Lights, Camera, Interaction: Studying Protein–Protein Interactions of the ER Protein Translocase in Living Cells
Abstract
1. Introduction
2. Results
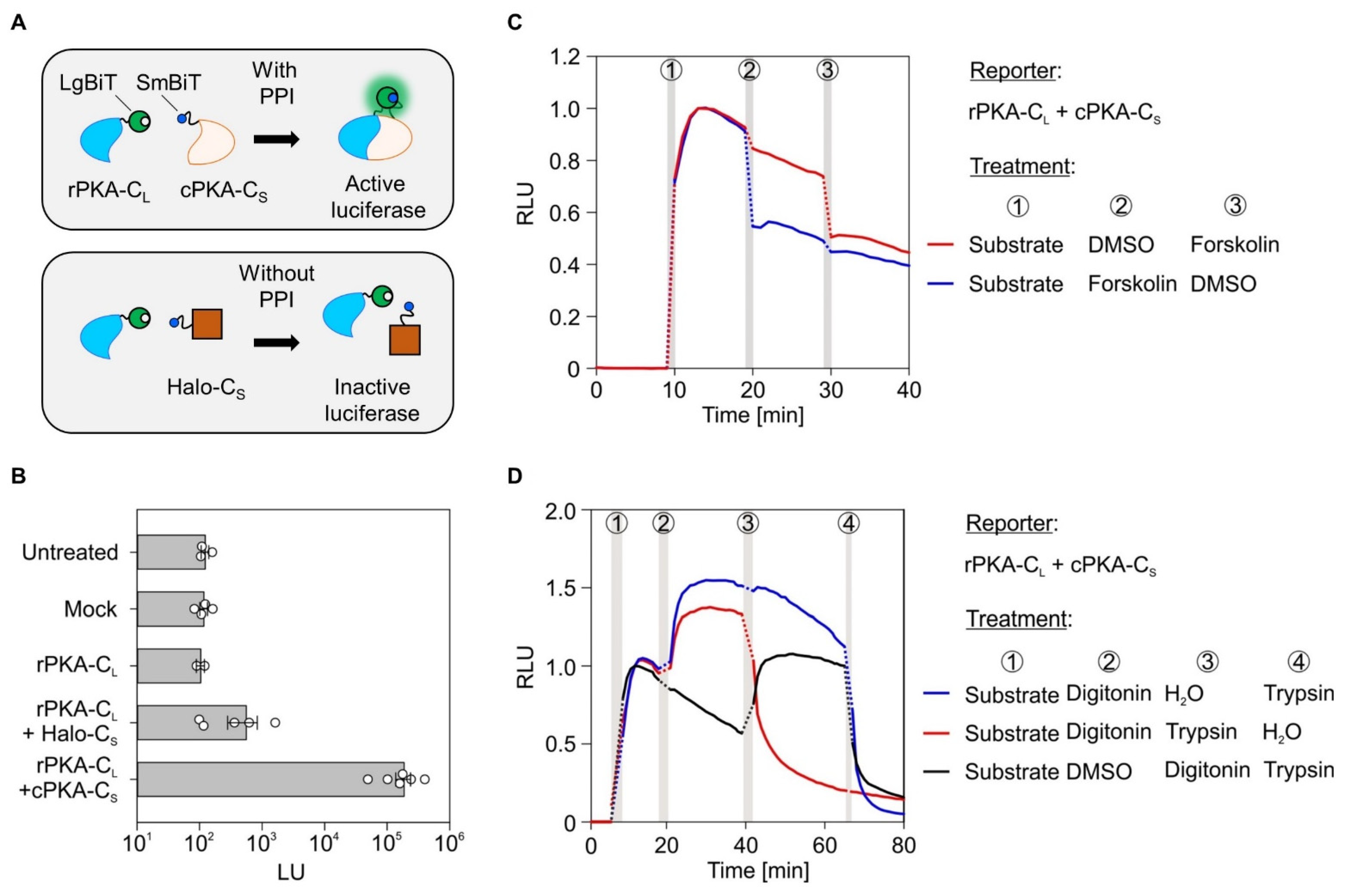
2.1. Establishing the NanoBiT Assay Based on the cAMP-Dependent Protein Kinase A, Forskolin, and Semi-Permeabilization
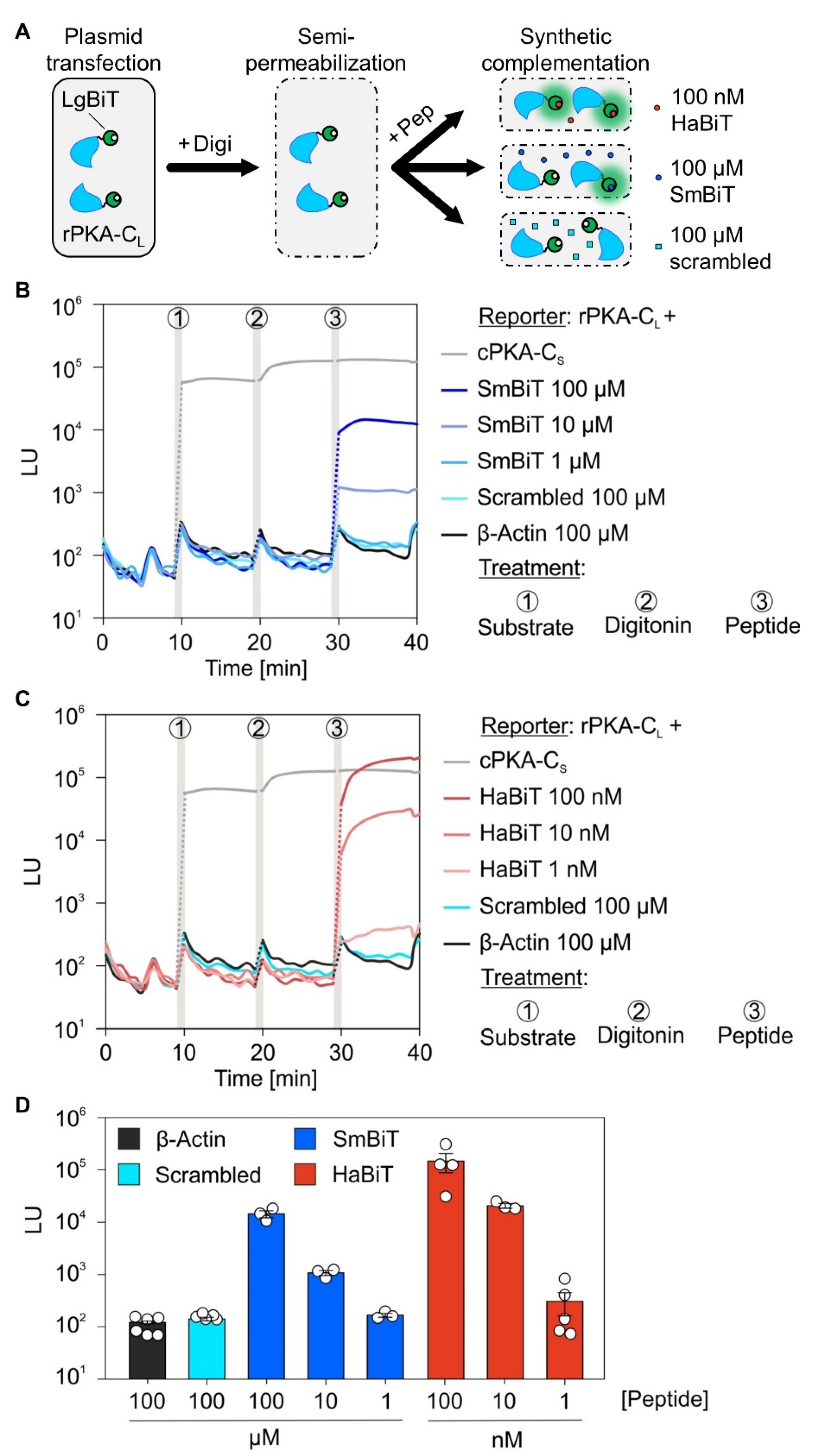
2.2. Expression Profiling via Synthetic Complementation Using Chemically Synthesized Low and High Affinity Oligopeptides
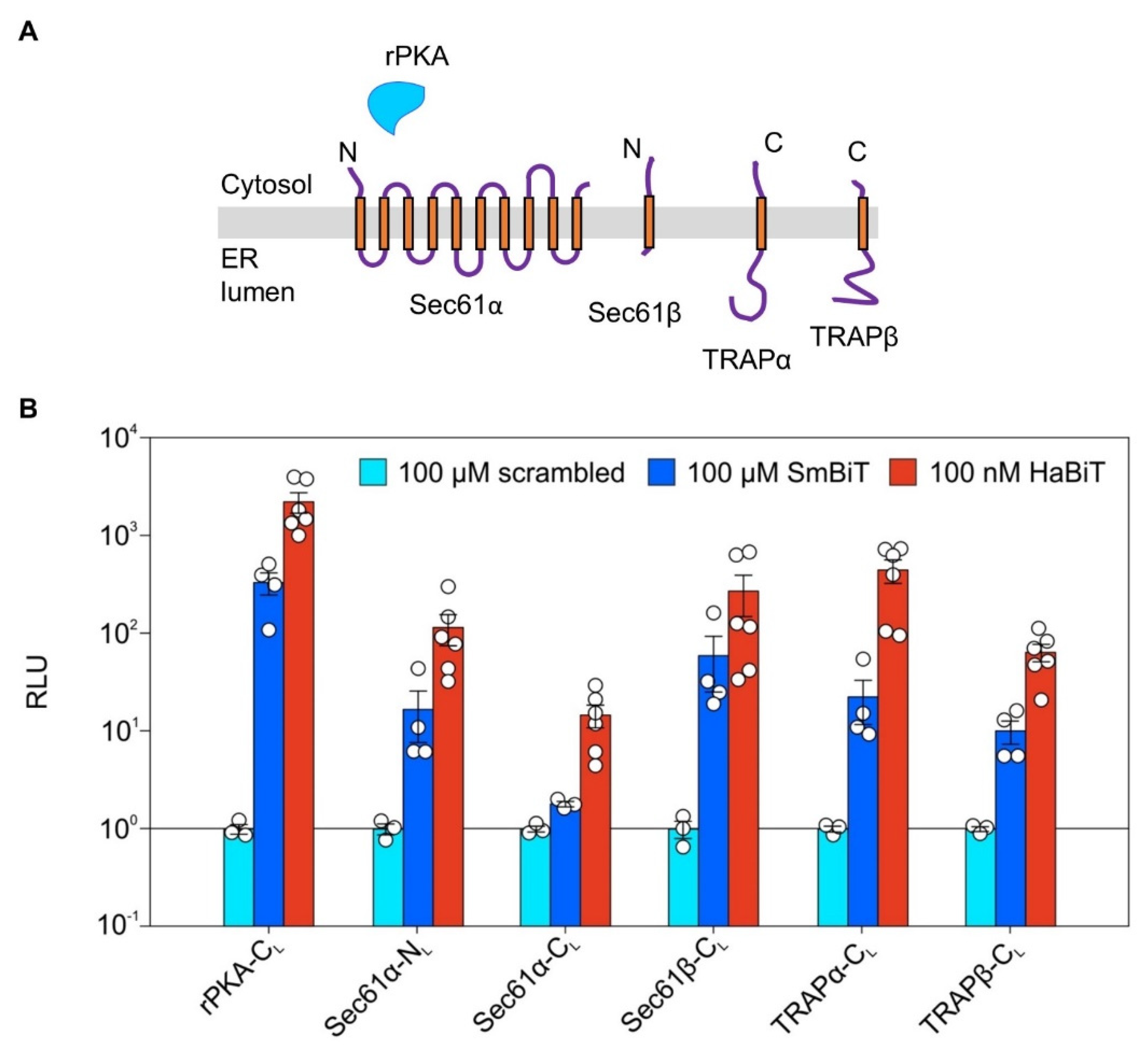
2.3. Verifying Expression of LgBit Fusion Constructs of ER Protein Translocase Subunits via Synthetic Peptide Complementation
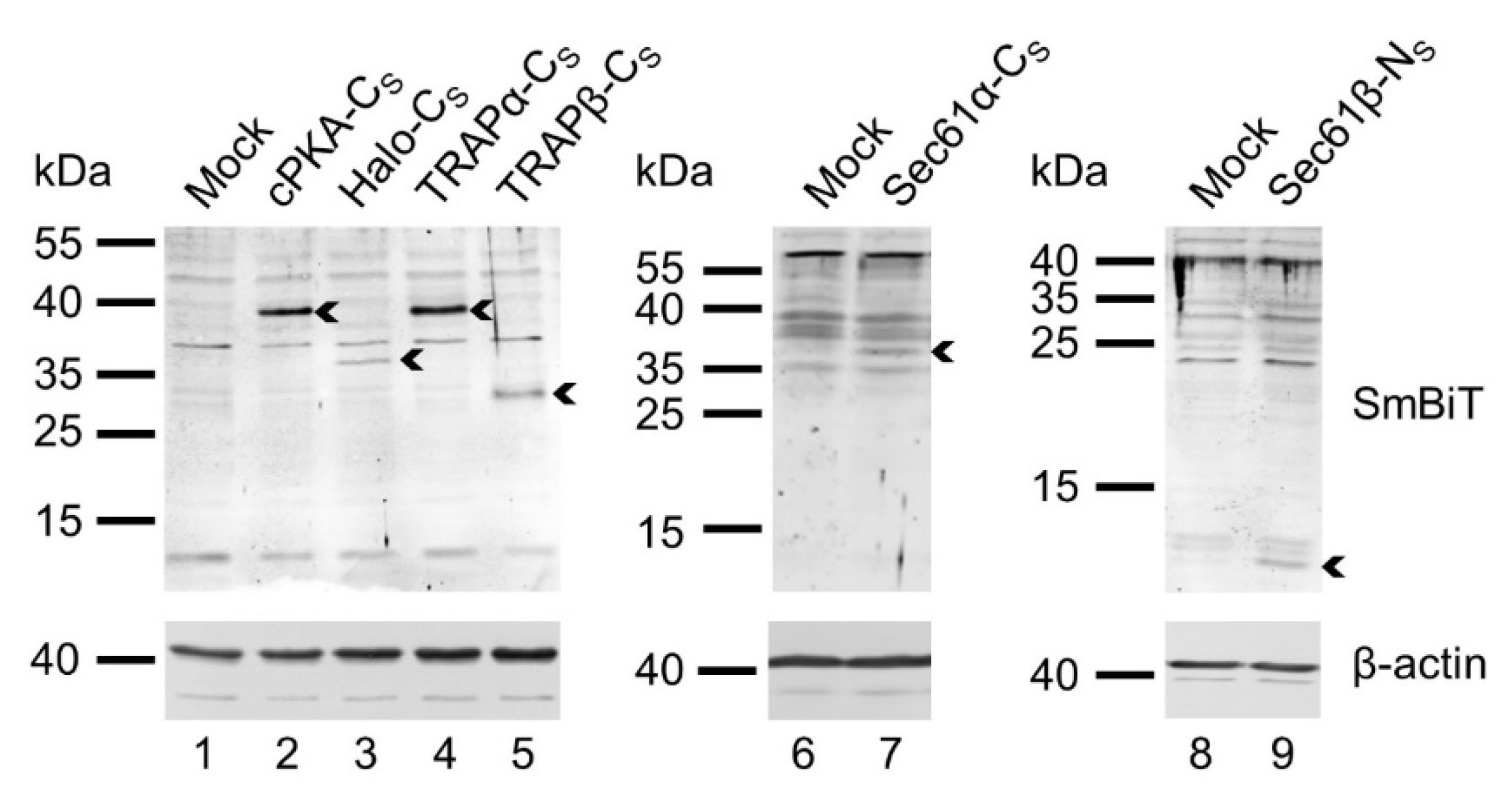
2.4. Validating Expression of SmBit Fusion Constructs via Western Blotting Using a Polyclonal Antibody Raised against the SmBiT
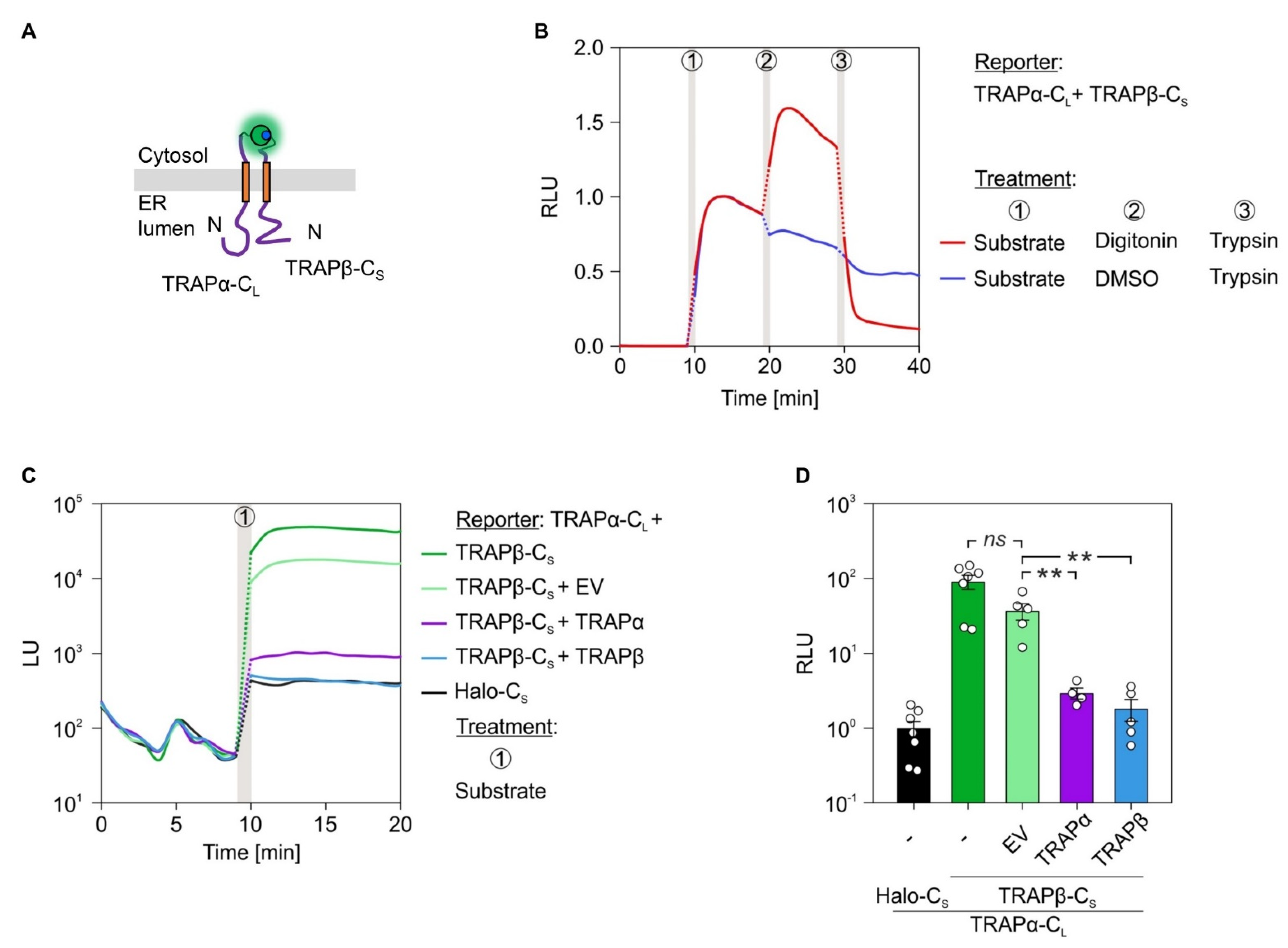
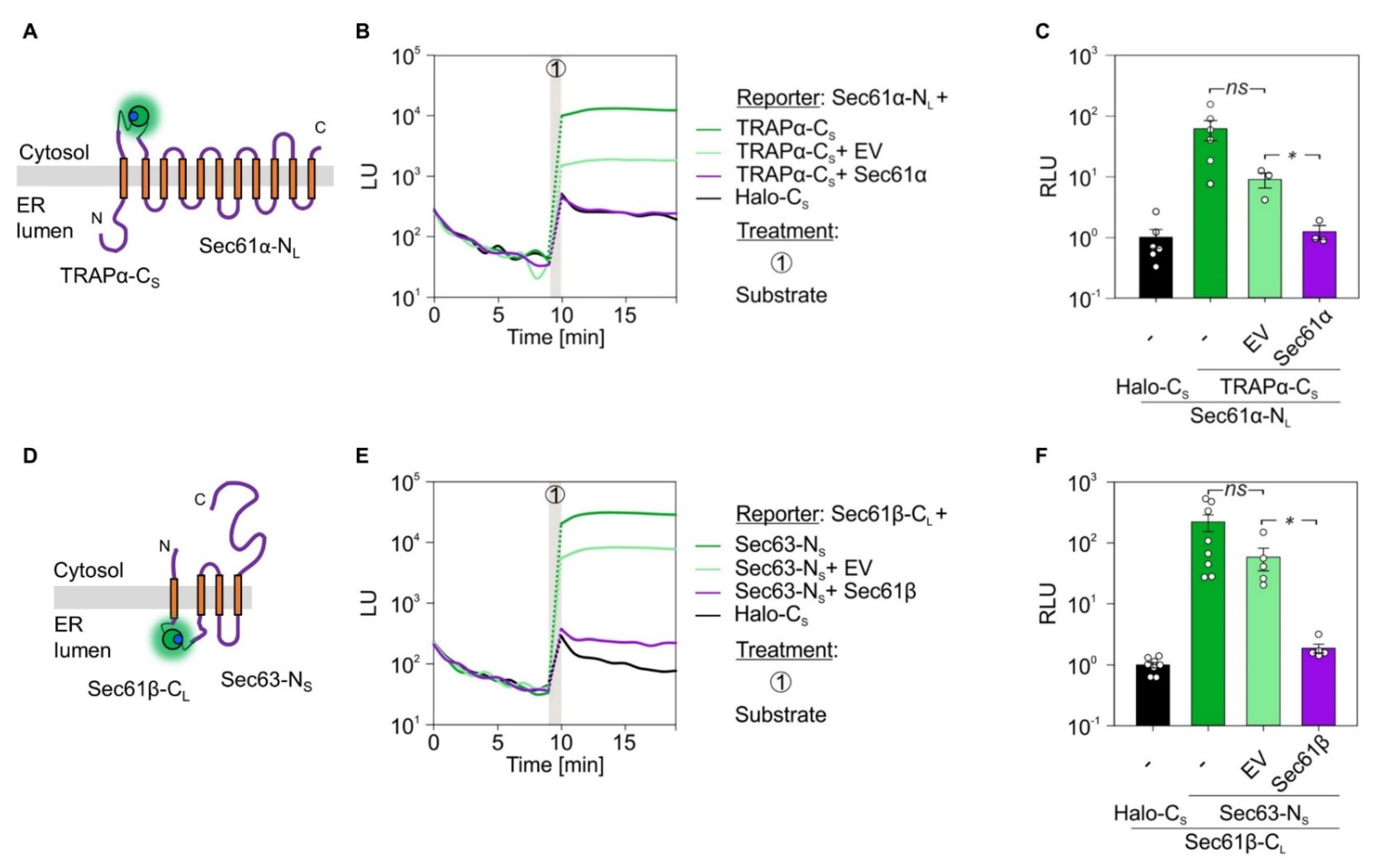
2.5. α- and β-Subunit Interactions of the Sec61 or TRAP Complex
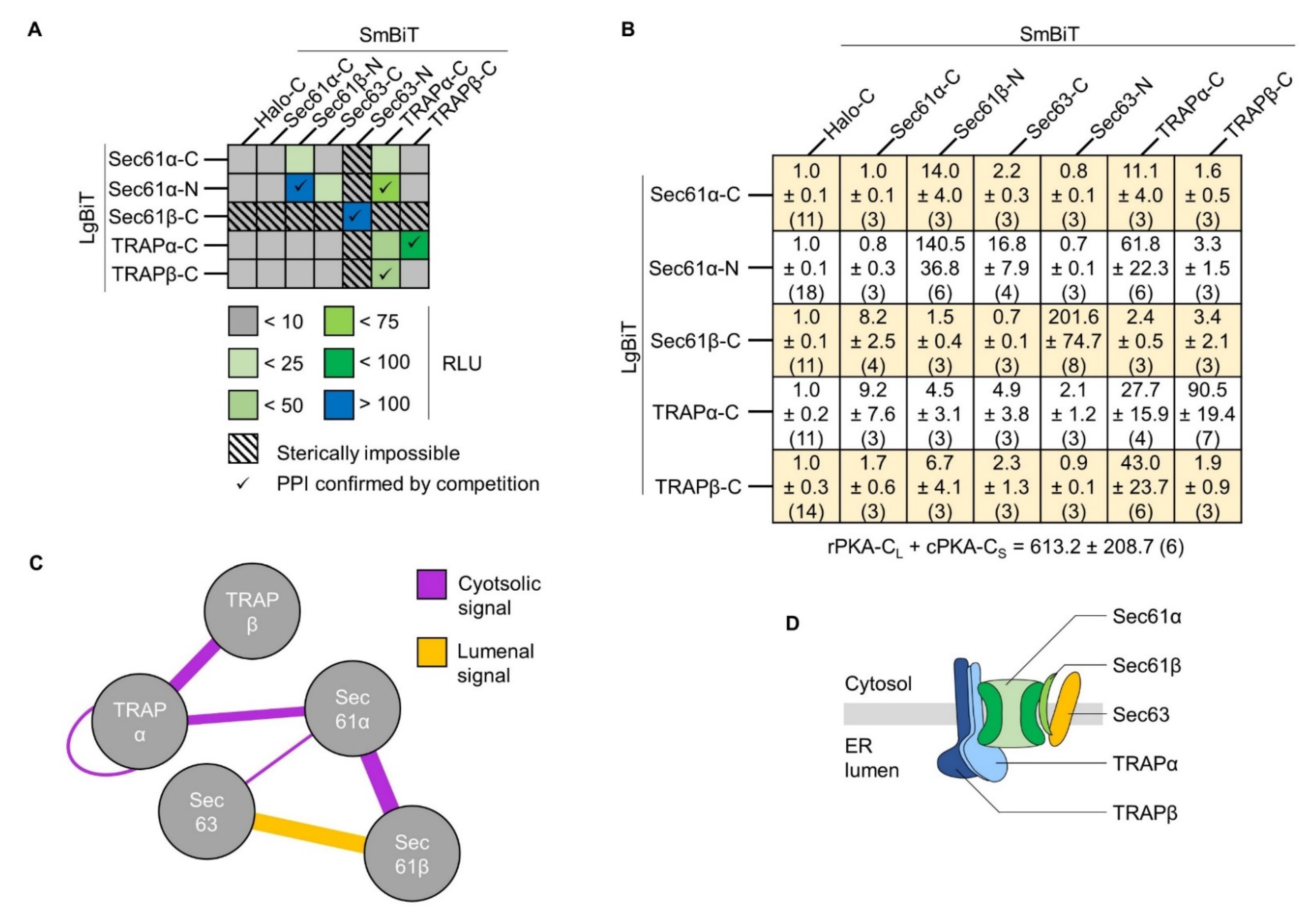
2.6. Inter-Complex Interactions between Sec61 and TRAP as well as Sec61 and Sec63
2.7. Sterically Impossible Complementations Set the Threshold for Authentic Protein Interactions
3. Discussion
4. Materials and Methods
4.1. Creation of a Plasmid Library
4.2. Cell Culture and Western Blot
4.3. NanoBiT Assay
4.4. Peptide Synthesis
4.5. Antibody Generation
4.6. Statistics and Graphical Representation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Palade, G.E. The endoplasmic reticulum. J. Biophys. Biochem. Cytol. 1956, 2, 85–98. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016, 73, 79–94. [Google Scholar] [CrossRef]
- Bravo, R.; Parra, V.; Gatica, D.; Rodriguez, A.E.; Torrealba, N.; Paredes, F.; Wang, Z.V.; Zorzano, A.; Hill, J.A.; Jaimovich, E.; et al. Chapter Five—Endoplasmic Reticulum and the Unfolded Protein Response: Dynamics and Metabolic Integration. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 301, pp. 215–290. [Google Scholar]
- Raffaello, A.; Mammucari, C.; Gherardi, G.; Rizzuto, R. Calcium at the Center of Cell Signaling: Interplay between Endoplasmic Reticulum, Mitochondria, and Lysosomes. Trends Biochem. Sci. 2016, 41, 1035–1049. [Google Scholar] [CrossRef]
- Lang, S.; Nguyen, D.; Pfeffer, S.; Förster, F.; Helms, V.; Zimmermann, R. Functions and Mechanisms of the Human Ribosome-Translocon Complex. In Macromolecular Protein Complexes II: Structure and Function; Harris, J.R., Marles-Wright, J., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 83–141. [Google Scholar]
- O′Keefe, S.; High, S. Membrane translocation at the ER: With a little help from my friends. FEBS J. 2020, 287, 4607–4611. [Google Scholar] [CrossRef] [PubMed]
- Gemmer, M.; Förster, F. A clearer picture of the ER translocon complex. J. Cell Sci. 2020, 133, jcs231340. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Aitblal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 26, 356. [Google Scholar] [CrossRef] [PubMed]
- Conti, B.J.; Devaraneni, P.K.; Yang, Z.; David, L.L.; Skach, W.R. Cotranslational Stabilization of Sec62/63 within the ER Sec61 Translocon Is Controlled by Distinct Substrate-Driven Translocation Events. Mol. Cell 2015, 58, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Dejgaard, K.; Theberge, J.-F.; Heath-Engel, H.; Chevet, E.; Tremblay, M.L.; Thomas, D.Y. Organization of the Sec61 Translocon, Studied by High Resolution Native Electrophoresis. J. Proteome Res. 2010, 9, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, B.; McKenna, M.; Johnson, N.; High, S.; Sinning, I.; Pool, M.R. Mammalian SRP receptor switches the Sec61 translocase from Sec62 to SRP-dependent translocation. Nat. Commun. 2015, 6, 133. [Google Scholar] [CrossRef]
- Wang, L.; Dobberstein, B. Oligomeric complexes involved in translocation of proteins across the membrane of the endoplasmic reticulum. FEBS Lett. 1999, 457, 316–322. [Google Scholar] [CrossRef]
- Kriegler, T.; Lang, S.; Notari, L.; Hessa, T. Prion Protein Translocation Mechanism Revealed by Pulling Force Studies. J. Mol. Biol. 2020, 432, 4447–4465. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Stutz, R.; Schorr, S.; Lang, S.; Pfeffer, S.; Freeze, H.H.; Förster, F.; Helms, V.; Dudek, J.; Zimmermann, R. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat. Commun. 2018, 9, 65. [Google Scholar] [CrossRef]
- Pfeffer, S.; Dudek, J.; Schaffer, M.; Ng, B.G.; Albert, S.; Plitzko, J.M.; Baumeister, W.; Zimmermann, R.; Freeze, H.H.; Engel, B.D.; et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Weng, T.-H.; Steinchen, W.; Beatrix, B.; Berninghausen, O.; Becker, T.; Bange, G.; Cheng, J.; Beckmann, R. Architecture of the active post-translational Sec translocon. EMBO J. 2021, 40, e105643. [Google Scholar] [CrossRef]
- Bhadra, P.; Yadhanapudi, L.; Römisch, K.; Helms, V. How does Sec63 affect the conformation of Sec61 in yeast? PLOS Comput. Biol. 2021, 17, e1008855. [Google Scholar] [CrossRef]
- Schorr, S.; Nguyen, D.; Haßdenteufel, S.; Nagaraj, N.; Cavalié, A.; Greiner, M.; Weissgerber, P.; Loi, M.; Paton, A.W.; Paton, J.C.; et al. Identification of signal peptide features for substrate specificity in human Sec62/Sec63-dependent ER protein import. FEBS J. 2020, 287, 4612–4640. [Google Scholar] [CrossRef]
- Ziska, A.; Tatzelt, J.; Dudek, J.; Paton, A.W.; Paton, J.C.; Zimmermann, R.; Haßdenteufel, S. The signal peptide plus a cluster of positive charges in prion protein dictate chaperone-mediated Sec61 channel gating. Biol. Open 2019, 4, 691. [Google Scholar] [CrossRef]
- Schuren, A.B.C.; Boer, I.G.J.; Bouma, E.; Lebbink, R.J.; Wiertz, E.J.H.J. Genetic editing of SEC61, SEC62, and SEC63 abrogates human cytomegalovirus US2 expression in a signal peptide-dependent manner. bioRxiv 2019, 9, 653857. [Google Scholar] [CrossRef]
- Lang, S.; Benedix, J.; Fedeles, S.V.; Schorr, S.; Schirra, C.; Schäuble, N.; Jalal, C.; Greiner, M.; Haßdenteufel, S.; Tatzelt, J.; et al. Different effects of Sec61α, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 2012, 125, 1958–1969. [Google Scholar] [CrossRef]
- Lang, S.; Pfeffer, S.; Lee, P.-H.; Cavalié, A.; Helms, V.; Förster, F.; Zimmermann, R. An Update on Sec61 Channel Functions, Mechanisms, and Related Diseases. Front. Physiol. 2017, 8, 887. [Google Scholar] [CrossRef]
- Walter, T.; Erdmann, R. Current Advances in Protein Import into Peroxisomes. Protein J. 2019, 38, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Li, H.-m.; Chiu, C.-C. Protein Transport into Chloroplasts. Annu. Rev. Plant Biol. 2010, 61, 157–180. [Google Scholar] [CrossRef]
- Wickner, W.; Schekman, R. Protein Translocation Across Biological Membranes. Science 2005, 310, 1452–1456. [Google Scholar] [CrossRef] [PubMed]
- Kunze, M.; Berger, J. The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front. Physiol. 2015, 6, 259. [Google Scholar] [CrossRef]
- Berggård, T.; Linse, S.; James, P. Methods for the detection and analysis of protein–protein interactions. Proteomics 2007, 7, 2833–2842. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N.S. Protein–protein interaction detection: Methods and analysis. Int. J. Proteom. 2014, 2014, 147648. [Google Scholar] [CrossRef]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent advances in the development of protein–protein interactions modulators: Mechanisms and clinical trials. Signal Transduct. Target. Ther. 2020, 5, 213. [Google Scholar] [CrossRef]
- Koh, G.C.K.W.; Porras, P.; Aranda, B.; Hermjakob, H.; Orchard, S.E. Analyzing Protein–Protein Interaction Networks. J. Proteome Res. 2012, 11, 2014–2031. [Google Scholar] [CrossRef]
- Nero, T.L.; Morton, C.J.; Holien, J.K.; Wielens, J.; Parker, M.W. Oncogenic protein interfaces: Small molecules, big challenges. Nat. Rev. Cancer 2014, 14, 248–262. [Google Scholar] [CrossRef]
- Braun, P. Interactome mapping for analysis of complex phenotypes: Insights from benchmarking binary interaction assays. Proteomics 2012, 12, 1499–1518. [Google Scholar] [CrossRef]
- Xing, S.; Wallmeroth, N.; Berendzen, K.W.; Grefen, C. Techniques for the Analysis of Protein–Protein Interactions In Vivo. Plant Physiol. 2016, 171, 727–758. [Google Scholar] [CrossRef]
- Milroy, L.-G.; Grossmann, T.N.; Hennig, S.; Brunsveld, L.; Ottmann, C. Modulators of Protein–Protein Interactions. Chem. Rev. 2014, 114, 4695–4748. [Google Scholar] [CrossRef]
- Michnick, S.W.; Ear, P.H.; Manderson, E.N.; Remy, I.; Stefan, E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat. Rev. Drug Discov. 2007, 6, 569–582. [Google Scholar] [CrossRef]
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408. [Google Scholar] [CrossRef]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef]
- Alasbahi, R.H.; Melzig, M.F. Forskolin and derivatives as tools for studying the role of cAMP. Die Pharm. Int. J. Pharm. Sci. 2012, 67, 5–13. [Google Scholar] [CrossRef]
- Hartmann, E.; Görlich, D.; Kostka, S.; Otto, A.; Kraft, R.; Knespel, S.; Bürger, E.; Rapoport, T.A.; Prehn, S. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur. J. Biochem. 1993, 214, 375–381. [Google Scholar] [CrossRef]
- Borgese, N.; Colombo, S.; Pedrazzini, E. The tale of tail-anchored proteins. J. Cell Biol. 2003, 161, 1013–1019. [Google Scholar] [CrossRef]
- Zimmermann, R.; Eyrisch, S.; Ahmad, M.; Helms, V. Protein translocation across the ER membrane. Biochim. Biophys. Acta 2011, 1808, 912–924. [Google Scholar] [CrossRef]
- Lehnert, E.; Tampé, R. Structure and Dynamics of Antigenic Peptides in Complex with TAP. Front. Immunol. 2017, 8, 10. [Google Scholar] [CrossRef]
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Förster, F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015, 6, 9403. [Google Scholar] [CrossRef]
- Pfeffer, S.; Dudek, J.; Zimmermann, R.; Förster, F. Organization of the native ribosome–translocon complex at the mammalian endoplasmic reticulum membrane. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2016, 1860, 2122–2129. [Google Scholar] [CrossRef]
- Ali, R.; Ramadurai, S.; Barry, F.; Nasheuer, H.P. Optimizing fluorescent protein expression for quantitative fluorescence microscopy and spectroscopy using herpes simplex thymidine kinase promoter sequences. FEBS Open Bio 2018, 8, 1043–1060. [Google Scholar] [CrossRef]
- Bañó-Polo, M.; Martínez-Garay, C.A.; Grau, B.; Martínez-Gil, L.; Mingarro, I. Membrane insertion and topology of the translocon-associated protein (TRAP) gamma subunit. Biochim. Biophys. Acta (BBA) Biomembr. 2017, 1859, 903–909. [Google Scholar] [CrossRef]
- Spiess, M.; Junne, T.; Janoschke, M. Membrane Protein Integration and Topogenesis at the ER. Protein J. 2019, 38, 306–316. [Google Scholar] [CrossRef]
- Junne, T.; Spiess, M. Integration of transmembrane domains is regulated by their downstream sequences. J. Cell Sci. 2017, 130, 372–381. [Google Scholar] [CrossRef]
- Brambillasca, S.; Yabal, M.; Makarow, M.; Borgese, N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J. Cell Biol. 2006, 175, 767–777. [Google Scholar] [CrossRef]
- Pfeffer, S.; Förster, F. Sec61: A static framework for membrane-protein insertion. Channels 2016, 10, 167–169. [Google Scholar] [CrossRef][Green Version]
- Snapp, E.L.; Reinhart, G.A.; Bogert, B.A.; Lippincott-Schwartz, J.; Hegde, R.S. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J. Cell Biol. 2004, 164, 997–1007. [Google Scholar] [CrossRef]
- Benedix, J.; Lajoie, P.; Jaiswal, H.; Burgard, C.; Greiner, M.; Zimmermann, R.; Rospert, S.; Snapp, E.L.; Dudek, J. BiP modulates the affinity of its co-chaperone ERj1 for ribosomes. J. Biol. Chem. 2010, 285, 36427–36433. [Google Scholar] [CrossRef]
- Van Puyenbroeck, V.; Vermeire, K. Inhibitors of protein translocation across membranes of the secretory pathway: Novel antimicrobial and anticancer agents. Cell. Mol. Life Sci. 2018, 75, 1541–1558. [Google Scholar] [CrossRef]
- Vermeire, K.; Bell, T.W.; Van Puyenbroeck, V.; Giraut, A.; Noppen, S.; Liekens, S.; Schols, D.; Hartmann, E.; Kalies, K.-U.; Marsh, M. Signal Peptide-Binding Drug as a Selective Inhibitor of Co-Translational Protein Translocation. PLoS Biol. 2014, 12, e1002011. [Google Scholar] [CrossRef]
- Gilles, A.; Frechin, L.; Natchiar, K.; Biondani, G.; Loeffelholz, O.v.; Holvec, S.; Malaval, J.-L.; Winum, J.-Y.; Klaholz, B.P.; Peyron, J.-F. Targeting the Human 80S Ribosome in Cancer: From Structure to Function and Drug Design for Innovative Adjuvant Therapeutic Strategies. Cells 2020, 9, 629. [Google Scholar] [CrossRef]
- Dmitriev, S.E.; Vladimirov, D.O.; Lashkevich, K.A. A Quick Guide to Small-Molecule Inhibitors of Eukaryotic Protein Synthesis. Biochemistry 2020, 85, 1389–1421. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, B.; Clemons, W.M., Jr.; Collinson, I.; Modis, Y.; Hartmann, E.; Harrison, S.C.; Rapoport, T.A. X-ray structure of a protein-conducting channel. Nature 2004, 427, 36–44. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Hegde, R.S. Structure of the Sec61 channel opened by a signal sequence. Science 2016, 351, 88–91. [Google Scholar] [CrossRef]
- Mothes, W.; Prehn, S.; Rapoport, T.A. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994, 13, 3973–3982. [Google Scholar] [CrossRef]
- Fons, R.D.; Bogert, B.A.; Hegde, R.S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003, 160, 529–539. [Google Scholar] [CrossRef]
- Ménétret, J.F.; Hegde, R.S.; Aguiar, M.; Gygi, S.P.; Park, E.; Rapoport, T.A.; Akey, C.W. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure 2008, 16, 1126–1137. [Google Scholar] [CrossRef]
- Meyer, H.-A.; Grau, H.; Kraft, R.; Kostka, S.; Prehn, S.; Kalies, K.-U.; Hartmann, E. Mammalian Sec61 Is Associated with Sec62 and Sec63. J. Biol. Chem. 2000, 275, 14550–14557. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cabanos, C.; Rapoport, T.A. Structure of the post-translational protein translocation machinery of the ER membrane. Nature 2019, 566, 136–139. [Google Scholar] [CrossRef]
- Itskanov, S.; Park, E. Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science 2018, 32, 6740. [Google Scholar] [CrossRef] [PubMed]
- Itskanov, S.; Kuo, K.M.; Gumbart, J.C.; Park, E. Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62. Nat. Struct. Mol. Biol. 2021, 28, 162–172. [Google Scholar] [CrossRef]
- O′Keefe, S.; Pool, M.R.; High, S. Membrane protein biogenesis at the ER: The highways and byways. FEBS J. 2021, 11, 51. [Google Scholar] [CrossRef]
- Kalies, K.-U.; Rapoport, T.A.; Hartmann, E. The beta-Subunit of the Sec61 Complex Facilitates Cotranslational Protein Transport and Interacts with the Signal Peptidase during Translocation. J. Cell Biol. 1998, 141, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Migliaccio, G.; Nicchitta, C.V.; Blobel, G. The signal sequence receptor, unlike the signal recognition particle receptor, is not essential for protein translocation. J. Cell Biol. 1992, 117, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Wiedmann, M.; Kurzchalia, T.V.; Hartmann, E.; Rapoport, T.A. A signal sequence receptor in the endoplasmic reticulum membrane. Nature 1987, 328, 830–833. [Google Scholar] [CrossRef]
- Dudek, J.; Lang, S.; Schorr, S.; Linxweiler, J.; Greiner, M.; Zimmermann, R. Analysis of Protein Translocation into the Endoplasmic Reticulum of Human Cells. In Membrane Biogenesis; Rapaport, D., Herrmann, J.M., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1033, pp. 285–299. [Google Scholar]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]

| Transfection Mix | One Plasmid Mix | Two Plasmid Mix | Three Plasmid Mix |
|---|---|---|---|
| Purpose | Synthetic complementation | PPI experiment | PPI competition experiment |
| Opti-MEM w/o phenol red | 7.36 | 6.72 | 6.08 |
| LgBit plasmid | 0.50 | 0.50 | 0.50 |
| SmBiT plasmid | - | 0.50 | 0.50 |
| Competition plasmid | - | - | 0.50 |
| FuGENE HD 1 | 0.14 | 0.28 | 0.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sicking, M.; Jung, M.; Lang, S. Lights, Camera, Interaction: Studying Protein–Protein Interactions of the ER Protein Translocase in Living Cells. Int. J. Mol. Sci. 2021, 22, 10358. https://doi.org/10.3390/ijms221910358
Sicking M, Jung M, Lang S. Lights, Camera, Interaction: Studying Protein–Protein Interactions of the ER Protein Translocase in Living Cells. International Journal of Molecular Sciences. 2021; 22(19):10358. https://doi.org/10.3390/ijms221910358
Chicago/Turabian StyleSicking, Mark, Martin Jung, and Sven Lang. 2021. "Lights, Camera, Interaction: Studying Protein–Protein Interactions of the ER Protein Translocase in Living Cells" International Journal of Molecular Sciences 22, no. 19: 10358. https://doi.org/10.3390/ijms221910358
APA StyleSicking, M., Jung, M., & Lang, S. (2021). Lights, Camera, Interaction: Studying Protein–Protein Interactions of the ER Protein Translocase in Living Cells. International Journal of Molecular Sciences, 22(19), 10358. https://doi.org/10.3390/ijms221910358






