Arabidopsis LSH8 Positively Regulates ABA Signaling by Changing the Expression Pattern of ABA-Responsive Proteins
Abstract
1. Introduction
2. Results
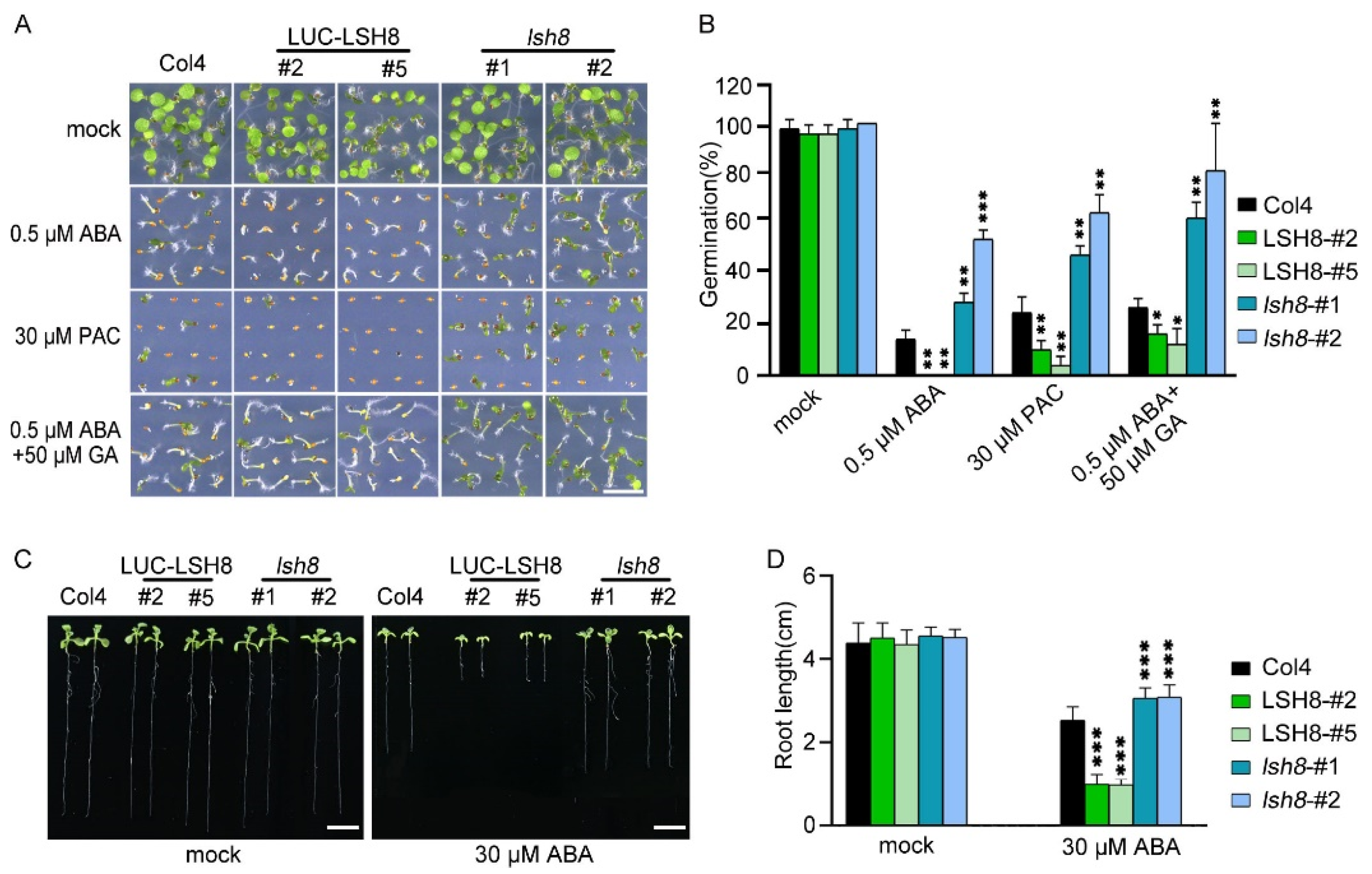
2.1. LSH8 Regulates Seed Germination and the Elongation of Primary and Lateral Root
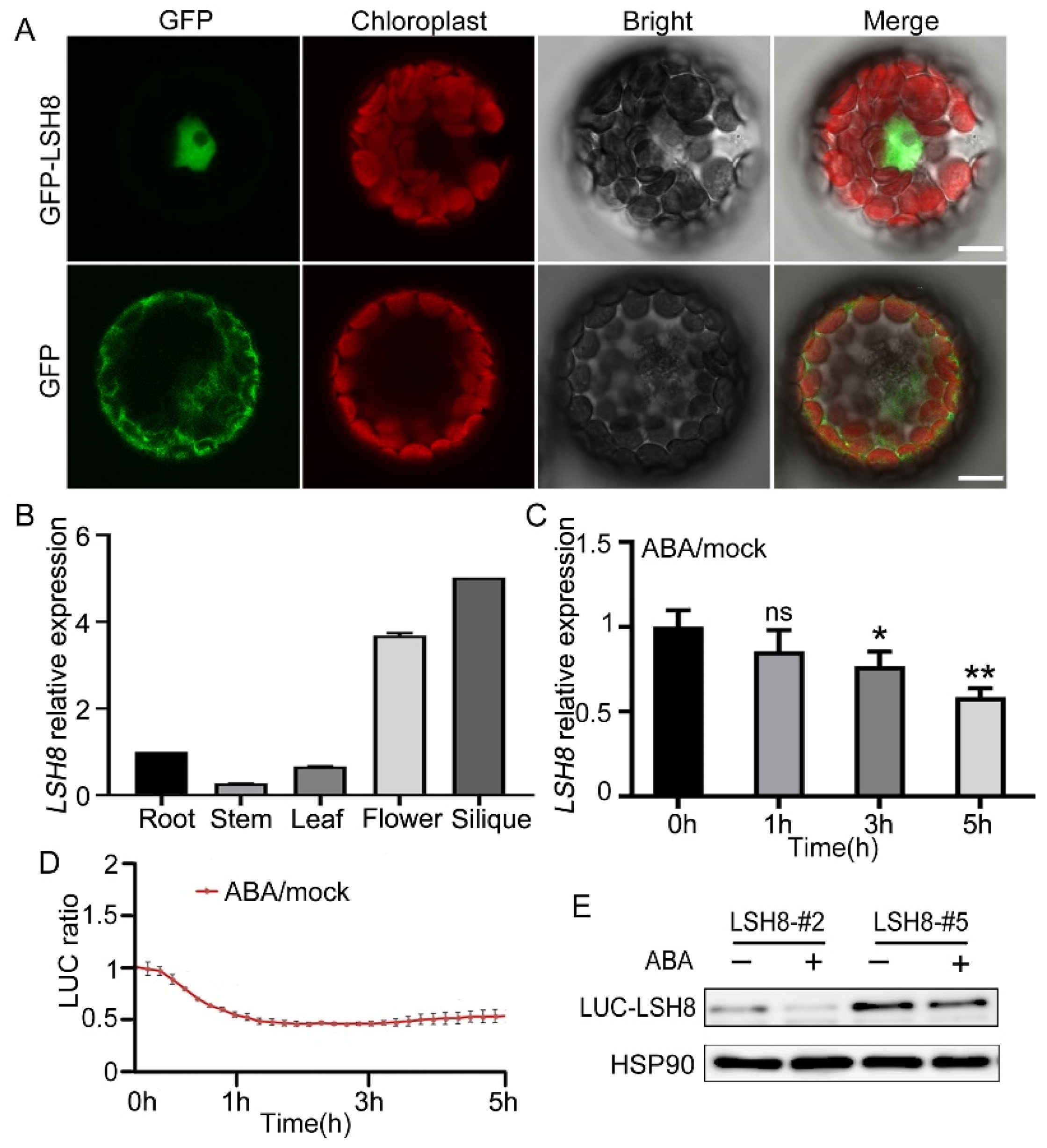
2.2. The Prediction of Upstream Element of LSH8 and the Expression of LSH8 Regulated by ABA
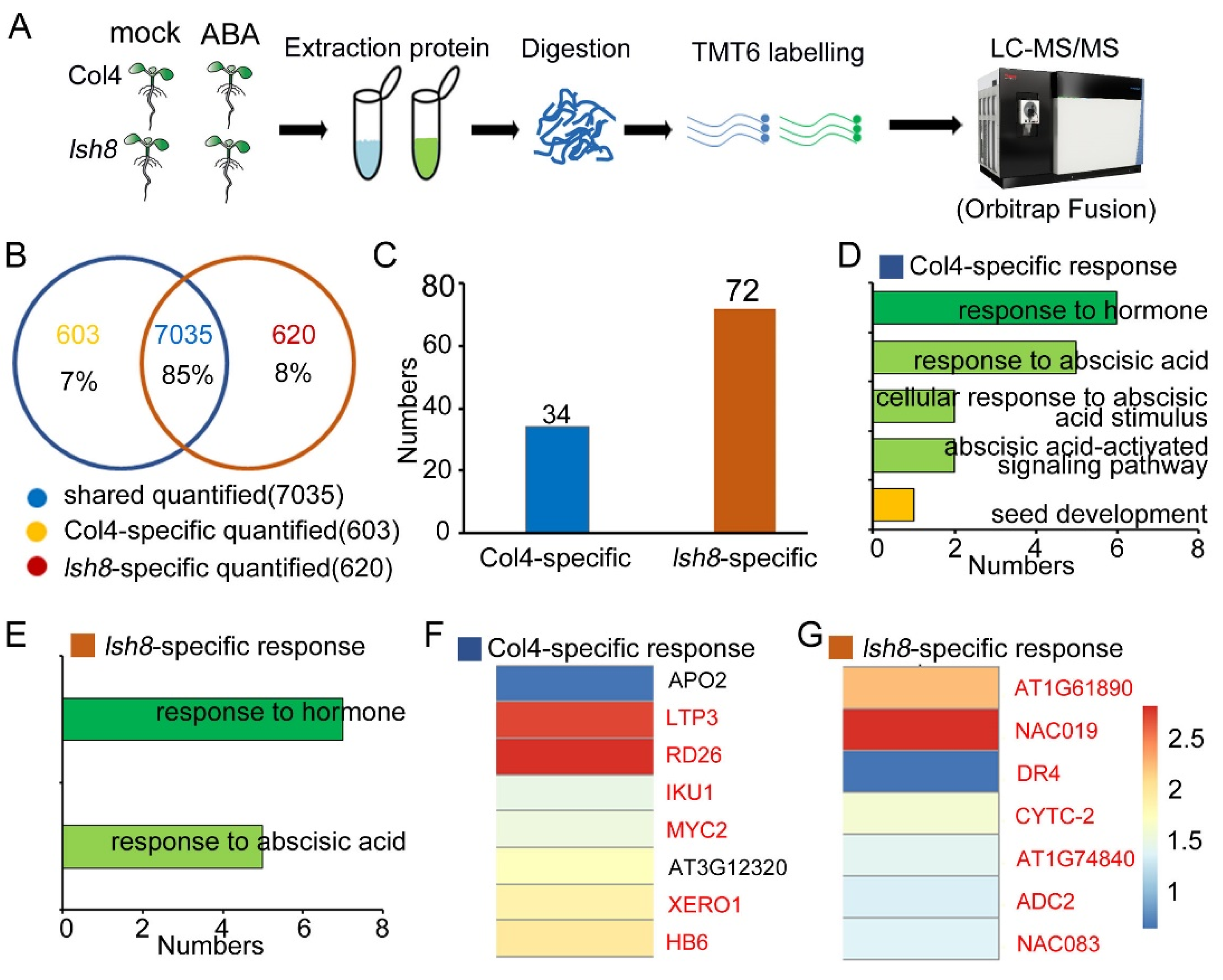
2.3. Quantitative Proteomic Analysis
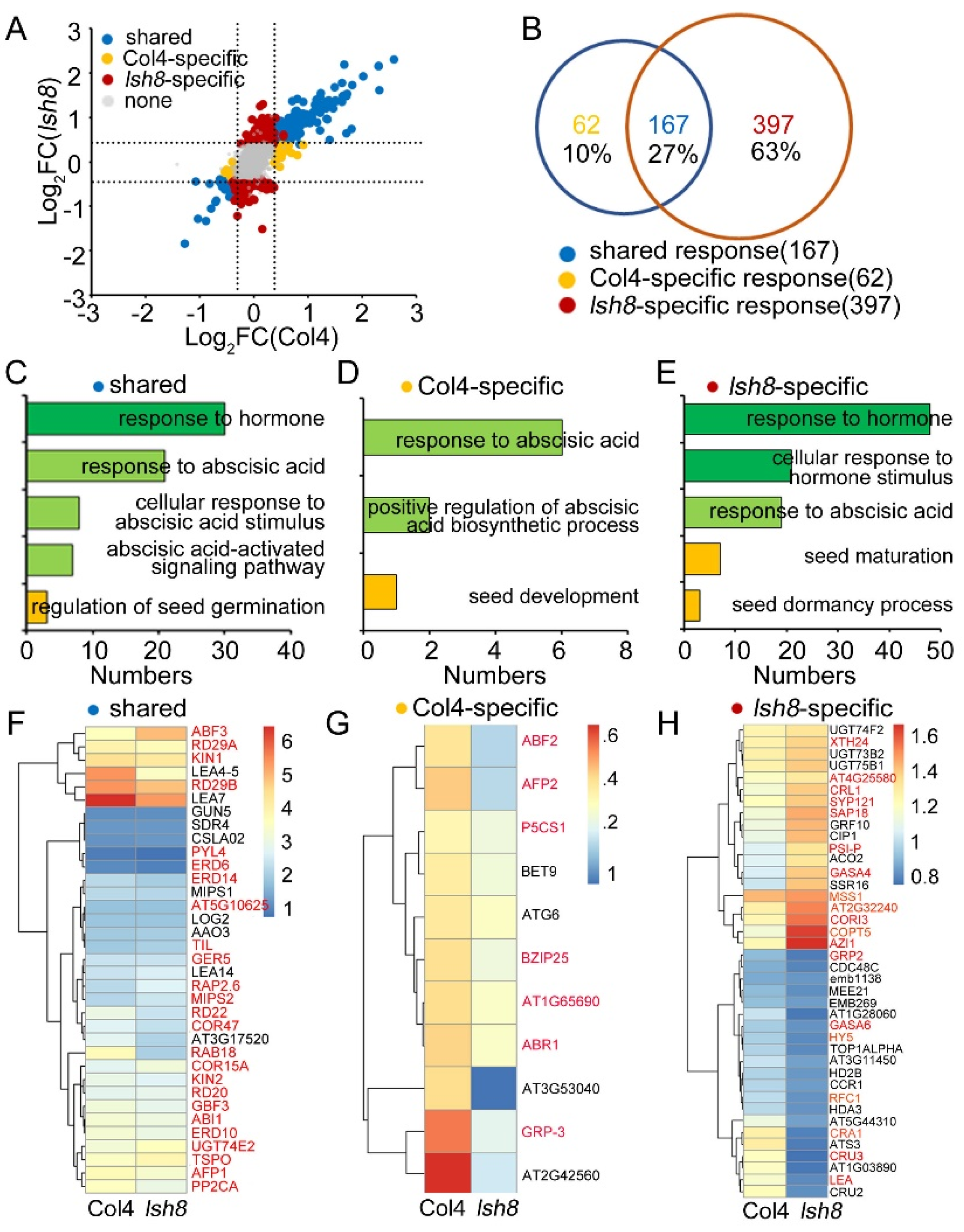
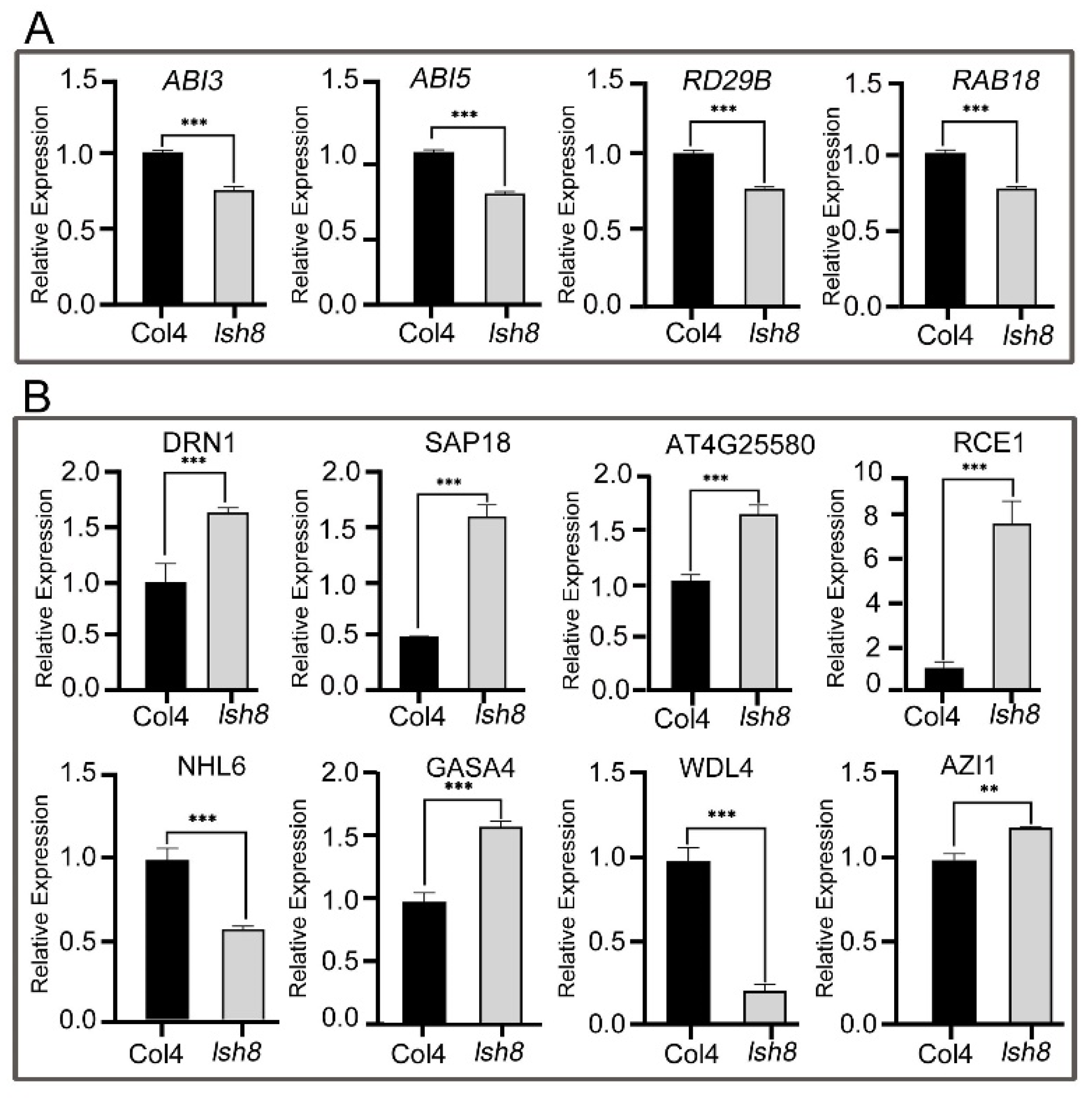
2.4. Functional Analysis of ARPs
2.5. Interaction Network of LSH8-Specific ARPs
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials and Growth Condition
5.2. Seed Germination Assays
5.3. Root Length Analysis
5.4. Confocal Microscopy
5.5. Luminescence Measurement
5.6. Quantitative Real-Time PCR (qPCR) Analysis
5.7. Protein Extraction, Specific-Antibody Preparation and Western Blotting
5.8. Protein Sample Preparation
5.9. TMT-Based Proteomics Analysis
5.10. GO Function Annotation Analysis
5.11. PPI Network Construction
5.12. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statements
Conflicts of Interest
References
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef]
- Verslues, P.E.; Zhu, J.K. New developments in abscisic acid perception and metabolism. Curr. Opin. Plant Biol. 2007, 10, 447–452. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic Acid synthesis and response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, W.; Wang, X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017, 15, 4–14. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol 2012, 160, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Giraudat, J.; Hauge, B.M.; Valon, C.; Smalle, J.; Parcy, F.; Goodman, H.M. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 1992, 4, 1251–1261. [Google Scholar]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Söderman, E.M.; Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000, 124, 1752–1765. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Xue, Q.; McCray, T.; Margavage, K.; Chen, F.; Lee, J.H.; Nezames, C.D.; Guo, L.; Terzaghi, W.; Wan, J.; et al. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 2013, 25, 517–534. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhang, P.; Ho, T.H. Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 1996, 8, 1107–1119. [Google Scholar] [PubMed]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription Factors and Plants Response to Drought Stress: Current Understanding and Future Directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. Cell Mol. Biol. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. Cell Mol. Biol. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Zhao, L.; Nakazawa, M.; Takase, T.; Manabe, K.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Matsui, M. Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J. Cell Mol. Biol. 2004, 37, 694–706. [Google Scholar] [CrossRef]
- Yoshida, A.; Suzaki, T.; Tanaka, W.; Hirano, H.Y. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 2009, 106, 20103–20108. [Google Scholar] [CrossRef]
- Chen, F.; Zhou, Q.; Wu, L.; Li, F.; Liu, B.; Zhang, S.; Zhang, J.; Bao, M.; Liu, G. Genome-wide identification and characterization of the ALOG gene family in Petunia. BMC Plant Biol. 2019, 19, 600. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. Cell Mol. Biol. 2011, 66, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Aravind, L. ALOG domains: Provenance of plant homeotic and developmental regulators from the DNA-binding domain of a novel class of DIRS1-type retroposons. Biol. Direct 2012, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Liu, L.; Fang, J.; Zhao, J.; Yuan, S.; Li, X. The rice TRIANGULAR HULL1 protein acts as a transcriptional repressor in regulating lateral development of spikelet. Sci. Rep. 2017, 7, 13712. [Google Scholar] [CrossRef]
- Hibara, K.; Takada, S.; Tasaka, M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. Cell Mol. Biol. 2003, 36, 687–696. [Google Scholar] [CrossRef]
- Kamiuchi, Y.; Yamamoto, K.; Furutani, M.; Tasaka, M.; Aida, M. The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Front. Plant Sci. 2014, 5, 165. [Google Scholar] [CrossRef]
- Press, M.O.; Queitsch, C. Variability in a Short Tandem Repeat Mediates Complex Epistatic Interactions in Arabidopsis thaliana. Genetics 2017, 205, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Schachtschabel, J.; Mishkind, M.; Munnik, T.; Arisz, S.A. Hot topic: Thermosensing in plants. Plant Cell Environ. 2021, 44, 2018–2033. [Google Scholar] [CrossRef]
- Yoshida, A.; Sasao, M.; Yasuno, N.; Takagi, K.; Daimon, Y.; Chen, R.; Yamazaki, R.; Tokunaga, H.; Kitaguchi, Y.; Sato, Y.; et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA 2013, 110, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ren, Z.; Cheng, C.; Wang, T.; Ji, H.; Zhao, Y.; Deng, Z.; Zhi, L.; Lu, J.; Wu, X.; et al. Counteraction of ABA-Mediated Inhibition of Seed Germination and Seedling Establishment by ABA Signaling Terminator in Arabidopsis. Mol. Plant 2020, 13, 1284–1297. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar] [PubMed]
- Zheng, J.; Chen, F.; Wang, Z.; Cao, H.; Li, X.; Deng, X.; Soppe, W.J.J.; Li, Y.; Liu, Y. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012, 193, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Han, X.; Yang, J.; Jiang, Y.; Hu, Y. The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell 2021. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef]
- Bunsick, M.; Toh, S.; Wong, C.; Xu, Z.; Ly, G.; McErlean, C.S.P.; Pescetto, G.; Nemrish, K.E.; Sung, P.; Li, J.D.; et al. SMAX1-dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat. Plants 2020, 6, 646–652. [Google Scholar] [CrossRef]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef]
- Wang, X.; Guo, C.; Peng, J.; Li, C.; Wan, F.; Zhang, S.; Zhou, Y.; Yan, Y.; Qi, L.; Sun, K.; et al. ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 2019, 221, 341–355. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C.P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020, 228, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Lång, V.; Palva, E.T. The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 1992, 20, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Wang, C.; Kong, X.; Chen, Q.; Yang, Y.; Hu, X. AFP2 as the novel regulator breaks high-temperature-induced seeds secondary dormancy through ABI5 and SOM in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 501, 232–238. [Google Scholar] [CrossRef]
- Costa, M.C.; Righetti, K.; Nijveen, H.; Yazdanpanah, F.; Ligterink, W.; Buitink, J.; Hilhorst, H.W. A gene co-expression network predicts functional genes controlling the re-establishment of desiccation tolerance in germinated Arabidopsis thaliana seeds. Planta 2015, 242, 435–449. [Google Scholar] [CrossRef]
- Mangeon, A.; Pardal, R.; Menezes-Salgueiro, A.D.; Duarte, G.L.; de Seixas, R.; Cruz, F.P.; Cardeal, V.; Magioli, C.; Ricachenevsky, F.K.; Margis, R.; et al. AtGRP3 Is Implicated in Root Size and Aluminum Response Pathways in Arabidopsis. PLoS ONE 2016, 11, e0150583. [Google Scholar] [CrossRef]
- Huang, K.C.; Lin, W.C.; Cheng, W.H. Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 2018, 18, 40. [Google Scholar] [CrossRef]
- Bouain, N.; Satbhai, S.B.; Korte, A.; Saenchai, C.; Desbrosses, G.; Berthomieu, P.; Busch, W.; Rouached, H. Natural allelic variation of the AZI1 gene controls root growth under zinc-limiting condition. PLoS Genet. 2018, 14, e1007304. [Google Scholar] [CrossRef]
- Becerra, C.; Puigdomenech, P.; Vicient, C.M. Computational and experimental analysis identifies Arabidopsis genes specifically expressed during early seed development. BMC Genom. 2006, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, B.C.; Xu, Y.; Zhu, Y.X. Systematic studies of 12S seed storage protein accumulation and degradation patterns during Arabidopsis seed maturation and early seedling germination stages. J. Biochem. Mol. Biol. 2007, 40, 373–381. [Google Scholar] [CrossRef]
- Ghelis, T.; Bolbach, G.; Clodic, G.; Habricot, Y.; Miginiac, E.; Sotta, B.; Jeannette, E. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 2008, 148, 1668–1680. [Google Scholar] [CrossRef]
- Hu, Y.; Zhou, L.; Yang, Y.; Zhang, W.; Chen, Z.; Li, X.; Qian, Q.; Kong, F.; Li, Y.; Liu, X.; et al. The gibberellin signaling negative regulator RGA-LIKE3 promotes seed storage protein accumulation. Plant Physiol. 2021, 185, 1697–1707. [Google Scholar] [CrossRef]
- Gao, C.; Qi, S.; Liu, K.; Li, D.; Jin, C.; Li, Z.; Huang, G.; Hai, J.; Zhang, M.; Chen, M. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 63–70. [Google Scholar] [CrossRef]
- Chibani, K.; Ali-Rachedi, S.; Job, C.; Job, D.; Jullien, M.; Grappin, P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol. 2006, 142, 1493–1510. [Google Scholar] [CrossRef] [PubMed]
- Savadogo, E.H.; Shiomi, Y.; Yasuda, J.; Akino, T.; Yamaguchi, M.; Yoshida, H.; Umegawachi, T.; Tanaka, R.; Suong, D.N.A.; Miura, K.; et al. Gene expression of PLAT and ATS3 proteins increases plant resistance to insects. Planta 2021, 253, 37. [Google Scholar] [CrossRef]
- Zhu, B.; Cai, G.; Hall, E.O.; Freeman, G.J. In-fusion assembly: Seamless engineering of multidomain fusion proteins, modular vectors, and mutations. BioTechniques 2007, 43, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, N.; Chen, P.; Gao, M.; Liu, J.; Wang, Y.; Zhao, T.; Li, Y.; Gai, J. Overexpression of a soybean ariadne-like ubiquitin ligase gene GmARI1 enhances aluminum tolerance in Arabidopsis. PLoS ONE 2014, 9, e111120. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Wang, W.; Chen, Q.; Xu, S.; Liu, W.C.; Zhu, X.; Song, C.P. Trehalose-6-phosphate phosphatase E modulates ABA-controlled root growth and stomatal movement in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1518–1534. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, J.; Xu, Y.; Jiang, H.; Wu, X.; Li, C. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 2007, 65, 655–665. [Google Scholar] [CrossRef]
- Xu, H.; Lantzouni, O.; Bruggink, T.; Benjamins, R.; Lanfermeijer, F.; Denby, K.; Schwechheimer, C.; Bassel, G.W. A Molecular Signal Integration Network Underpinning Arabidopsis Seed Germination. Curr. Biol. 2020, 30, 3703–3712.e3704. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.L.; Chen, F.; Zou, X.L.; Shen, S.S.; Wang, X.G.; Yao, G.X.; Xu, B.B. Graphene oxide and ABA cotreatment regulates root growth of Brassica napus L. by regulating IAA/ABA. J. Plant Physiol. 2019, 240, 153007. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, G.; Li, Y.; Kong, X.; Zhang, L.; Wang, J.; Li, X.; Yang, Y. ABA Receptor Subfamily III Enhances Abscisic Acid Sensitivity and Improves the Drought Tolerance of Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1938. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.J.; Gao, J.; Wang, P.; Duan, C.G.; Zhu, X.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Asai, S.; Furzer, O.J.; Cevik, V.; Kim, D.S.; Ishaque, N.; Goritschnig, S.; Staskawicz, B.J.; Shirasu, K.; Jones, J.D.G. A downy mildew effector evades recognition by polymorphism of expression and subcellular localization. Nat. Commun. 2018, 9, 5192. [Google Scholar] [CrossRef]
- Wang, Y.; Kang, H.; Liu, X.; Tong, Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J. Med. Virol. 2020, 92, 538–539. [Google Scholar] [CrossRef]
- Guo, R.; Guo, H.; Zhang, Q.; Guo, M.; Xu, Y.; Zeng, M.; Lv, P.; Chen, X.; Yang, M. Evaluation of reference genes for RT-qPCR analysis in wild and cultivated Cannabis. Biosci. Biotechnol. Biochem. 2018, 82, 1902–1910. [Google Scholar] [CrossRef]
- Ludwig, K.R.; Schroll, M.M.; Hummon, A.B. Comparison of In-Solution, FASP, and S-Trap Based Digestion Methods for Bottom-Up Proteomic Studies. J. Proteome Res. 2018, 17, 2480–2490. [Google Scholar] [CrossRef] [PubMed]
- Cen, W.; Liu, J.; Lu, S.; Jia, P.; Yu, K.; Han, Y.; Li, R.; Luo, J. Comparative proteomic analysis of QTL CTS-12 derived from wild rice (Oryza rufipogon Griff.), in the regulation of cold acclimation and de-acclimation of rice (Oryza sativa L.) in response to severe chilling stress. BMC Plant Biol. 2018, 18, 163. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zhang, M.; Li, T.; Qin, X. Serum Proteomic Analysis by Tandem Mass Tags (TMT) Based Quantitative Proteomics in Gastric Cancer Patients. Clin. Lab. 2018, 64, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, J.; Li, Z.; Tang, H.; Zhang, L.; Li, J.; Li, Y.; Yao, N.; Li, Y.; Yang, D.; Zuo, Z. Arabidopsis LSH8 Positively Regulates ABA Signaling by Changing the Expression Pattern of ABA-Responsive Proteins. Int. J. Mol. Sci. 2021, 22, 10314. https://doi.org/10.3390/ijms221910314
Zou J, Li Z, Tang H, Zhang L, Li J, Li Y, Yao N, Li Y, Yang D, Zuo Z. Arabidopsis LSH8 Positively Regulates ABA Signaling by Changing the Expression Pattern of ABA-Responsive Proteins. International Journal of Molecular Sciences. 2021; 22(19):10314. https://doi.org/10.3390/ijms221910314
Chicago/Turabian StyleZou, Jinpeng, Zhifang Li, Haohao Tang, Li Zhang, Jingdu Li, Yuhong Li, Nan Yao, Yaxing Li, Deguang Yang, and Zecheng Zuo. 2021. "Arabidopsis LSH8 Positively Regulates ABA Signaling by Changing the Expression Pattern of ABA-Responsive Proteins" International Journal of Molecular Sciences 22, no. 19: 10314. https://doi.org/10.3390/ijms221910314
APA StyleZou, J., Li, Z., Tang, H., Zhang, L., Li, J., Li, Y., Yao, N., Li, Y., Yang, D., & Zuo, Z. (2021). Arabidopsis LSH8 Positively Regulates ABA Signaling by Changing the Expression Pattern of ABA-Responsive Proteins. International Journal of Molecular Sciences, 22(19), 10314. https://doi.org/10.3390/ijms221910314





