; Pierson, D.; Venkateswaran, K.; Mehta, S. The influence of spaceflight on the astronaut salivary microbiome and the search for a microbiome biomarker for viral reactivation. Microbiome 2020, 8, 56. [Google Scholar] [CrossRef]
- Wang, X.; Abraham, S.; McKenzie, J.A.G.; Jeffs, N.; Swire, M.; Tripathi, V.B.; Luhmann, U.F.O.; Lange, C.A.K.; Zhai, Z.; Arthur, H.M.; et al. LRG1 promotes angiogenesis by modulating endothelial TGF-beta signalling. Nature 2013, 499, 306–311. [Google Scholar] [CrossRef]
- Zwart, S.R.; Gregory, J.F.; Zeisel, S.H.; Gibson, C.R.; Mader, T.H.; Kinchen, J.M.; Ueland, P.M.; Ploutz-Snyder, R.; Heer, M.A.; Smith, S.M. Genotype, B-vitamin status, and androgens affect spaceflight-induced ophthalmic changes. FASEB J. 2016, 30, 141–148. [Google Scholar] [CrossRef]
- Mognato, M.; Burdak-Rothkamm, S.; Rothkamm, K. Interplay between DNA replication stress, chromatin dynamics and DNA-damage response for the maintenance of genome stability. Mutat. Res. 2021, 787, 108346. [Google Scholar] [CrossRef]
- Wang, N.; Tytell, J.D.; Ingber, D.E. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. [Google Scholar] [CrossRef]
- Vassy, J.; Portet, S.; Beil, M.; Millot, G.; Fauvel-Lafeve, F.; Gasset, G.; Schoevaert, D. Weightlessness acts on human breast cancer cell line MCF-7. Adv. Space Res. 2003, 32, 1595–1603. [Google Scholar] [CrossRef]
- Durante, M.; Snigiryova, G.; Akaeva, E.; Bogomazova, A.; Druzhinin, S.; Fedorenko, B.; Greco, O.; Novitskaya, N.; Rubanovich, A.; Shevchenko, V.; et al. Chromosome aberration dosimetry in cosmonauts after single or multiple space flights. Cytogenet Genome Res. 2003, 103, 40–46. [Google Scholar] [CrossRef]
- George, K.; Durante, M.; Cucinotta, F.A. Chromosome aberrations in astronauts. Adv. Space Res. 2007, 40, 483–490. [Google Scholar] [CrossRef][Green Version]
- George, K.; Durante, M.; Wu, H.; Willingham, V.; Badhwar, G.; Cucinotta, F.A. Chromosome aberrations in the blood lymphocytes of astronauts after space flight. Radiat. Res. 2001, 156, 731–738. [Google Scholar] [CrossRef]
- George, K.; Willingham, V.; Cucinotta, F.A. Stability of chromosome aberrations in the blood lymphocytes of astronauts measured after space flight by FISH chromosome painting. Radiat. Res. 2005, 164 Pt 2, 474–480. [Google Scholar] [CrossRef]
- George, K.; Rhone, J.; Beitman, A.; Cucinotta, F.A. Cytogenetic damage in the blood lymphocytes of astronauts: Effects of repeat long-duration space missions. Mutat. Res. 2013, 756, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Feiveson, A.; George, K.; Shavers, M.; Moreno-Villanueva, M.; Zhang, Y.; Babiak-Vazquez, A.; Crucian, B.; Semones, E.; Wu, H. Predicting chromosome damage in astronauts participating in international space station missions. Sci. Rep. 2021, 11, 5293. [Google Scholar] [CrossRef]
- Alpen, E.L.; Powers-Risius, P.; Curtis, S.B.; DeGuzman, R. Tumorigenic potential of high-Z, high-LET charged-particle radiations. Radiat. Res. 1993, 136, 382–391. [Google Scholar] [CrossRef]
- Bonassi, S.; Norppa, H.; Ceppi, M.; Stromberg, U.; Vermeulen, R.; Znaor, A.; Cebulska-Wasilewska, A.; Fabianova, E.; Fucic, A.; Gundy, S.; et al. Chromosomal aberration frequency in lymphocytes predicts the risk of cancer: Results from a pooled cohort study of 22 358 subjects in 11 countries. Carcinogenesis 2008, 29, 1178–1183. [Google Scholar] [CrossRef] [PubMed]
- Hada, M.; Ikeda, H.; Rhone, J.R.; Beitman, A.J.; Plante, I.; Souda, H.; Yoshida, Y.; Held, K.D.; Fujiwara, K.; Saganti, P.B.; et al. Increased Chromosome Aberrations in Cells Exposed Simultaneously to Simulated Microgravity and Radiation. Int. J. Mol. Sci. 2018, 20, 43. [Google Scholar] [CrossRef]
- Yamanouchi, S.; Adachi, T.; Yoshida, Y.; Rhone, J.R.; Mao, J.H.; Fujiwara, K.; Saganti, P.B.; Takahashi, A.; Hada, M. The combined effect of simulated microgravity and radiation on chromosome aberrations in human peripheral blood lymphocytes. Biol. Sci. Space 2021, 35, 15–23. [Google Scholar] [CrossRef]
- Yamanouchi, S.; Rhone, J.; Mao, J.H.; Fujiwara, K.; Saganti, P.B.; Takahashi, A.; Hada, M. Simultaneous Exposure of Cultured Human Lymphoblastic Cells to Simulated Microgravity and Radiation Increases Chromosome Aberrations. Life 2020, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Gambacurta, A.; Merlini, G.; Ruggiero, C.; Diedenhofen, G.; Battista, N.; Bari, M.; Balsamo, M.; Piccirillo, S.; Valentini, G.; Mascetti, G.; et al. Human osteogenic differentiation in Space: Proteomic and epigenetic clues to better understand osteoporosis. Sci. Rep. 2019, 9, 8343. [Google Scholar] [CrossRef]
- Tauber, S.; Hauschild, S.; Crescio, C.; Secchi, C.; Paulsen, K.; Pantaleo, A.; Saba, A.; Buttron, I.; Thiel, C.S.; Cogoli, A.; et al. Signal transduction in primary human T lymphocytes in altered gravity—Results of the MASER-12 suborbital space flight mission. Cell Commun. Signal. 2013, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Stanbouly, S.; Nishiyama, N.C.; Chen, X.; Delp, M.D.; Qiu, H.; Mao, X.W.; Wang, C. Spaceflight decelerates the epigenetic clock orchestrated with a global alteration in DNA methylome and transcriptome in the mouse retina. Precis Clin. Med. 2021, 4, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr Bioinf. 2019, 16. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Mognato, M.; Celotti, L. MicroRNAs Used in Combination with Anti-Cancer Treatments Can Enhance Therapy Efficacy. Mini Rev. Med. Chem. 2015, 15, 1052–1062. [Google Scholar] [CrossRef]
- Vanderburg, C.; Beheshti, A. MicroRNAs (miRNAs), the Final Frontier: The Hidden Master Regulators Impacting Biological Response in All Organisms Due to Spaceflight. 2020. Available online: https://three.jsc.nasa.gov/articles/miRNA_Beheshti.pdf (accessed on 23 September 2021).
- Hughes-Fulford, M.; Chang, T.T.; Martinez, E.M.; Li, C.F. Spaceflight alters expression of microRNA during T-cell activation. FASEB J. 2015, 29, 4893–4900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, T.; Wong, M.; Wang, X.; Stodieck, L.; Karouia, F.; Story, M.; Wu, H. Transient gene and microRNA expression profile changes of confluent human fibroblast cells in spaceflight. FASEB J. 2016, 30, 2211–2224. [Google Scholar] [CrossRef]
- Beheshti, A.; McDonald, J.T.; Miller, J.; Grabham, P.; Costes, S.V. GeneLab Database Analyses Suggest Long-Term Impact of Space Radiation on the Cardiovascular System by the Activation of FYN Through Reactive Oxygen Species. Int. J. Mol. Sci. 2019, 20, 661. [Google Scholar] [CrossRef]
- Hammond, T.G.; Lewis, F.C.; Goodwin, T.J.; Linnehan, R.M.; Wolf, D.A.; Hire, K.P.; Campbell, W.C.; Benes, E.; O’Reilly, K.C.; Globus, R.K.; et al. Gene expression in space. Nat. Med. 1999, 5, 359. [Google Scholar] [CrossRef]
- Versari, S.; Longinotti, G.; Barenghi, L.; Maier, J.A.; Bradamante, S. The challenging environment on board the International Space Station affects endothelial cell function by triggering oxidative stress through thioredoxin interacting protein overexpression: The ESA-SPHINX experiment. FASEB J. 2013, 27, 4466–4475. [Google Scholar] [CrossRef]
- Camberos, V.; Baio, J.; Mandujano, A.; Martinez, A.F.; Bailey, L.; Hasaniya, N.; Kearns-Jonker, M. The Impact of Spaceflight and Microgravity on the Human Islet-1+ Cardiovascular Progenitor Cell Transcriptome. Int. J. Mol. Sci. 2021, 22, 3577. [Google Scholar] [CrossRef]
- Thiel, C.S.; Tauber, S.; Christoffel, S.; Huge, A.; Lauber, B.A.; Polzer, J.; Paulsen, K.; Lier, H.; Engelmann, F.; Schmitz, B.; et al. Rapid coupling between gravitational forces and the transcriptome in human myelomonocytic U937 cells. Sci. Rep. 2018, 8, 13267. [Google Scholar] [CrossRef]
- Chang, T.T.; Walther, I.; Li, C.F.; Boonyaratanakornkit, J.; Galleri, G.; Meloni, M.A.; Pippia, P.; Cogoli, A.; Hughes-Fulford, M. The Rel/NF-kappaB pathway and transcription of immediate early genes in T cell activation are inhibited by microgravity. J. Leukoc. Biol. 2012, 92, 1133–1145. [Google Scholar] [CrossRef]
- Barrila, J.; Ott, C.M.; LeBlanc, C.; Mehta, S.K.; Crabbe, A.; Stafford, P.; Pierson, D.L.; Nickerson, C.A. Spaceflight modulates gene expression in the whole blood of astronauts. NPJ Microgravity 2016, 2, 16039. [Google Scholar] [CrossRef] [PubMed]
- Grosse, J.; Wehland, M.; Pietsch, J.; Ma, X.; Ulbrich, C.; Schulz, H.; Saar, K.; Hubner, N.; Hauslage, J.; Hemmersbach, R.; et al. Short-term weightlessness produced by parabolic flight maneuvers altered gene expression patterns in human endothelial cells. FASEB J. 2012, 26, 639–655. [Google Scholar] [CrossRef] [PubMed]
- Hammond, T.G.; Benes, E.; O’Reilly, K.C.; Wolf, D.A.; Linnehan, R.M.; Taher, A.; Kaysen, J.H.; Allen, P.L.; Goodwin, T.J. Mechanical culture conditions effect gene expression: Gravity-induced changes on the space shuttle. Physiol Genom. 2000, 3, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, E. Transcriptional analysis of normal human fibroblast responses to microgravity stress. Genom. Proteom. Bioinform. 2008, 6, 29–41. [Google Scholar] [CrossRef]
- Wnorowski, A.; Sharma, A.; Chen, H.; Wu, H.; Shao, N.Y.; Sayed, N.; Liu, C.; Countryman, S.; Stodieck, L.S.; Rubins, K.H.; et al. Effects of Spaceflight on Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Structure and Function. Stem Cell Rep. 2019, 13, 960–969. [Google Scholar] [CrossRef]
- Curry, J.; Bebb, G.; Moffat, J.; Young, D.; Khaidakov, M.; Mortimer, A.; Glickman, B.W. Similar mutant frequencies observed between pairs of monozygotic twins. Hum. Mutat. 1997, 9, 445–451. [Google Scholar] [CrossRef]
- Mognato, M.; Celotti, L. Modeled microgravity affects cell survival and HPRT mutant frequency, but not the expression of DNA repair genes in human lymphocytes irradiated with ionising radiation. Mutat. Res. 2005, 578, 417–429. [Google Scholar] [CrossRef]
- Canova, S.; Fiorasi, F.; Mognato, M.; Grifalconi, M.; Reddi, E.; Russo, A.; Celotti, L. “Modeled microgravity” affects cell response to ionizing radiation and increases genomic damage. Radiat. Res. 2005, 163, 191–199. [Google Scholar] [CrossRef]
- Khaidakov, M.; Young, D.; Erfle, H.; Mortimer, A.; Voronkov, Y.; Glickman, B.W. Molecular analysis of mutations in T-lymphocytes from experienced Soviet cosmonauts. Environ. Mol. Mutagen. 1997, 30, 21–30. [Google Scholar] [CrossRef]
- Niazi, Y.; Thomsen, H.; Smolkova, B.; Vodickova, L.; Vodenkova, S.; Kroupa, M.; Vymetalkova, V.; Kazimirova, A.; Barancokova, M.; Volkovova, K.; et al. DNA Repair Gene Polymorphisms and Chromosomal Aberrations in Exposed Populations. Front. Genet. 2021, 12, 691947. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.S.; Gil, O.M.; Silva, S.N.; Gomes, B.C.; Ferreira, T.C.; Limbert, E.; Rueff, J. Micronuclei Formation upon Radioiodine Therapy for Well-Differentiated Thyroid Cancer: The Influence of DNA Repair Genes Variants. Genes 2020, 11, 1083. [Google Scholar] [CrossRef] [PubMed]
- Toprani, S.M.; Das, B. Radio-adaptive response, individual radio-sensitivity and correlation of base excision repair gene polymorphism (hOGG1, APE1, XRCC1, and LIGASE1) in human peripheral blood mononuclear cells exposed to gamma radiation. Environ. Mol. Mutagen. 2020, 61, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Sassa, A.; Odagiri, M. Understanding the sequence and structural context effects in oxidative DNA damage repair. DNA Repair 2020, 93, 102906. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, Y.; Qiu, C.; Zhao, J.; Gong, Y.; Nie, C.; Wu, B.; Yang, Y.; Wang, F.; Luo, L. Effects of Simulated Microgravity on Ultrastructure and Apoptosis of Choroidal Vascular Endothelial Cells. Front. Physiol. 2020, 11, 577325. [Google Scholar] [CrossRef]
- Neelam, S.; Richardson, B.; Barker, R.; Udave, C.; Gilroy, S.; Cameron, M.J.; Levine, H.G.; Zhang, Y. Changes in Nuclear Shape and Gene Expression in Response to Simulated Microgravity Are LINC Complex-Dependent. Int. J. Mol. Sci. 2020, 21, 6762. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Zhang, X.; Zhang, F.; Lu, D.; Shangguan, B.; Gao, Y.; Long, M. Flow-enhanced priming of hESCs through H2B acetylation and chromatin decondensation. Stem Cell Res. Ther. 2019, 10, 349. [Google Scholar] [CrossRef]
- Garcia-Gimenez, J.L.; Garces, C.; Roma-Mateo, C.; Pallardo, F.V. Oxidative stress-mediated alterations in histone post-translational modifications. Free Radic. Biol. Med. 2021, 170, 6–18. [Google Scholar] [CrossRef]
- Koaykul, C.; Kim, M.H.; Kawahara, Y.; Yuge, L.; Kino-Oka, M. Maintenance of Neurogenic Differentiation Potential in Passaged Bone Marrow-Derived Human Mesenchymal Stem Cells Under Simulated Microgravity Conditions. Stem Cells Dev. 2019, 28, 1552–1561. [Google Scholar] [CrossRef]
- Singh, K.P.; Kumari, R.; Dumond, J.W. Simulated microgravity-induced epigenetic changes in human lymphocytes. J. Cell Biochem. 2010, 111, 123–129. [Google Scholar] [CrossRef]
- Vidyasekar, P.; Shyamsunder, P.; Arun, R.; Santhakumar, R.; Kapadia, N.K.; Kumar, R.; Verma, R.S. Genome Wide Expression Profiling of Cancer Cell Lines Cultured in Microgravity Reveals Significant Dysregulation of Cell Cycle and MicroRNA Gene Networks. PLoS ONE 2015, 10, e0135958. [Google Scholar] [CrossRef]
- Van Houten, B.; Santa-Gonzalez, G.A.; Camargo, M. DNA repair after oxidative stress: Current challenges. Curr. Opin. Toxicol. 2018, 7, 9–16. [Google Scholar] [CrossRef]
- Fuentes, T.I.; Appleby, N.; Raya, M.; Bailey, L.; Hasaniya, N.; Stodieck, L.; Kearns-Jonker, M. Simulated Microgravity Exerts an Age-Dependent Effect on the Differentiation of Cardiovascular Progenitors Isolated from the Human Heart. PLoS ONE 2015, 10, e0132378. [Google Scholar] [CrossRef]
- Iosim, S.; MacKay, M.; Westover, C.; Mason, C.E. Translating current biomedical therapies for long duration, deep space missions. Precis. Clin. Med. 2019, 2, 259–269. [Google Scholar] [CrossRef]
- Chowdhury, B.; Seetharam, A.; Wang, Z.; Liu, Y.; Lossie, A.C.; Thimmapuram, J.; Irudayaraj, J. A Study of Alterations in DNA Epigenetic Modifications (5mC and 5hmC) and Gene Expression Influenced by Simulated Microgravity in Human Lymphoblastoid Cells. PLoS ONE 2016, 11, e0147514. [Google Scholar] [CrossRef]
- Jin, Y.; Tan, Y.; Chen, L.; Liu, Y.; Ren, Z. Reactive Oxygen Species Induces Lipid Droplet Accumulation in HepG2 Cells by Increasing Perilipin 2 Expression. Int. J. Mol. Sci. 2018, 19, 3445. [Google Scholar] [CrossRef]
- Girardi, C.; De Pitta, C.; Casara, S.; Sales, G.; Lanfranchi, G.; Celotti, L.; Mognato, M. Analysis of miRNA and mRNA expression profiles highlights alterations in ionizing radiation response of human lymphocytes under modeled microgravity. PLoS ONE 2012, 7, e31293. [Google Scholar] [CrossRef]
- Mognato, M.; Girardi, C.; Fabris, S.; Celotti, L. DNA repair in modeled microgravity: Double strand break rejoining activity in human lymphocytes irradiated with gamma-rays. Mutat. Res. 2009, 663, 32–39. [Google Scholar] [CrossRef]
- Fu, H.; Su, F.; Zhu, J.; Zheng, X.; Ge, C. Effect of simulated microgravity and ionizing radiation on expression profiles of miRNA, lncRNA, and mRNA in human lymphoblastoid cells. Life Sci. Space Res. 2020, 24, 1–8. [Google Scholar] [CrossRef]
- Mangala, L.S.; Zhang, Y.; He, Z.; Emami, K.; Ramesh, G.T.; Story, M.; Rohde, L.H.; Wu, H. Effects of simulated microgravity on expression profile of microRNA in human lymphoblastoid cells. J. Biol. Chem. 2011, 286, 32483–32490. [Google Scholar] [CrossRef]
- Girardi, C.; De Pitta, C.; Casara, S.; Calura, E.; Romualdi, C.; Celotti, L.; Mognato, M. Integration analysis of microRNA and mRNA expression profiles in human peripheral blood lymphocytes cultured in modeled microgravity. Biomed. Res. Int. 2014, 2014, 296747. [Google Scholar] [CrossRef]
- Kasiviswanathan, D.; Chinnasamy Perumal, R.; Bhuvaneswari, S.; Kumar, P.; Sundaresan, L.; Philip, M.; Puthenpurackal Krishnankutty, S.; Chatterjee, S. Interactome of miRNAs and transcriptome of human umbilical cord endothelial cells exposed to short-term simulated microgravity. NPJ Microgravity 2020, 6, 18. [Google Scholar] [CrossRef]
- Pan, L.X.; Ding, W. LncRNA HAGLR accelerates femoral neck fracture healing through negatively regulating miRNA-19a-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4080–4087. [Google Scholar]
- Sheyn, D.; Pelled, G.; Netanely, D.; Domany, E.; Gazit, D. The effect of simulated microgravity on human mesenchymal stem cells cultured in an osteogenic differentiation system: A bioinformatics study. Tissue Eng. Part. A 2010, 16, 3403–3412. [Google Scholar] [CrossRef]
- Hybel, T.E.; Dietrichs, D.; Sahana, J.; Corydon, T.J.; Nassef, M.Z.; Wehland, M.; Kruger, M.; Magnusson, N.E.; Bauer, J.; Utpatel, K.; et al. Simulated Microgravity Influences VEGF, MAPK, and PAM Signaling in Prostate Cancer Cells. Int. J. Mol. Sci. 2020, 21, 1263. [Google Scholar] [CrossRef]
- Han, Y.; Zeger, L.; Tripathi, R.; Egli, M.; Ille, F.; Lockowandt, C.; Florin, G.; Atic, E.; Redwan, I.N.; Fredriksson, R.; et al. Molecular genetic analysis of neural stem cells after space flight and simulated microgravity on earth. Biotechnol. Bioeng 2021, 118, 3832–3846. [Google Scholar] [CrossRef]
- Patel, Z.S.; Brunstetter, T.J.; Tarver, W.J.; Whitmire, A.M.; Zwart, S.R.; Smith, S.M.; Huff, J.L. Red risks for a journey to the red planet: The highest priority human health risks for a mission to Mars. NPJ Microgravity 2020, 6, 33. [Google Scholar] [CrossRef]
- Pariset, E.; Bertucci, A.; Petay, M.; Malkani, S.; Lopez Macha, A.; Paulino Lima, I.G.; Gomez Gonzalez, V.; Tin, A.S.; Tang, J.; Plante, I.; et al. DNA Damage Baseline Predicts Resilience to Space Radiation and Radiotherapy. Cell Rep. 2020, 33, 108434. [Google Scholar] [CrossRef]
- Saini, D.; Sudheer, K.R.; Kumar, P.R.V.; Soren, D.C.; Jain, V.; Koya, P.K.M.; Jaikrishan, G.; Das, B. Evaluation of the influence of chronic low-dose radiation on DNA repair gene polymorphisms [XRCC1, XRCC3, PRKDC (XRCC7), LIG1, NEIL1] in individuals from normal and high level natural radiation areas of Kerala Coast. Int. J. Radiat. Biol. 2020, 96, 734–739. [Google Scholar] [CrossRef]
- Matsuno, Y.; Hyodo, M.; Suzuki, M.; Tanaka, Y.; Horikoshi, Y.; Murakami, Y.; Torigoe, H.; Mano, H.; Tashiro, S.; Yoshioka, K.I. Replication-stress-associated DSBs induced by ionizing radiation risk genomic destabilization and associated clonal evolution. iScience 2021, 24, 102313. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; Smirnova, O.A. Dependence of the human leukemia risk on the dose and dose rate of continuous irradiation: Modeling study. Life Sci. Space Res. 2018, 19, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Horie, K.; Hinoi, E.; Hiraiwa, M.; Kato, A.; Maekawa, Y.; Takahashi, A.; Furukawa, S. How does spaceflight affect the acquired immune system? NPJ Microgravity 2020, 6, 14. [Google Scholar] [CrossRef]
- Lee, E.H.; Ding, W.; Kulkarni, A.D.; Granstein, R.D. Tumor growth and immune function in mice during hind-limb unloading. Aviat. Space Environ. Med. 2005, 76, 536–540. [Google Scholar] [PubMed]
- Takahashi, A.; Wakihata, S.; Ma, L.; Adachi, T.; Hirose, H.; Yoshida, Y.; Ohira, Y. Temporary Loading Prevents Cancer Progression and Immune Organ Atrophy Induced by Hind-Limb Unloading in Mice. Int. J. Mol. Sci. 2018, 19, 3959. [Google Scholar] [CrossRef] [PubMed]
- Mencia-Trinchant, N.; MacKay, M.J.; Chin, C.; Afshinnekoo, E.; Foox, J.; Meydan, C.; Butler, D.; Mozsary, C.; Vernice, N.A.; Darby, C.; et al. Clonal hematopoiesis before, during, and after human spaceflight. Cell Rep. 2021, 34, 108740. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gonon, G.; Buonanno, M.; Autsavapromporn, N.; de Toledo, S.M.; Pain, D.; Azzam, E.I. Health risks of space exploration: Targeted and nontargeted oxidative injury by high-charge and high-energy particles. Antioxid. Redox Signal. 2014, 20, 1501–1523. [Google Scholar] [CrossRef]
- Kennedy, A.R. Biological Effects of Space Radiation and Development of Effective Countermeasures. Life Sci. Space Res. 2014, 1, 10–43. [Google Scholar] [CrossRef]
- Villasana, L.E.; Rosenthal, R.A.; Doctrow, S.R.; Pfankuch, T.; Zuloaga, D.G.; Garfinkel, A.M.; Raber, J. Effects of alpha-lipoic acid on associative and spatial memory of sham-irradiated and 56Fe-irradiated C57BL/6J male mice. Pharmacol. Biochem. Behav. 2013, 103, 487–493. [Google Scholar] [CrossRef][Green Version]
- Burns, F.J.; Tang, M.S.; Frenkel, K.; Nadas, A.; Wu, F.; Uddin, A.; Zhang, R. Induction and prevention of carcinogenesis in rat skin exposed to space radiation. Radiat. Environ. Biophys. 2007, 46, 195–199. [Google Scholar] [CrossRef]
- Guan, J.; Wan, X.S.; Zhou, Z.; Ware, J.; Donahue, J.J.; Biaglow, J.E.; Kennedy, A.R. Effects of dietary supplements on space radiation-induced oxidative stress in Sprague-Dawley rats. Radiat. Res. 2004, 162, 572–579. [Google Scholar] [CrossRef]
- Davis, J.G.; Wan, X.S.; Ware, J.H.; Kennedy, A.R. Dietary supplements reduce the cataractogenic potential of proton and HZE-particle radiation in mice. Radiat. Res. 2010, 173, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Schreurs, A.S.; Shirazi-Fard, Y.; Shahnazari, M.; Alwood, J.S.; Truong, T.A.; Tahimic, C.G.; Limoli, C.L.; Turner, N.D.; Halloran, B.; Globus, R.K. Dried plum diet protects from bone loss caused by ionizing radiation. Sci. Rep. 2016, 6, 21343. [Google Scholar] [CrossRef] [PubMed]
- Steczina, S.; Tahimic, C.G.T.; Pendleton, M.; M’Saad, O.; Lowe, M.; Alwood, J.S.; Halloran, B.P.; Globus, R.K.; Schreurs, A.S. Dietary countermeasure mitigates simulated spaceflight-induced osteopenia in mice. Sci. Rep. 2020, 10, 6484. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.M.; Dolan, L.B.; Norton, L.; Charles, J.B.; Jones, L.W. Multisystem Toxicity in Cancer: Lessons from NASA’s Countermeasures Program. Cell 2019, 179, 1003–1009. [Google Scholar] [CrossRef]
- Kim, D.S.; Weber, T.; Straube, U.; Hellweg, C.E.; Nasser, M.; Green, D.A.; Fogtman, A. The Potential of Physical Exercise to Mitigate Radiation Damage-A Systematic Review. Front. Med. 2021, 8, 585483. [Google Scholar] [CrossRef]
- Carnell, L.S. Spaceflight medical countermeasures: A strategic approach for mitigating effects from solar particle events. Int. J. Radiat. Biol. 2020, 1–7. [Google Scholar] [CrossRef]
- Romero-Weaver, A.L.; Wan, X.S.; Diffenderfer, E.S.; Lin, L.; Kennedy, A.R. Kinetics of neutrophils in mice exposed to radiation and/or granulocyte colony-stimulating factor treatment. Radiat. Res. 2013, 180, 177–188. [Google Scholar] [CrossRef]
- Mehta, H.M.; Malandra, M.; Corey, S.J. G-CSF and GM-CSF in Neutropenia. J. Immunol. 2015, 195, 1341–1349. [Google Scholar] [CrossRef]
- Hughson, R.L.; Helm, A.; Durante, M. Heart in space: Effect of the extraterrestrial environment on the cardiovascular system. Nat. Rev. Cardiol. 2018, 15, 167–180. [Google Scholar] [CrossRef]
- Vernice, N.A.; Meydan, C.; Afshinnekoo, E.; Mason, C.E. Long-term spaceflight and the cardiovascular system. Precis. Clin. Med. 2020, 3, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Rosi, S.; Costes, S.V. Central Nervous System Responses to Simulated Galactic Cosmic Rays. Int. J. Mol. Sci. 2018, 19, 3669. [Google Scholar] [CrossRef] [PubMed]
- Crucian, B.E.; Chouker, A.; Simpson, R.J.; Mehta, S.; Marshall, G.; Smith, S.M.; Zwart, S.R.; Heer, M.; Ponomarev, S.; Whitmire, A.; et al. Immune System Dysregulation During Spaceflight: Potential Countermeasures for Deep Space Exploration Missions. Front. Immunol. 2018, 9, 1437. [Google Scholar] [CrossRef] [PubMed]
- Hughson, R.L.; Robertson, A.D.; Arbeille, P.; Shoemaker, J.K.; Rush, J.W.; Fraser, K.S.; Greaves, D.K. Increased postflight carotid artery stiffness and inflight insulin resistance resulting from 6-mo spaceflight in male and female astronauts. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H628–H638. [Google Scholar] [CrossRef] [PubMed]
- Boerma, M.; Roberto, K.A.; Hauer-Jensen, M. Prevention and treatment of functional and structural radiation injury in the rat heart by pentoxifylline and alpha-tocopherol. Int. J. Radiat. Oncol. Biol. Phys. 2008, 72, 170–177. [Google Scholar] [CrossRef] [PubMed]
| Space Environment | Duration | Type of Cells | Methods | Molecular Alterations | Pathway | References |
|---|---|---|---|---|---|---|
| International Space Station (ISS) | 1 year | Human peripheral blood mononuclear cells (PBMCs) | Whole genome sequencing (WGBS) | MiRNA expression Gene expression | Immune/Inflammation-related pathways DNA-damage response | [3] |
| International Space Station (ISS) | 3–14 days | Human normal foreskin fibroblasts (AG1522) | Microarray | MiRNA expression Gene expression | NF-kB pathway Growth-related pathways | [90] |
| International Space Station (ISS) | 1 month | Human normal foreskin fibroblasts (AG1522) | Microarray | Gene expression | DNA-damage response pathway | [46] |
| Spaceflight (STS-93 mission) | 4 days and 23 h | Human normal fibroblasts (WI-38) | cDNA libraries | Gene expression | Apoptosis, senescence | [100] |
| Soyuz 13S (TMA-09) + ISS | 11 days | Human T cells | Microarray | MiRNA expression Gene expression | Immune system | [89] |
| Space transportation system flight STS-90 | 6 days | Primary human renal cell cultures | Microarray | Gene expression | Transcription factors | [99] |
| Soyuz 13S (TMA-09) + ISS | 11 days | Human T cells | Microarray | Gene expression | Rel-NF-kB pathway | [96] |
| SpaceX CRS-11 + ISS | 30 days | Human adult and neonatal cardiovascular progenitors | RNA sequencing | MiRNA expression Gene expression | Senescence, stemness, cell proliferation, survival, oxidative stress | [94] |
| International Space Station (ISS) | 5.5 weeks | Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) | RNA sequencing | Gene expression | Mitochondrial metabolism; DNA damage and repair | [101] |
| International Space Station (ISS) | 10 days | Human Umbilical Vein Endothelial Cells (HUVECs) | Microarray | Gene expression | Cell adhesion, oxidative phosphorylation, stress responses, cell cycle, apoptosis | [93] |
| Parabolic flight and suborbital ballistic rocket experiments | Human myelomonocytic cells (U937), Jurkat T cells, primary T lymphocytes | ROS metabolism, antioxidative systems, oxidative stress response | [43,83] | |||
| Parabolic flight | 22 s | Human endothelial cells (EA.hy926) | Microarray and qRT-PCR | Gene expression | [98] | |
| Space shuttle | 10–13 days | Human whole blood | Microarray | Gene expression | [97] | |
| International Space Station (ISS) | 48–72 h | Human blood-derived stem cells (BDSCs) | Histone H3 PTMs | Cell differentiation | [82] |
| Condition | Duration | Type of Cells | Methods | Molecular Alterations | Signalling Pathway | References |
|---|---|---|---|---|---|---|
| γ-irradiation (0.2–2 Gy) + microgravity (simulated with the RWV) | 4 and 24 h | Human peripheral blood lymphocytes (PBLs) | Microarray | MiRNA expression Gene expression | DNA-damage response, p53 pathway | [122] |
| γ-irradiation (2 Gy) + microgravity (simulated with the RWV) | 24 h | Human lymphoblastoid cells (TK6) | Microarray | Gene expression NcRNAs expression MiRNA expression | Apoptosis, immune/inflammatory response, NF-kB pathway, p53 pathway | [124] |
| γ-irradiation (2 Gy) + microgravity (simulated with the RWV) | 24 h | Human PBLs; human lymphoblastoid cells (TK6) | T-cell clonal assay | Mutant frequency | Hypoxanthine-phosphorybosil transferase gene | [103,104] |
| Microgravity (simulated with the HARV bioreactor) | 72 h | Human lymphoblastoid cells (TK6) | Microarray | MiRNA expression | Immune response, NF-kB pathway, apoptosis, survival | [125] |
| Microgravity (simulated with the RWV) | 24 h | Human peripheral blood lymphocytes (PBLs) | Microarray | MiRNA expression Gene expression | Immune system/Inflammation | [126] |
| Microgravity (simulated with the RWV) | 6 days | Primary human renal cell cultures | Microarray | Gene expression | Shear stress response, adhesion, apoptosis, cytoskeleton, differentiation, | [99] |
| Microgravity (simulated with a Clinostat) | 2 h | Human endothelial cells (HUVEC) | Next-Generation Sequencing (NGS) | MiRNA expression Gene expression | NF-kB pathway, inflammation, cell cycle, proliferation, angiogenesis | [127] |
| Microgravity (simulated with a RPM) | 3–5 days | Human prostate cancer cells | qRT-PCR | Gene expression | VEGF, MAPK and PAM signalling pathways | [130] |
| Microgravity (simulated with a RCCS) | 72 h–7 days | Human T-lymphocyte cells | qRT-PCR | Gene expression | DNA methylation, histone acetylation | [115] |
| Microgravity (simulated with a RCCS) | 24–48 h | Human colorectal cancer cells Human lymphoblast leukemia cells | Microarray | Gene expression | Cell cycle regulation, apoptosis, Notch signalling pathway | [116] |
| Microgravity (simulated with a RCCS) | 3 days | Human choroidal vascular endothelial cells | qRT-PCR TEM | Gene expression; chromatin condensation/margination; mitochondria vacuolization | Apoptosis, PI3K/AKT pathway | [110] |
| Microgravity (simulated with a 2D-clinostat) | 48 h | Human HUVEC cells | RNA seq qRT-PCR | MiRNA expression Gene expression | Apoptosis | [128] |
| Microgravity (simulated with a 3D-clinostat) | 2–20 h | Human breast epithelial cells | RNA seq | Gene expression | Cell cycle, cell adhesion, cytoskeleton | [111] |
| Microgravity (simulated with a RCCS) | 48 h | Human lymphoblastoid cells (TK6) | Next-Generation Sequencing (NGS) | DNA methylation; gene expression | Response to oxidative stress, ion transport, DNA-dependent transcription, carbohydrate metabolic processes | [120] |
| Microgravity (simulated with a RCCS) | 1 week | Human mesenchymal stem cells | Microarray | Gene expression | Osteogenic differentiation, cell adhesion/communication, cell cycle, cytoskeleton, immune response | [129] |
| Microgravity (simulated with a 2D-clinostat) | 6–7 days | Human cardiac progenitors | RT-PCR | Gene expression | Telomerase maintenance | [118] |
| Microgravity (simulated with a 3D Clinostat) | 7 days | Human mesenchymal stem cells | Western blotting qRT-PCR | Histone modification | Cytoskeleton, histone modification | [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beheshti, A.; McDonald, J.T.; Hada, M.; Takahashi, A.; Mason, C.E.; Mognato, M. Genomic Changes Driven by Radiation-Induced DNA Damage and Microgravity in Human Cells. Int. J. Mol. Sci. 2021, 22, 10507. https://doi.org/10.3390/ijms221910507
Beheshti A, McDonald JT, Hada M, Takahashi A, Mason CE, Mognato M. Genomic Changes Driven by Radiation-Induced DNA Damage and Microgravity in Human Cells. International Journal of Molecular Sciences. 2021; 22(19):10507. https://doi.org/10.3390/ijms221910507
Chicago/Turabian StyleBeheshti, Afshin, J. Tyson McDonald, Megumi Hada, Akihisa Takahashi, Christopher E. Mason, and Maddalena Mognato. 2021. "Genomic Changes Driven by Radiation-Induced DNA Damage and Microgravity in Human Cells" International Journal of Molecular Sciences 22, no. 19: 10507. https://doi.org/10.3390/ijms221910507
APA StyleBeheshti, A., McDonald, J. T., Hada, M., Takahashi, A., Mason, C. E., & Mognato, M. (2021). Genomic Changes Driven by Radiation-Induced DNA Damage and Microgravity in Human Cells. International Journal of Molecular Sciences, 22(19), 10507. https://doi.org/10.3390/ijms221910507









