Circulating Exosomal miR-221 from Maternal Obesity Inhibits Angiogenesis via Targeting Angptl2
Abstract
:1. Introduction
2. Results
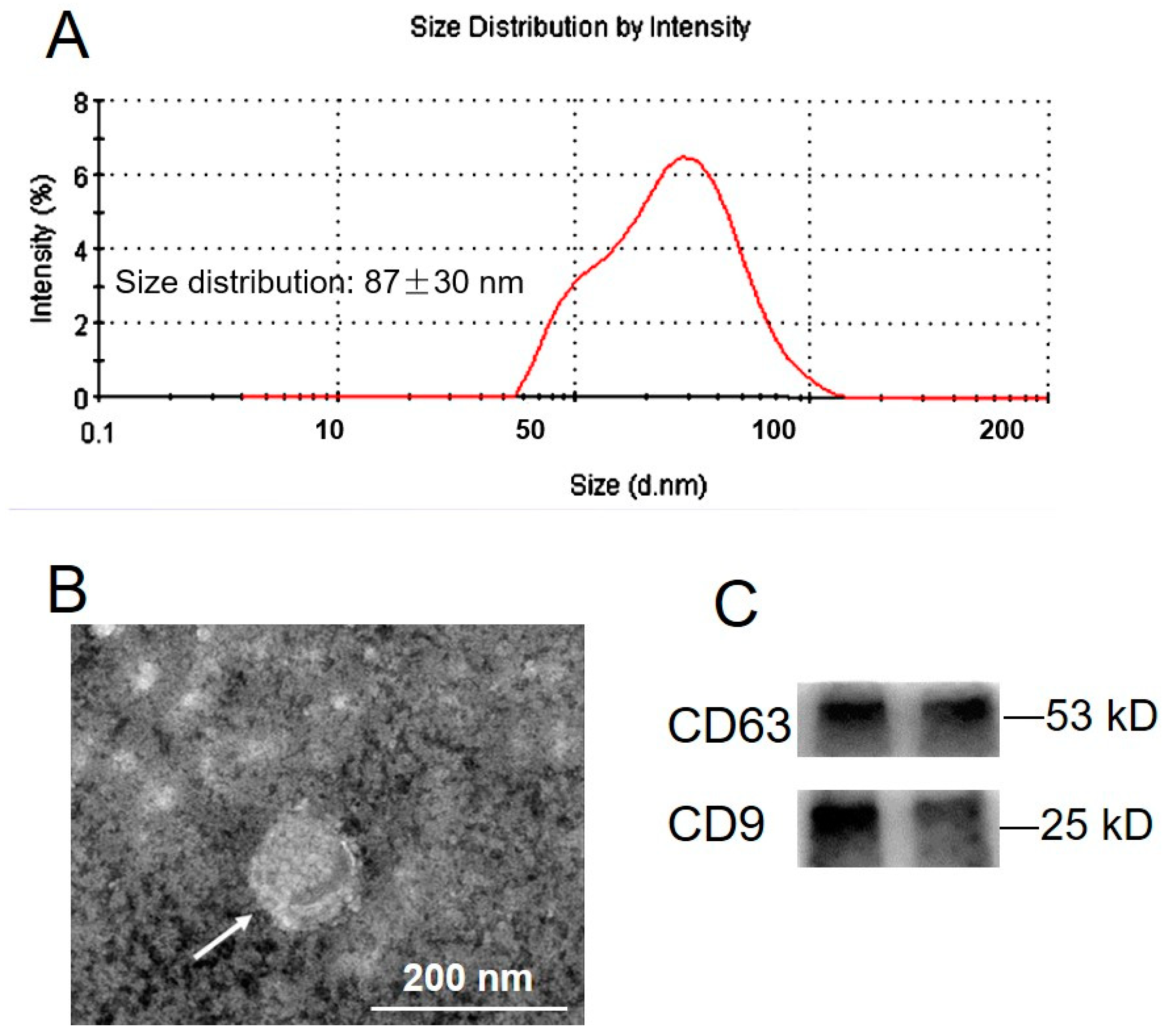
2.1. Exosome Isolation and Characterization
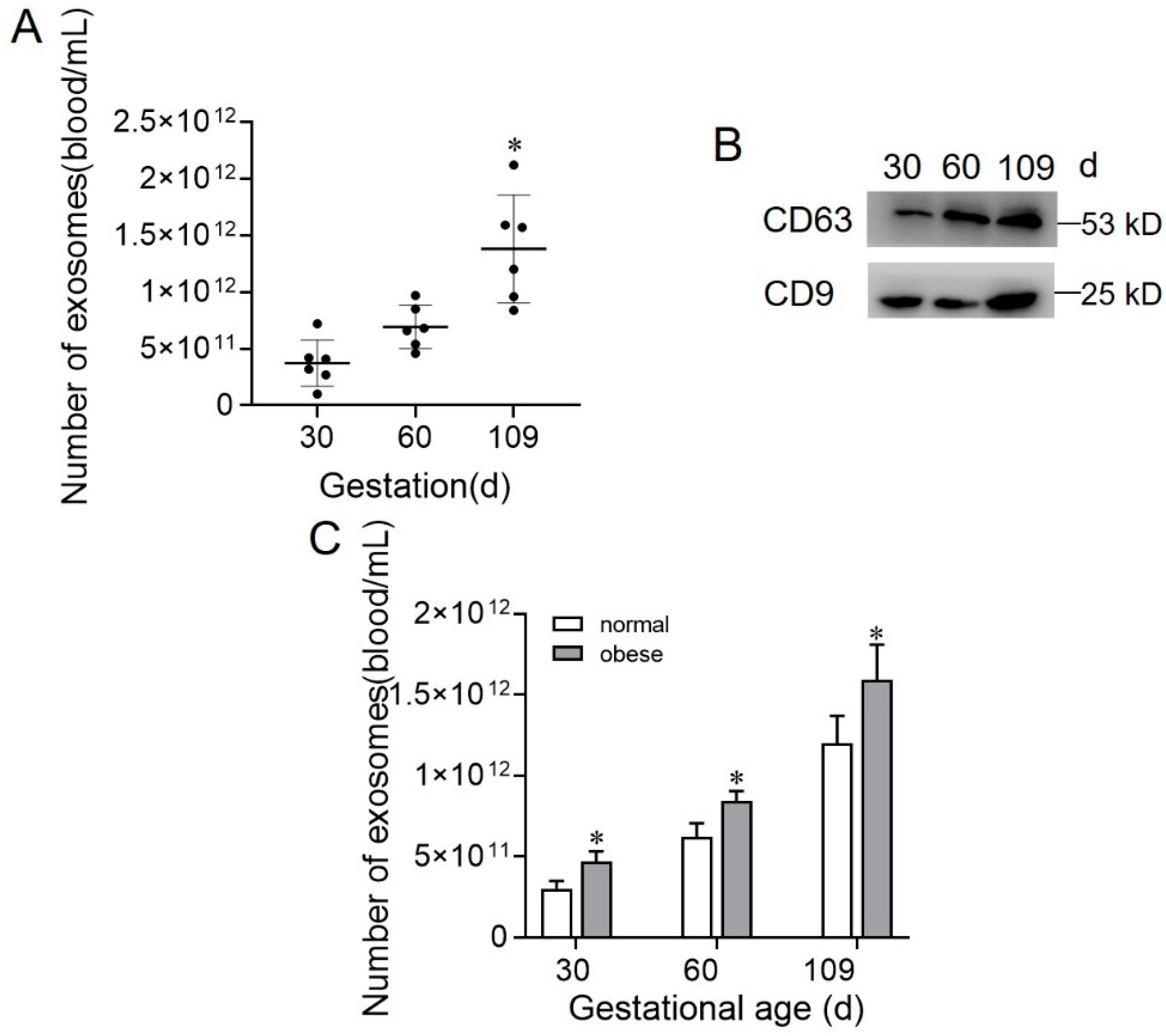
2.2. Exosome Concentration during Gestation Stage in Normal and Obese Sows
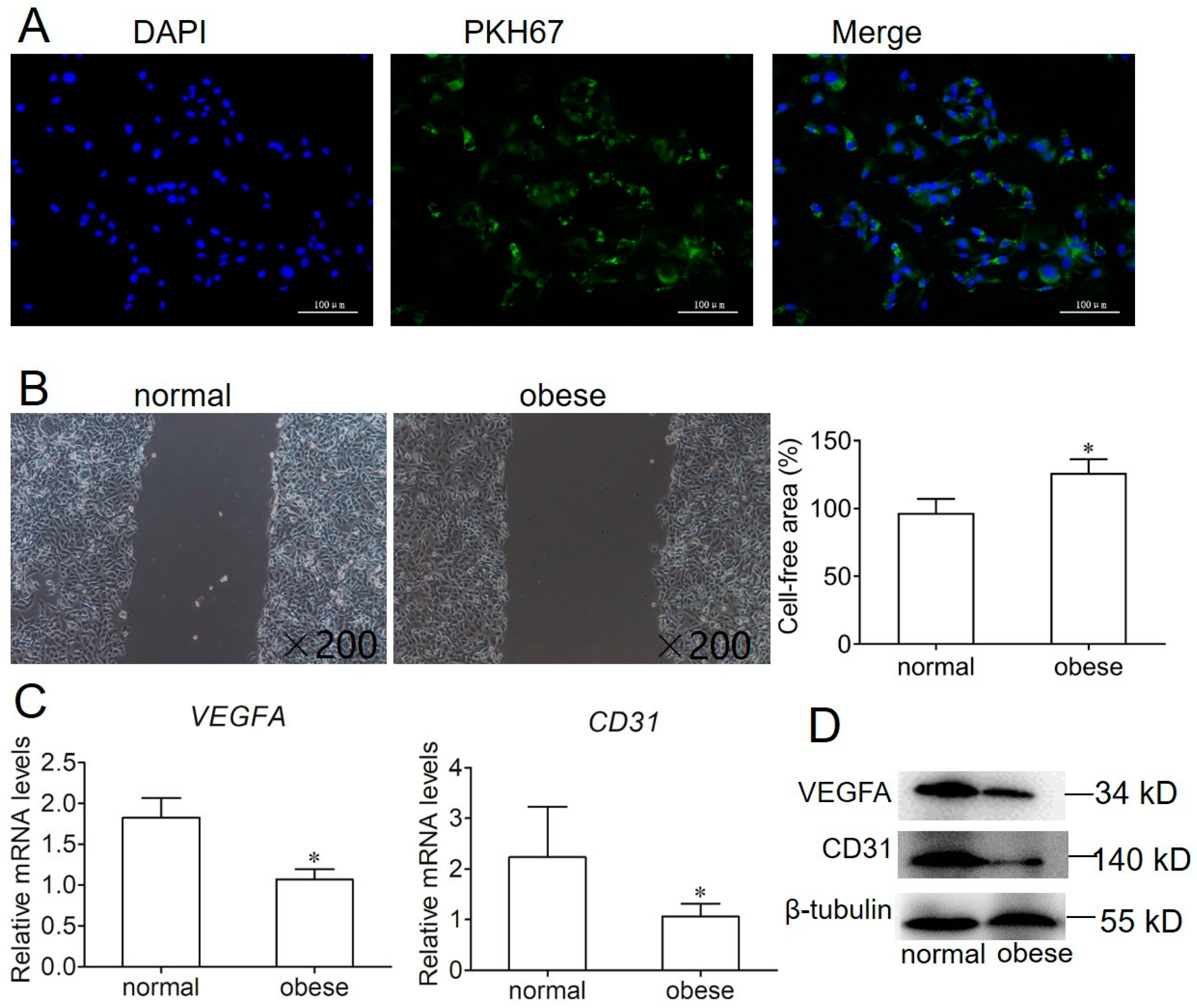
2.3. Effect of Exosome on Endothelial Cell Migration and Angiogenesis
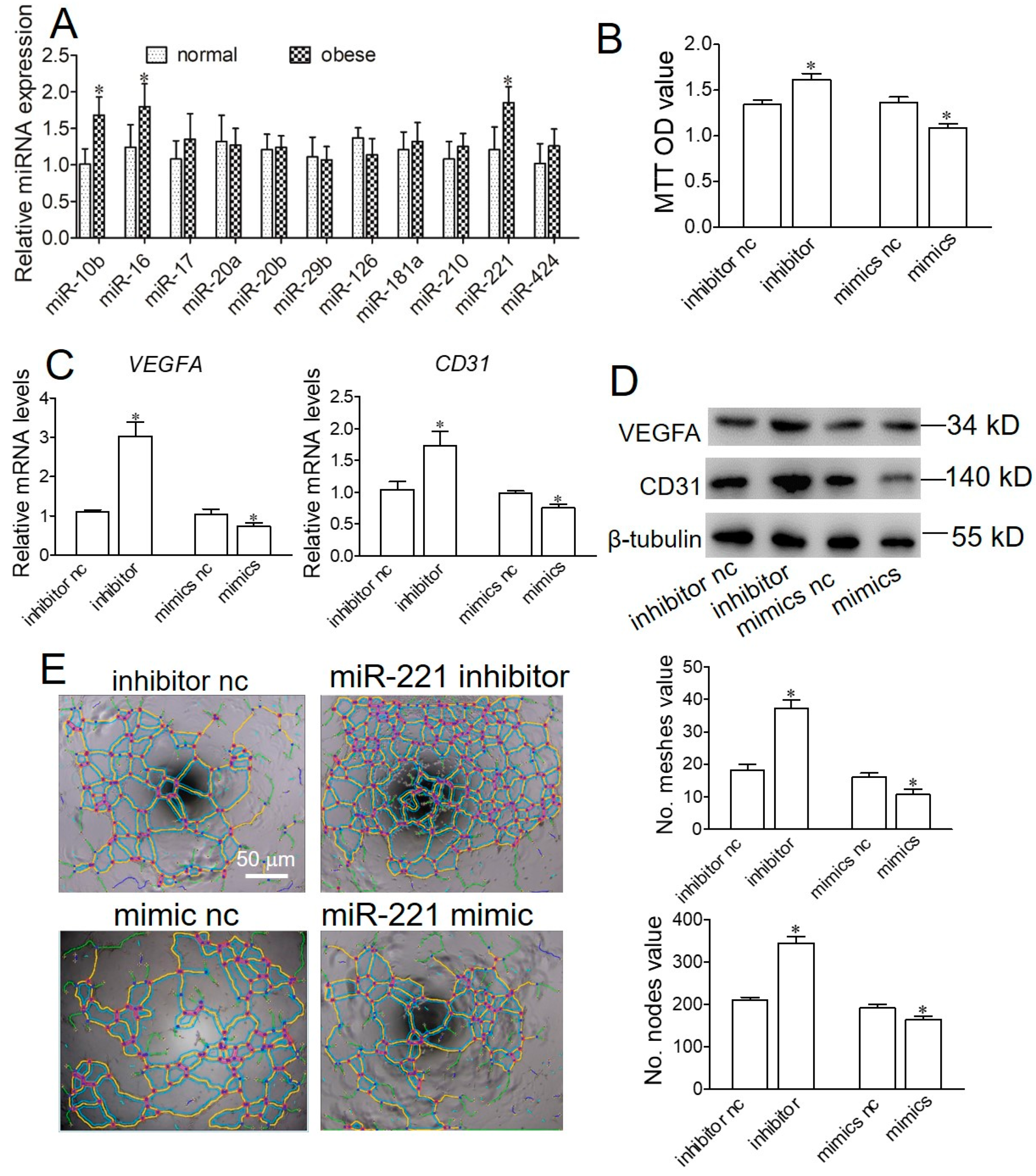
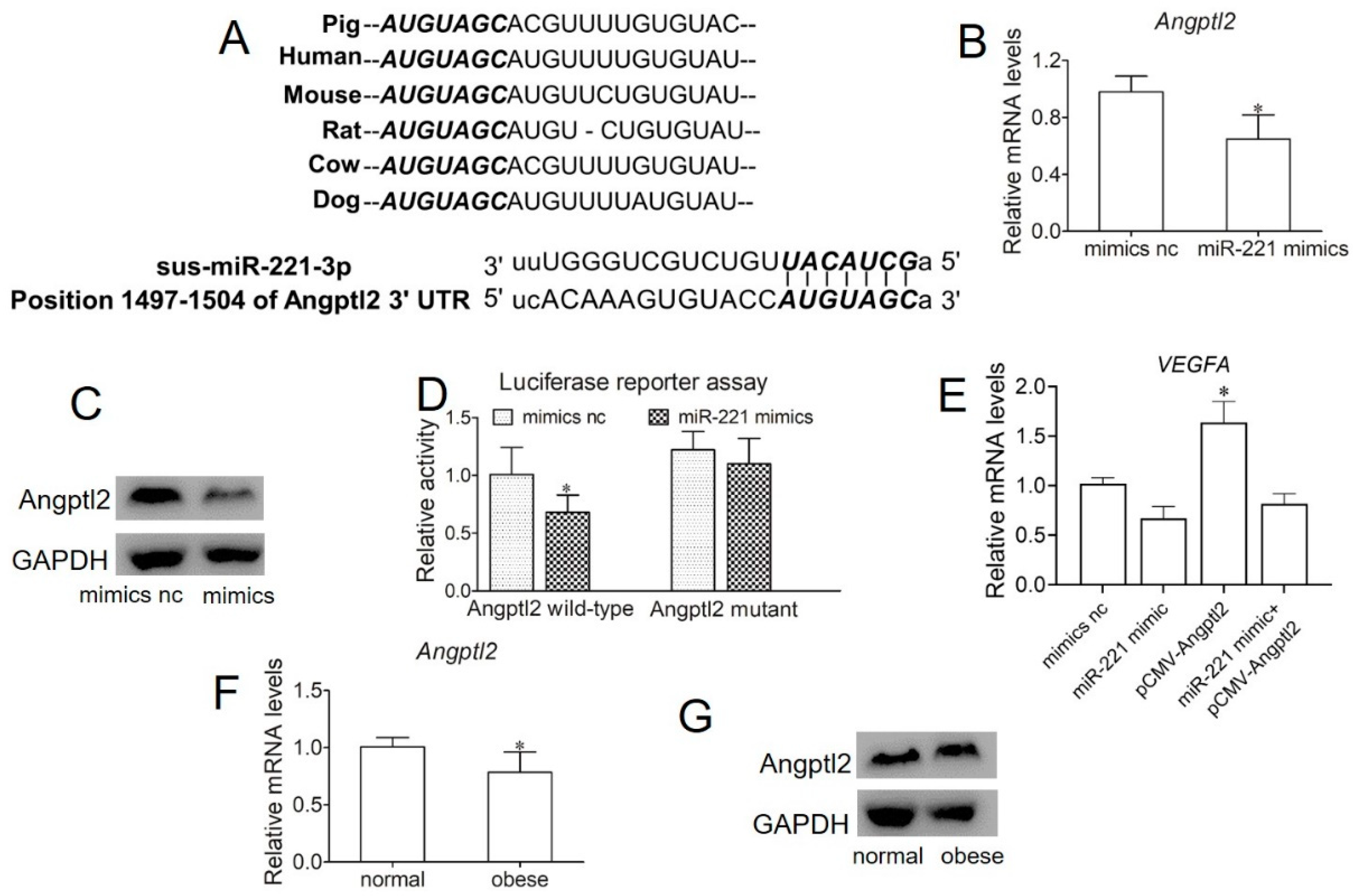
2.4. Obese Sow Exosomal Mir-221 Is Upregulated and Impairs Angiogenesis
2.5. miR-221 Impairs Angiogenesis in Endothelial Cells by Targeting Angptl2
3. Discussion
4. Materials and Methods
4.1. Animal Group and Samples
4.2. Isolation of Exosomes from Plasma Samples
4.3. Exosome Characterization
4.4. Endothelial Cell Culture
4.5. Endothelial Cell Migration
4.6. Cell Proliferation Assay
4.7. miRNA and Plasmid Transfection
4.8. Luciferase Reporter Assay
4.9. Tube Formation Assay
4.10. RNA Extraction, Reverse Transcriptase (RT)-PCR, and Real-Time RT-PCR
4.11. Western Blotting
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Angpl2 | Angiopoietin-like protein 2 |
| IUGR | Intrauterine growth restriction |
| LBW | Low birth weight |
| LGA | Large for gestational age |
| miRNAs | microRNAs |
| NTA | Nanoparticle tracking analysis |
| PPARγ | Peroxisome proliferator-activated receptor γ |
| SGA | Small for gestational age |
| SUVECs | Swine umbilicus veins endothelial cells |
| TEM | Transmission electron microscopy |
| VEGFA | Vascular endothelial growth factor A |
References
- Stang, J.; Huffman, L.G. Position of the academy of nutrition and dietetics: Obesity, reproduction, and pregnancy outcomes. J. Acda. Nutr. Diet. 2016, 116, 677–691. [Google Scholar] [CrossRef] [Green Version]
- Higgins, L.; Greenwood, S.L.; Wareing, M.; Sibley, C.P.; Mills, T.A. Obesity and the placenta: A consideration of nutrient exchange mechanisms in relation to aberrant fetal growth. Placenta 2011, 32, 1–7. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, T.; Cai, A.; Wu, Y.; Wei, H.; Jiang, S.; Peng, J. Excessive backfat of sows at 109 d of gestation induces lipotoxic placental environment and is associated with declining reproductive performance. J. Anim. Sci. 2018, 96, 250–257. [Google Scholar] [CrossRef]
- Kim, J.S.; Yang, X.; Pangeni, D.; Baidoo, S.K. Relationship between backfat thickness of sows during late gestation and reproductive efficiency at different parities. ACTA Agric. Scand. A—Anim. 2015, 65, 1–8. [Google Scholar] [CrossRef]
- Leddy, M.A.; Power, M.L.; Schulkin, J. The impact of maternal obesity on maternal and fetal health. Rev. Obstet. Gynecol. 2008, 1, 170. [Google Scholar] [PubMed]
- Weckman, A.M.; Ngai, M.; Wright, J.; McDonald, C.R.; Kain, K.C. The impact of infection in pregnancy on placental vascular development and adverse birth outcomes. Front. Microbiol. 2019, 10, 1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarvie, E.; Hauguel-de-Mouzon, S.; Nelson, S.M.; Sattar, N.; Catalano, P.M.; Freeman, D.J. Lipotoxicity in obese pregnancy and its potential role in adverse pregnancy outcome and obesity in the offspring. Clin. Sci. 2010, 119, 123–129. [Google Scholar] [CrossRef] [Green Version]
- Saben, J.; Lindsey, F.; Zhong, Y.; Thakali, K.; Badger, T.M.; Andres, A.; Gomez-Acevedo, H.; Shankar, K. Maternal obesity is associated with a lipotoxic placental environment. Placenta 2014, 35, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Lai, Y.; Cao, L.; Li, Y.; Chen, G.; Chen, L. Human umbilical cord mesenchymal stem cells-derived exosomal microRNA-451a represses epithelial–mesenchymal transition of hepatocellular carcinoma cells by inhibiting ADAM10. RNA. Biol. 2020, 1–16. [Google Scholar] [CrossRef]
- Ahima, R.S.; Flier, J.S. Adipose tissue as an endocrine organ. Trends Endocrin. Met. 2000, 11, 327–332. [Google Scholar] [CrossRef]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell 2017, 171, 372–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crewe, C.; Joffin, N.; Rutkowski, J.M.; Kim, M.; Zhang, F.; Towler, D.A.; Gordillo, R.; Scherer, P.E. An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell 2018, 175, 695–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-derived exosomal MiR-27a induces insulin resistance in skeletal muscle through repression of PPARγ. Theranostics 2018, 8, 2171. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.R.; Robbins, E.P.; Vemula, S.; Critser, P.J.; Whittington, C.; Voytik-Harbin, S.L.; Yoder, M.C. Angiopoietin-like protein 2 regulates endothelial colony forming cell vasculogenesis. Angiogenesis 2014, 17, 675–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, S.Y.; Leng, Y.; Wang, Z.X.; Xiao, X.; Zhang, X.; Wen, T.; Gong, H.Z.; Hong, A.; Ma, Y. Exosomal transfer of obesity adipose tissue for decreased miR-141-3p mediate insulin resistance of hepatocytes. Int. J. Biol. Sci. 2019, 15, 351. [Google Scholar] [CrossRef]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef] [PubMed]
- Salomon, C.; Scholz-Romero, K.; Sarker, S.; Sweeney, E.; Kobayashi, M.; Correa, P.; Longo, S.; Duncombe, G.; Mitchell, M.D.; Rice, G.E.; et al. Gestational diabetes mellitus is associated with changes in the concentration and bioactivity of placenta-derived exosomes in maternal circulation across gestation. Diabetes 2016, 65, 598–609. [Google Scholar] [CrossRef] [Green Version]
- Barranco, I.; Padilla, L.; Parrilla, I.; Álvarez-Barrientos, A.; Pérez-Patiño, C.; Peña, F.J.; Martínez, E.A.; Rodriguez-Martínez, H.; Roca, J. Extracellular vesicles isolated from porcine seminal plasma exhibit different tetraspanin expression profiles. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Jayabalan, N.; Nair, S.; Nuzhat, Z.; Rice, G.E.; Zuñiga, F.A.; Sobrevia, L.; Leiva, A.; Sanhueza, C.; Gutiérrez, J.A.; Lappas, M.; et al. Cross Talk between Adipose Tissue and Placenta in Obese and Gestational Diabetes Mellitus Pregnancies via Exosomes. Front. Endocrinol. 2017, 8, 239. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.W.; Lee, M.; Oh, K.J. Adipose tissue-derived signatures for obesity and type 2 diabetes: Adipokines, batokines and microRNAs. J. Clin. Med. 2019, 8, 854. [Google Scholar] [CrossRef] [Green Version]
- Kita, S.; Maeda, N.; Shimomura, I. Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J. Clin. Invest. 2019, 129, 4041–4049. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Invest. 2019, 129, 834–849. [Google Scholar] [CrossRef] [Green Version]
- Otero-Ortega, L.; Laso-García, F.; Gómez-de Frutos, M.; Fuentes, B.; Diekhorst, L.; Díez-Tejedor, E.; Gutiérrez-Fernández, M. Role of exosomes as a treatment and potential biomarker for stroke. Transl. Stroke Res. 2019, 10, 241–249. [Google Scholar] [CrossRef]
- Guanzon, D.; Lai, A.; Scholz-Romero, K.; Zuniga, F.; Diaz, E.; McIntyre, H.D.; Lappas, M.; Salomón, C. Circulating Exosomal miRNA Signature in Pregnancies with Gestational Diabetes Mellitus across Gestation. Diabetes 2018, 67 (Suppl. S1). [Google Scholar] [CrossRef]
- Nicoli, S.; Knyphausen, C.P.; Zhu, L.J.; Lakshmanan, A.; Lawson, N.D. miR-221 is required for endothelial tip cell behaviors during vascular development. Dev. Cell 2012, 22, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Chiu, S.C.; Chiang, E.P.; Tsai, S.Y.; Wang, F.Y.; Pai, M.H.; Syu, J.N.; Cheng, C.C.; Rodriguez, R.L.; Tang, F.Y. Eicosapentaenoic acid induces neovasculogenesis in human endothelial progenitor cells by modulating c-kit protein and PI3-K/Akt/eNOS signaling pathways. J. Nutr. Biochem. 2014, 25, 934–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.G.; Zhou, C.F.; Zhang, Y.M.; Yan, R.M.; Wei, W.F.; Chen, X.J.; Yi, H.Y.; Liang, L.J.; Fan, L.S.; Liang, L. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis 2019, 22, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Yamamoto, G.; Hayashi, J.I.; Nishida, E.; Goto, H.; Sasaki, Y.; Kikuchi, T.; Fukuda, M.; Hasegawa, Y.; Mogi, M. Angiopoietin-like protein 2 regulates Porphyromonas gingivalis lipopolysaccharide-induced inflammatory response in human gingival epithelial cells. PLoS ONE 2017, 12, e0184825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadomatsu, T.; Endo, M.; Miyata, K.; Oike, Y. Diverse roles of ANGPTL2 in physiology and pathophysiology. Trends. Endocrinol. Metab. 2014, 25, 245–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Hu, Z.; Wang, Z.; Cui, Y.; Cui, X. Angiopoietin-like protein 2 is an important facilitator of tumor proliferation, metastasis, angiogenesis and glycolysis in osteosarcoma. Am. J. Transl. Res. 2019, 11, 6341. [Google Scholar] [PubMed]
- Oike, Y.; Urano, T.; Ito, Y.; Fukuhara, S.; Akao, M.; Mochizuki, N. Angiopoietin-Like Protein 2 Induces Angiogenesis, Lymphangiogenesis, and Leukocytes Recruitment, Resulting in Enhancement of Inflammation. Circulation 2006, 114, S18. [Google Scholar]
- He, X.X.; Guo, A.Y.; Xu, C.R.; Chang, Y.; Xiang, G.Y.; Gong, J.; Dan, Z.L.; Tian, D.A.; Liao, J.Z.; Lin, J.S. Bioinformatics analysis identifies miR-221 as a core regulator in hepatocellular carcinoma and its silencing suppresses tumor properties. Oncol. Rep. 2014, 32, 1200–1210. [Google Scholar] [CrossRef]
- Salomon, C.; Torres, M.J.; Kobayashi, M.; Scholz-Romero, K.; Sobrevia, L.; Dobierzewska, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS ONE 2014, 9, e98667. [Google Scholar] [CrossRef] [Green Version]
- Ning, P.; Zhang, Y.; Guo, K.; Chen, R.; Liang, W.; Lin, Z.; Li, H. Discovering up-regulated VEGF-C expression in swine umbilical vein endothelial cells by classical swine fever virus Shimen. Vet. Res. 2014, 45, 48. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.R.; Kim, G.; Tak, W.Y.; Jang, S.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Chun, J.M.; Park, S.Y. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 2019, 144, 1444–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, S.H.; Lee, B.H.; Choi, S.H.; Kim, H.J.; Won, K.J.; Lee, H.M.; Rhim, H.; Kim, H.C.; Nah, S.Y. Effects of gintonin on the proliferation, migration, and tube formation of human umbilical-vein endothelial cells: Involvement of lysophosphatidic-acid receptors and vascular-endothelial-growth-factor signaling. J. Ginseng Res. 2016, 40, 325–333. [Google Scholar] [CrossRef] [Green Version]
- DeCicco-Skinner, K.L.; Henry, G.H.; Cataisson, C.; Tabib, T.; Gwilliam, J.C.; Watson, N.J.; Bullwinkle, E.M.; Falkenburg, L.; O’Neill, R.C.; Morin, A. Endothelial cell tube formation assay for the in vitro study of angiogenesis. J. Vis. Exp. 2014, 91, e51312. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhou, Y.; Deng, Z.; Zhang, H.; Wu, Y.; Song, T.; Yang, Y.; Wei, H.; Peng, J. miR-221 negatively regulates inflammation and insulin sensitivity in white adipose tissue by repression of sirtuin-1 (SIRT1). J. Cell. Biochem. 2018, 119, 6418–6428. [Google Scholar] [CrossRef]
- Zhou, Y.F.; Ren, J.; Song, T.X.; Peng, J.; Wei, H.K. Methionine Regulates mTORC1 via the T1R1/T1R3-PLCβ-Ca2+-ERK1/2 Signal Transduction Process in C2C12 Cells. Int. J. Mol. Sci. 2016, 17, 1684. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Xia, M.; Cui, C.; Wei, H.; Jiang, S.; Peng, J. Circulating Exosomal miR-221 from Maternal Obesity Inhibits Angiogenesis via Targeting Angptl2. Int. J. Mol. Sci. 2021, 22, 10343. https://doi.org/10.3390/ijms221910343
Zhou Y, Xia M, Cui C, Wei H, Jiang S, Peng J. Circulating Exosomal miR-221 from Maternal Obesity Inhibits Angiogenesis via Targeting Angptl2. International Journal of Molecular Sciences. 2021; 22(19):10343. https://doi.org/10.3390/ijms221910343
Chicago/Turabian StyleZhou, Yuanfei, Mao Xia, Chenbin Cui, Hongkui Wei, Siwen Jiang, and Jian Peng. 2021. "Circulating Exosomal miR-221 from Maternal Obesity Inhibits Angiogenesis via Targeting Angptl2" International Journal of Molecular Sciences 22, no. 19: 10343. https://doi.org/10.3390/ijms221910343
APA StyleZhou, Y., Xia, M., Cui, C., Wei, H., Jiang, S., & Peng, J. (2021). Circulating Exosomal miR-221 from Maternal Obesity Inhibits Angiogenesis via Targeting Angptl2. International Journal of Molecular Sciences, 22(19), 10343. https://doi.org/10.3390/ijms221910343





