Identification of Estrogen Signaling in a Prioritization Study of Intraocular Pressure-Associated Genes
Abstract
1. Introduction
2. Results
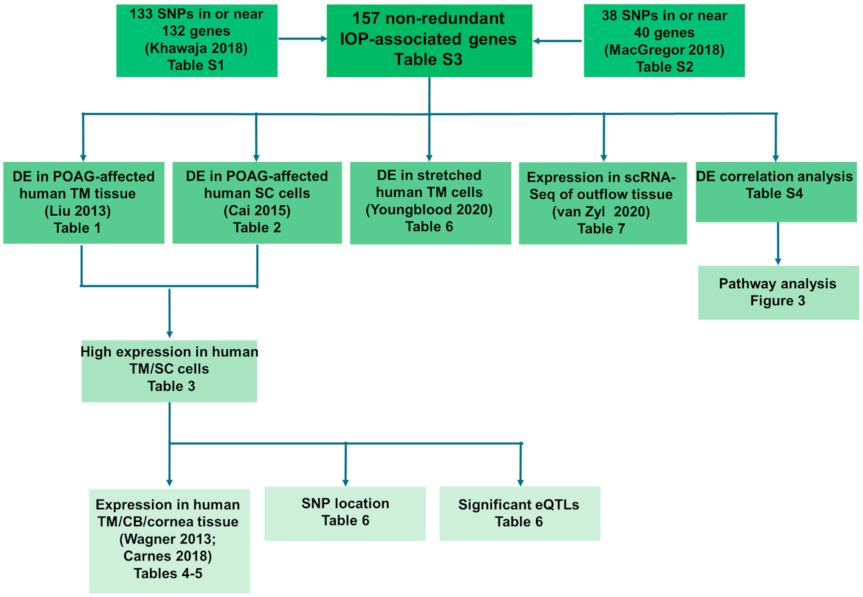
2.1. Selection of Genes for Prioritization
2.2. Gene Prioritization
2.2.1. Differential Expression in POAG-Affected Human TM Tissue or SC Cells
2.2.2. High Expression in RNA-Seq of Primary Human TM and SC Cells
2.2.3. High Expression in Human Ocular Tissue
2.2.4. Variant Impacts on Expression Due to Location in Regulatory Regions
2.2.5. Differential Expression in Cyclically Stretched Primary Human TM Cells
2.2.6. Differential Expression in Outflow Pathway scRNA-Seq
2.2.7. Differential Expression Correlation Analysis
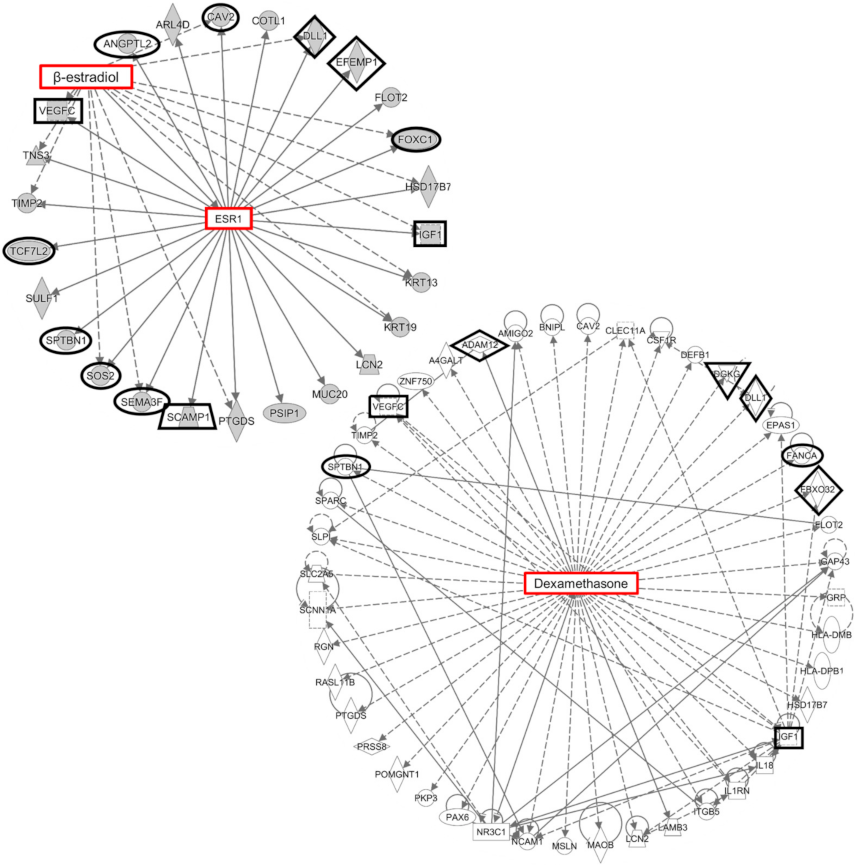
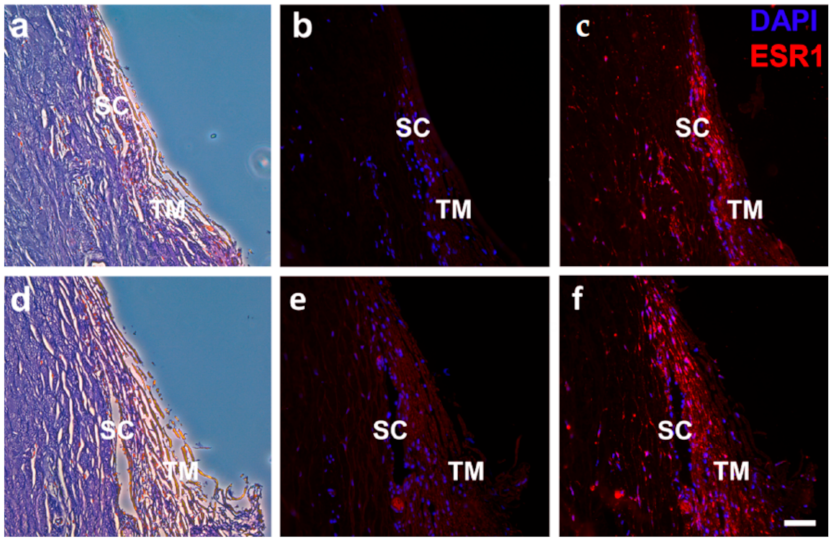
2.3. Estrogen Signaling in the Human Outflow Pathway
3. Discussion
3.1. Overview
3.2. IOP-Associated Gene Prioritization
3.3. Estrogen Signaling in IOP Regulation
3.4. Limitations and Future Work
3.5. Conclusions
4. Materials and Methods
4.1. Gene Prioritization Using RNA-Sequencing and Previously Published Datasets
4.2. Differential Expression Correlation and Network Analyses
4.3. Reagents
4.4. RNA-Sequencing
4.5. Aqueous Humor Estradiol Quantification
4.6. Quantitative Real-Time PCR
4.7. Immunofluorescence
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quigley, H.A.; Broman, A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 2006, 90, 262–267. [Google Scholar] [CrossRef]
- Tham, Y.C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef]
- Jonas, J.B.; Aung, T.; Bourne, R.R.; Bron, A.M.; Ritch, R.; Panda-Jonas, S. Glaucoma. Lancet 2017, 390, 2183–2193. [Google Scholar] [CrossRef]
- Weinreb, R.N.; Khaw, P.T. Primary open-angle glaucoma. Lancet 2004, 363, 1711–1720. [Google Scholar] [CrossRef]
- Allingham, R.R.; Liu, Y.; Rhee, D.J. The genetics of primary open-angle glaucoma: A review. Exp. Eye Res. 2009, 88, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Quigley, H.A. Glaucoma. Lancet 2011, 377, 1367–1377. [Google Scholar] [CrossRef]
- Weinreb, R.N.; Aung, T.; Medeiros, F.A. The pathophysiology and treatment of glaucoma: A review. JAMA 2014, 311, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.J.; Wiggs, J.L. Glaucoma: Genes, phenotypes, and new directions for therapy. J. Clin. Investig. 2010, 120, 3064–3072. [Google Scholar] [CrossRef]
- Liu, Y.; Allingham, R.R. Molecular genetics in glaucoma. Exp. Eye Res. 2011, 93, 331–339. [Google Scholar] [CrossRef]
- Liu, Y.; Allingham, R.R. Major review: Molecular genetics of primary open-angle glaucoma. Exp. Eye Res. 2017, 160, 62–84. [Google Scholar] [CrossRef]
- Brubaker, R.F. The flow of aqueous humor in the human eye. Trans. Am. Ophthalmol. Soc. 1982, 80, 391–474. [Google Scholar]
- Bill, A. Editorial: The drainage of aqueous humor. Investig. Ophthalmol. 1975, 14, 1–3. [Google Scholar]
- Stamer, W.D. The cell and molecular biology of glaucoma: Mechanisms in the conventional outflow pathway. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2470–2472. [Google Scholar] [CrossRef] [PubMed]
- Stamer, W.D.; Acott, T.S. Current understanding of conventional outflow dysfunction in glaucoma. Curr. Opin. Ophthalmol. 2012, 23, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Stamer, W.D.; Clark, A.F. The many faces of the trabecular meshwork cell. Exp. Eye Res. 2017, 158, 112–123. [Google Scholar] [CrossRef] [PubMed]
- van Koolwijk, L.M.; Ramdas, W.D.; Ikram, M.K.; Jansonius, N.M.; Pasutto, F.; Hysi, P.G.; Macgregor, S.; Janssen, S.F.; Hewitt, A.W.; Viswanathan, A.C.; et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012, 8, e1002611. [Google Scholar] [CrossRef] [PubMed]
- Blue Mountains Eye Study (BMES); Wellcome Trust Case Control Consortium 2 (WTCCC2). Genome-wide association study of intraocular pressure identifies the GLCCI1/ICA1 region as a glaucoma susceptibility locus. Hum. Mol. Genet. 2013, 22, 4653–4660. [Google Scholar] [CrossRef]
- Hysi, P.G.; Cheng, C.Y.; Springelkamp, H.; Macgregor, S.; Bailey, J.N.C.; Wojciechowski, R.; Vitart, V.; Nag, A.; Hewitt, A.W.; Höhn, R.; et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat. Genet. 2014, 46, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Nag, A.; Venturini, C.; Small, K.S.; Young, T.L.; Viswanathan, A.C.; Mackey, D.A.; Hysi, P.G.; Hammond, C. A genome-wide association study of intra-ocular pressure suggests a novel association in the gene FAM125B in the TwinsUK cohort. Hum. Mol. Genet. 2014, 23, 3343–3348. [Google Scholar] [CrossRef]
- Ozel, A.B.; Moroi, S.E.; Reed, D.M.; Nika, M.; Schmidt, C.M.; Akbari, S.; Scott, K.; Rozsa, F.; Pawar, H.; Musch, D.C.; et al. Genome-wide association study and meta-analysis of intraocular pressure. Hum. Genet. 2014, 133, 41–57. [Google Scholar] [CrossRef]
- Chen, Y.; Hughes, G.; Chen, X.; Qian, S.; Cao, W.; Wang, L.; Wang, M.; Sun, X. Genetic Variants Associated With Different Risks for High Tension Glaucoma and Normal Tension Glaucoma in a Chinese Population. Investig. Ophthalmol. Vis. Sci. 2015, 56, 2595–2600. [Google Scholar] [CrossRef]
- Springelkamp, H.; Iglesias, A.I.; Cuellar-Partida, G.; Amin, N.; Burdon, K.P.; van Leeuwen, E.M.; Gharahkhani, P.; Mishra, A.; van der Lee, S.J.; Hewitt, A.W.; et al. ARHGEF12 influences the risk of glaucoma by increasing intraocular pressure. Hum. Mol. Genet. 2015, 24, 2689–2699. [Google Scholar] [CrossRef]
- Springelkamp, H.; Iglesias, A.I.; Mishra, A.; Höhn, R.; Wojciechowski, R.; Khawaja, A.P.; Nag, A.; Wang, Y.X.; Wang, J.J.; Cuellar-Partida, G.; et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum. Mol. Genet. 2017, 26, 438–453. [Google Scholar] [CrossRef]
- Khawaja, A.P.; Cooke Bailey, J.N.; Wareham, N.J.; Scott, R.A.; Simcoe, M.; Igo, R.P., Jr.; Song, Y.E.; Wojciechowski, R.; Cheng, C.Y.; Khaw, P.T.; et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet. 2018, 50, 778–782. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, S.; Ong, J.S.; An, J.; Han, X.; Zhou, T.; Siggs, O.M.; Law, M.H.; Souzeau, E.; Sharma, S.; Lynn, D.J.; et al. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat. Genet. 2018, 50, 1067–1071. [Google Scholar] [CrossRef] [PubMed]
- Gharahkhani, P.; Jorgenson, E.; Hysi, P.; Khawaja, A.P.; Pendergrass, S.; Han, X.; Ong, J.S.; Hewitt, A.W.; Segrè, A.V.; Rouhana, J.M.; et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun 2021, 12, 1258. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Allingham, R.R.; Qin, X.; Layfield, D.; Dellinger, A.E.; Gibson, J.; Wheeler, J.; Ashley-Koch, A.E.; Stamer, W.D.; Hauser, M.A. Gene expression profile in human trabecular meshwork from patients with primary open-angle glaucoma. Investig. Ophthalmol. Vis. Sci. 2013, 54, 6382–6389. [Google Scholar] [CrossRef]
- Cai, J.; Perkumas, K.M.; Qin, X.; Hauser, M.A.; Stamer, W.D.; Liu, Y. Expression Profiling of Human Schlemm’s Canal Endothelial Cells From Eyes with and Without Glaucoma. Investig. Ophthalmol. Vis. Sci. 2015, 56, 6747–6753. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.H.; Anand, V.N.; Wang, W.H.; Chatterton, J.E.; Sun, D.; Shepard, A.R.; Jacobson, N.; Pang, I.H.; Deluca, A.P.; Casavant, T.L.; et al. Exon-level expression profiling of ocular tissues. Exp. Eye Res. 2013, 111, 105–111. [Google Scholar] [CrossRef][Green Version]
- Carnes, M.U.; Allingham, R.R.; Ashley-Koch, A.; Hauser, M.A. Transcriptome analysis of adult and fetal trabecular meshwork, cornea, and ciliary body tissues by RNA sequencing. Exp. Eye Res. 2018, 167, 91–99. [Google Scholar] [CrossRef]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Battle, A.; Brown, C.D.; Engelhardt, B.E.; Montgomery, S.B. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [PubMed]
- eGTEx Project. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat. Genet. 2017, 49, 1664–1670. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Youngblood, H.; Cai, J.; Drewry, M.D.; Helwa, I.; Hu, E.; Liu, S.; Yu, H.; Mu, H.; Hu, Y.; Perkumas, K.; et al. Expression of mRNAs, miRNAs, and lncRNAs in Human Trabecular Meshwork Cells Upon Mechanical Stretch. Investig. Ophthalmol. Vis. Sci. 2020, 61, 2. [Google Scholar] [CrossRef] [PubMed]
- van Zyl, T.; Yan, W.; McAdams, A.; Peng, Y.R.; Shekhar, K.; Regev, A.; Juric, D.; Sanes, J.R. Cell atlas of aqueous humor outflow pathways in eyes of humans and four model species provides insight into glaucoma pathogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 10339–10349. [Google Scholar] [CrossRef]
- Ramos, R.F.; Hoying, J.B.; Witte, M.H.; Stamer, D.W. Schlemm’s canal endothelia, lymphatic, or blood vasculature? J. Glaucoma 2007, 16, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Allingham, R.R.; de Kater, A.W.; Ethier, C.R. Schlemm’s canal and primary open angle glaucoma: Correlation between Schlemm’s canal dimensions and outflow facility. Exp. Eye Res. 1996, 62, 101–109. [Google Scholar] [CrossRef]
- Herndon, L.W.; Weizer, J.S.; Stinnett, S.S. Central corneal thickness as a risk factor for advanced glaucoma damage. Arch. Ophthalmol. 2004, 122, 17–21. [Google Scholar] [CrossRef]
- Susanna, B.N.; Ogata, N.G.; Jammal, A.A.; Susanna, C.N.; Berchuck, S.I.; Medeiros, F.A. Corneal Biomechanics and Visual Field Progression in Eyes with Seemingly Well-Controlled Intraocular Pressure. Ophthalmology 2019, 126, 1640–1646. [Google Scholar] [CrossRef]
- Johnstone, M.A.; Grant, W.G. Pressure-dependent changes in structures of the aqueous outflow system of human and monkey eyes. Am. J. Ophthalmol. 1973, 75, 365–383. [Google Scholar] [CrossRef]
- Ramos, R.F.; Sumida, G.M.; Stamer, W.D. Cyclic mechanical stress and trabecular meshwork cell contractility. Investig. Ophthalmol. Vis. Sci. 2009, 50, 3826–3832. [Google Scholar] [CrossRef]
- Coleman, D.J.; Trokel, S. Direct-recorded intraocular pressure variations in a human subject. Arch. Ophthalmol. 1969, 82, 637–640. [Google Scholar] [CrossRef]
- Perkins, E.S. The ocular pulse. Curr. Eye Res. 1981, 1, 19–23. [Google Scholar] [CrossRef]
- Johnstone, M.; Martin, E.; Jamil, A. Pulsatile flow into the aqueous veins: Manifestations in normal and glaucomatous eyes. Exp. Eye Res. 2011, 92, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.; Fury, W.; Yang, H.; Gomez-Caraballo, M.; Bai, Y.; Yang, T.; Adler, C.; Wei, Y.; Ni, M.; Schmitt, H.; et al. Molecular taxonomy of human ocular outflow tissues defined by single-cell transcriptomics. Proc. Natl. Acad. Sci. USA 2020, 117, 12856–12867. [Google Scholar] [CrossRef]
- Clark, A.F.; Wilson, K.; McCartney, M.D.; Miggans, S.T.; Kunkle, M.; Howe, W. Glucocorticoid-induced formation of cross-linked actin networks in cultured human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 1994, 35, 281–294. [Google Scholar]
- Gasiorowski, J.Z.; Russell, P. Biological properties of trabecular meshwork cells. Exp. Eye Res. 2009, 88, 671–675. [Google Scholar] [CrossRef]
- Raghunathan, V.K.; Morgan, J.T.; Park, S.A.; Weber, D.; Phinney, B.S.; Murphy, C.J.; Russell, P. Dexamethasone Stiffens Trabecular Meshwork, Trabecular Meshwork Cells, and Matrix. Investig. Ophthalmol. Vis. Sci. 2015, 56, 4447–4459. [Google Scholar] [CrossRef]
- Wang, K.; Read, A.T.; Sulchek, T.; Ethier, C.R. Trabecular meshwork stiffness in glaucoma. Exp. Eye Res. 2017, 158, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Johar Sr, K.; Nagpal, K.; Vasavada, A.J.S. Sex hormone receptors in the human eye. Surv. Ophthalmol. 2005, 50, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, S.; Acharya, M.; Banerjee, D.; Bhattacharjee, A.; Ray, K. Molecular basis for involvement of CYP1B1 in MYOC upregulation and its potential implication in glaucoma pathogenesis. PLoS ONE 2012, 7, e45077. [Google Scholar] [CrossRef]
- Iqbal, Z.; Midgley, J.M.; Watson, D.G. Determination of oestrone, 17alpha- and 17beta-oestradiol in bovine aqueous humor using gas chromatography-negative ion chemical ionization mass spectrometry. Arch. Pharm. Res. 1997, 20, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Colitz, C.M.; Lu, P.; Sugimoto, Y.; Barden, C.A.; Chandler, H.L. Estradiol biosynthesis in canine lens epithelial cells. Curr. Eye Res. 2015, 40, 541–548. [Google Scholar] [CrossRef]
- Stárka, L.; Hampl, R.; Bicíková, M.; Obenberger, J. Indentification and radioimmunologic estimation of sexual steroid hormones in aqueous humor and vitreous of rabbit eye. Albrecht Von Graefes Arch. Klin. Exp. Ophthalmol. 1976, 199, 261–266. [Google Scholar] [CrossRef]
- Zhang, X.H.; Sun, H.M.; Ji, J.; Zhang, H.; Ma, W.J.; Jin, Z.; Yuan, J.Q. Sex hormones and their receptors in patients with age-related cataract. J. Cataract. Refract. Surg. 2003, 29, 71–77. [Google Scholar] [CrossRef]
- Tobias, E.S.; Hurlstone, A.F.; MacKenzie, E.; McFarlane, R.; Black, D.M. The TES gene at 7q31.1 is methylated in tumours and encodes a novel growth-suppressing LIM domain protein. Oncogene 2001, 20, 2844–2853. [Google Scholar] [CrossRef] [PubMed]
- Coutts, A.S.; MacKenzie, E.; Griffith, E.; Black, D.M. TES is a novel focal adhesion protein with a role in cell spreading. J. Cell Sci. 2003, 116, 897–906. [Google Scholar] [CrossRef]
- Garvalov, B.K.; Higgins, T.E.; Sutherland, J.D.; Zettl, M.; Scaplehorn, N.; Köcher, T.; Piddini, E.; Griffiths, G.; Way, M. The conformational state of Tes regulates its zyxin-dependent recruitment to focal adhesions. J. Cell Biol. 2003, 161, 33–39. [Google Scholar] [CrossRef]
- Sala, S.; Oakes, P.W. Stress fiber strain recognition by the LIM protein testin is cryptic and mediated by RhoA. Mol. Biol. Cell 2021, 32, 1758–1771. [Google Scholar] [CrossRef]
- Tumminia, S.J.; Mitton, K.P.; Arora, J.; Zelenka, P.; Epstein, D.L.; Russell, P. Mechanical stretch alters the actin cytoskeletal network and signal transduction in human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 1998, 39, 1361–1371. [Google Scholar]
- Clark, A.F.; Brotchie, D.; Read, A.T.; Hellberg, P.; English-Wright, S.; Pang, I.H.; Ethier, C.R.; Grierson, I. Dexamethasone alters F-actin architecture and promotes cross-linked actin network formation in human trabecular meshwork tissue. Cell Motil. Cytoskelet. 2005, 60, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, X.; Zhong, Y. Rho/Rho-associated kinase pathway in glaucoma (Review). Int. J. Oncol. 2013, 43, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Hill, L.J.; Mead, B.; Thomas, C.N.; Foale, S.; Feinstein, E.; Berry, M.; Blanch, R.J.; Ahmed, Z.; Logan, A. TGF-β-induced IOP elevations are mediated by RhoA in the early but not the late fibrotic phase of open angle glaucoma. Mol. Vis. 2018, 24, 712–726. [Google Scholar]
- Wernert, N.; Raes, M.B.; Lassalle, P.; Dehouck, M.P.; Gosselin, B.; Vandenbunder, B.; Stehelin, D. c-ets1 proto-oncogene is a transcription factor expressed in endothelial cells during tumor vascularization and other forms of angiogenesis in humans. Am. J. Pathol. 1992, 140, 119–127. [Google Scholar]
- Bedford, F.K.; Ashworth, A.; Enver, T.; Wiedemann, L.M. HEX: A novel homeobox gene expressed during haematopoiesis and conserved between mouse and human. Nucleic Acids Res. 1993, 21, 1245–1249. [Google Scholar] [CrossRef] [PubMed]
- Marfil, V.; Moya, M.; Pierreux, C.E.; Castell, J.V.; Lemaigre, F.P.; Real, F.X.; Bort, R. Interaction between Hhex and SOX13 modulates Wnt/TCF activity. J. Biol. Chem. 2010, 285, 5726–5737. [Google Scholar] [CrossRef]
- Mao, W.; Millar, J.C.; Wang, W.H.; Silverman, S.M.; Liu, Y.; Wordinger, R.J.; Rubin, J.S.; Pang, I.H.; Clark, A.F. Existence of the canonical Wnt signaling pathway in the human trabecular meshwork. Investig. Ophthalmol. Vis. Sci. 2012, 53, 7043–7051. [Google Scholar] [CrossRef]
- Wang, X.; Huai, G.; Wang, H.; Liu, Y.; Qi, P.; Shi, W.; Peng, J.; Yang, H.; Deng, S.; Wang, Y. Mutual regulation of the Hippo/Wnt/LPA/TGF-β signaling pathways and their roles in glaucoma (Review). Int. J. Mol. Med. 2018, 41, 1201–1212. [Google Scholar] [CrossRef]
- Werner, E.; Kowalczyk, A.P.; Faundez, V. Anthrax toxin receptor 1/tumor endothelium marker 8 mediates cell spreading by coupling extracellular ligands to the actin cytoskeleton. J. Biol. Chem. 2006, 281, 23227–23236. [Google Scholar] [CrossRef]
- Muragaki, Y.; Jacenko, O.; Apte, S.; Mattei, M.G.; Ninomiya, Y.; Olsen, B.R. The alpha 2(VIII) collagen gene. A novel member of the short chain collagen family located on the human chromosome 1. J. Biol. Chem. 1991, 266, 7721–7727. [Google Scholar] [CrossRef]
- Saharinen, J.; Hyytiäinen, M.; Taipale, J.; Keski-Oja, J. Latent transforming growth factor-beta binding proteins (LTBPs)—structural extracellular matrix proteins for targeting TGF-beta action. Cytokine Growth Factor Rev. 1999, 10, 99–117. [Google Scholar] [CrossRef]
- Oklü, R.; Hesketh, R. The latent transforming growth factor beta binding protein (LTBP) family. Biochem. J. 2000, 352, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Mok, A.; Miskie, B.; Hegele, R.A. Single-nucleotide polymorphisms of the proprotein convertase subtilisin/ kexin type 5 (PCSK5) gene. J. Hum. Genet. 2001, 46, 730–732. [Google Scholar] [CrossRef]
- Dittmer, J.; Vetter, M.; Blumenthal, S.G.; Lindemann, R.K.; Kölbl, H. Importance of ets1 proto-oncogene for breast cancer progression. Zent. Gynakol. 2004, 126, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Bai, X.H.; Lodyga, M.; Xu, J.; Yang, B.B.; Keshavjee, S.; Post, M.; Liu, M. Conversion of mechanical force into biochemical signaling. J. Biol. Chem. 2004, 279, 54793–54801. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Kostka, G.; Garbe, J.H.; Keene, D.R.; Bächinger, H.P.; Hanisch, F.G.; Markova, D.; Tsuda, T.; Timpl, R.; Chu, M.L.; et al. A comparative analysis of the fibulin protein family. Biochemical characterization, binding interactions, and tissue localization. J. Biol. Chem. 2007, 282, 11805–11816. [Google Scholar] [CrossRef]
- Bielenberg, D.R.; Shimizu, A.; Klagsbrun, M. Semaphorin-induced cytoskeletal collapse and repulsion of endothelial cells. Methods Enzymol. 2008, 443, 299–314. [Google Scholar]
- Khoshnoodi, J.; Pedchenko, V.; Hudson, B.G. Mammalian collagen IV. Microsc. Res. Tech. 2008, 71, 357–370. [Google Scholar] [CrossRef]
- Gaur, P.; Bielenberg, D.R.; Samuel, S.; Bose, D.; Zhou, Y.; Gray, M.J.; Dallas, N.A.; Fan, F.; Xia, L.; Lu, J.; et al. Role of class 3 semaphorins and their receptors in tumor growth and angiogenesis. Clin. Cancer Res. 2009, 15, 6763–6770. [Google Scholar] [CrossRef] [PubMed]
- Nievergall, E.; Janes, P.W.; Stegmayer, C.; Vail, M.E.; Haj, F.G.; Teng, S.W.; Neel, B.G.; Bastiaens, P.I.; Lackmann, M. PTP1B regulates Eph receptor function and trafficking. J. Cell Biol. 2010, 191, 1189–1203. [Google Scholar] [CrossRef] [PubMed]
- Paule, S.; Aljofan, M.; Simon, C.; Rombauts, L.J.; Nie, G. Cleavage of endometrial α-integrins into their functional forms is mediated by proprotein convertase 5/6. Hum. Reprod. 2012, 27, 2766–2774. [Google Scholar] [CrossRef]
- Ye, K.; Ouyang, X.; Wang, Z.; Yao, L.; Zhang, G. SEMA3F Promotes Liver Hepatocellular Carcinoma Metastasis by Activating Focal Adhesion Pathway. DNA Cell Biol. 2020, 39, 474–483. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, S.R.; Yue, B.Y. Adhesion of human trabecular meshwork cells to extracellular matrix proteins. Roles and distribution of integrin receptors. Investig. Ophthalmol. Vis. Sci. 1996, 37, 104–113. [Google Scholar]
- Acott, T.S.; Kelley, M.J. Extracellular matrix in the trabecular meshwork. Exp. Eye Res. 2008, 86, 543–561. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, J.; Luo, X. Integrins in trabecular meshwork and optic nerve head: Possible association with the pathogenesis of glaucoma. Biomed. Res. Int. 2013, 2013, 202905. [Google Scholar] [CrossRef]
- Acott, T.S.; Vranka, J.A.; Keller, K.E.; Raghunathan, V.; Kelley, M.J. Normal and glaucomatous outflow regulation. Prog. Retin. Eye Res. 2021, 82, 100897. [Google Scholar] [CrossRef]
- Liton, P.B.; Gonzalez, P. Stress response of the trabecular meshwork. J. Glaucoma 2008, 17, 378–385. [Google Scholar] [CrossRef] [PubMed]
- WuDunn, D. Mechanobiology of trabecular meshwork cells. Exp. Eye Res. 2009, 88, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Lakk, M.; Križaj, D. TRPV4-Rho signaling drives cytoskeletal and focal adhesion remodeling in trabecular meshwork cells. Am. J. Physiol. Cell Physiol. 2021, 320, C1013–C1030. [Google Scholar] [CrossRef]
- Nakatsuka, H.; Sokabe, T.; Yamamoto, K.; Sato, Y.; Hatakeyama, K.; Kamiya, A.; Ando, J. Shear stress induces hepatocyte PAI-1 gene expression through cooperative Sp1/Ets-1 activation of transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 291, G26–G34. [Google Scholar] [CrossRef]
- Milkiewicz, M.; Uchida, C.; Gee, E.; Fudalewski, T.; Haas, T.L. Shear stress-induced Ets-1 modulates protease inhibitor expression in microvascular endothelial cells. J. Cell Physiol. 2008, 217, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Chumley, P.; Allon, M.; George, J.; Scott, D.W.; Patel, R.P.; Litovsky, S.; Jaimes, E.A. The transcription factor E26 transformation-specific sequence-1 mediates neointima formation in arteriovenous fistula. J. Am. Soc. Nephrol. 2014, 25, 475–487. [Google Scholar] [CrossRef]
- Yang, J.H.; Briggs, W.H.; Libby, P.; Lee, R.T. Small mechanical strains selectively suppress matrix metalloproteinase-1 expression by human vascular smooth muscle cells. J. Biol. Chem. 1998, 273, 6550–6555. [Google Scholar] [CrossRef]
- Wang, P.; Yang, L.; You, X.; Singh, G.K.; Zhang, L.; Yan, Y.; Sung, K.L. Mechanical stretch regulates the expression of matrix metalloproteinase in rheumatoid arthritis fibroblast-like synoviocytes. Connect. Tissue Res. 2009, 50, 98–109. [Google Scholar] [CrossRef]
- Colorado, P.C.; Torre, A.; Kamphaus, G.; Maeshima, Y.; Hopfer, H.; Takahashi, K.; Volk, R.; Zamborsky, E.D.; Herman, S.; Sarkar, P.K.; et al. Anti-angiogenic cues from vascular basement membrane collagen. Cancer Res. 2000, 60, 2520–2526. [Google Scholar]
- Lelièvre, E.; Lionneton, F.; Soncin, F.; Vandenbunder, B. The Ets family contains transcriptional activators and repressors involved in angiogenesis. Int. J. Biochem. Cell Biol. 2001, 33, 391–407. [Google Scholar] [CrossRef]
- Sato, Y. Role of ETS family transcription factors in vascular development and angiogenesis. Cell Struct. Funct. 2001, 26, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Oike, Y.; Yasunaga, K.; Suda, T. Angiopoietin-related/angiopoietin-like proteins regulate angiogenesis. Int. J. Hematol. 2004, 80, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Adiguzel, E.; Hou, G.; Mulholland, D.; Hopfer, U.; Fukai, N.; Olsen, B.; Bendeck, M. Migration and growth are attenuated in vascular smooth muscle cells with type VIII collagen-null alleles. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, K.A.; Basile, C.M.; Spring, S.C.; Bonuccelli, G.; Lisanti, M.P.; Terman, B.I. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell-matrix interactions on collagen. Exp. Cell Res. 2005, 305, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.J.; Murphy, M.O.; Kielty, C.M.; Shuttleworth, C.A.; Black, R.A.; Humphries, M.J.; Walker, M.G.; Canfield, A.E. Alpha2(VIII) collagen substrata enhance endothelial cell retention under acute shear stress flow via an alpha2beta1 integrin-dependent mechanism: An in vitro and in vivo study. Circulation 2006, 114, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Plaisier, E.; Gribouval, O.; Alamowitch, S.; Mougenot, B.; Prost, C.; Verpont, M.C.; Marro, B.; Desmettre, T.; Cohen, S.Y.; Roullet, E.; et al. COL4A1 mutations and hereditary angiopathy, nephropathy, aneurysms, and muscle cramps. N. Engl. J. Med. 2007, 357, 2687–2695. [Google Scholar] [CrossRef]
- Nyberg, P.; Xie, L.; Sugimoto, H.; Colorado, P.; Sund, M.; Holthaus, K.; Sudhakar, A.; Salo, T.; Kalluri, R. Characterization of the anti-angiogenic properties of arresten, an alpha1beta1 integrin-dependent collagen-derived tumor suppressor. Exp. Cell Res. 2008, 314, 3292–3305. [Google Scholar] [CrossRef] [PubMed]
- Plaisier, E.; Chen, Z.; Gekeler, F.; Benhassine, S.; Dahan, K.; Marro, B.; Alamowitch, S.; Paques, M.; Ronco, P. Novel COL4A1 mutations associated with HANAC syndrome: A role for the triple helical CB3[IV] domain. Am. J. Med. Genet. A 2010, 152, 2550–2555. [Google Scholar] [CrossRef]
- Staton, C.A. Class 3 semaphorins and their receptors in physiological and pathological angiogenesis. Biochem. Soc. Trans. 2011, 39, 1565–1570. [Google Scholar] [CrossRef]
- Rosenquist, R.; Epstein, D.; Melamed, S.; Johnson, M.; Grant, W.M. Outflow resistance of enucleated human eyes at two different perfusion pressures and different extents of trabeculotomy. Curr. Eye Res. 1989, 8, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Carreon, T.; van der Merwe, E.; Fellman, R.L.; Johnstone, M.; Bhattacharya, S.K. Aqueous outflow—A continuum from trabecular meshwork to episcleral veins. Prog. Retin. Eye Res. 2017, 57, 108–133. [Google Scholar] [CrossRef]
- Gonzalez, J.M., Jr.; Ko, M.K.; Hong, Y.K.; Weigert, R.; Tan, J.C.H. Deep tissue analysis of distal aqueous drainage structures and contractile features. Sci. Rep. 2017, 7, 17071. [Google Scholar] [CrossRef]
- Waxman, S.; Wang, C.; Dang, Y.; Hong, Y.; Esfandiari, H.; Shah, P.; Lathrop, K.L.; Loewen, R.T.; Loewen, N.A. Structure-Function Changes of the Porcine Distal Outflow Tract in Response to Nitric Oxide. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4886–4895. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Harris, A.; Toris, C.; Tobe, L.; Lang, M.; Belamkar, A.; Ng, A.; Verticchio Vercellin, A.C.; Mathew, S.; Siesky, B. Effects of Sex Hormones on Ocular Blood Flow and Intraocular Pressure in Primary Open-angle Glaucoma: A Review. J. Glaucoma 2018, 27, 1037–1041. [Google Scholar] [CrossRef]
- Sator, M.O.; Joura, E.A.; Frigo, P.; Kurz, C.; Metka, M.; Hommer, A.; Huber, J.C. Hormone replacement therapy and intraocular pressure. Maturitas 1997, 28, 55–58. [Google Scholar] [CrossRef]
- Sator, M.O.; Akramian, J.; Joura, E.A.; Nessmann, A.; Wedrich, A.; Gruber, D.; Metka, M.; Huber, J.C. Reduction of intraocular pressure in a glaucoma patient undergoing hormone replacement therapy. Maturitas 1998, 29, 93–95. [Google Scholar] [CrossRef]
- Sorrentino, C.; Affinito, P.; Mattace Raso, F.; Loffredo, M.; Merlino, P.; Loffredo, A.; Palomba, S.; Nappi, C. Effect of hormone replacement therapy on postmenopausal ocular function. Minerva Ginecol. 1998, 50, 19–24. [Google Scholar] [PubMed]
- Lang, Y.; Lang, N.; Ben-Ami, M.; Garzozi, H. The effects of hormone replacement therapy (HRT) on the human eye. Harefuah 2002, 141, 287–291. [Google Scholar]
- Altintas, O.; Caglar, Y.; Yuksel, N.; Demirci, A.; Karabas, L. The effects of menopause and hormone replacement therapy on quality and quantity of tear, intraocular pressure and ocular blood flow. Ophthalmologica 2004, 218, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Uncu, G.; Avci, R.; Uncu, Y.; Kaymaz, C.; Develioğlu, O.J.G.E. The effects of different hormone replacement therapy regimens on tear function, intraocular pressure and lens opacity. Gynecol. Endocrinol. 2006, 22, 501–505. [Google Scholar] [CrossRef]
- Wojtowiec, A.; Wojtowiec, P.; Misiuk-Hojto, M. The role of estrogen in the development of glacoma tous optic neuropathy. Klin. Oczna 2016, 117, 275–277. [Google Scholar]
- Harris, A.; Harris, M.; Biller, J.; Garzozi, H.; Zarfty, D.; Ciulla, T.A.; Martin, B. Aging affects the retrobulbar circulation differently in women and men. Arch. Ophthalmol. 2000, 118, 1076–1080. [Google Scholar] [CrossRef]
- Yucel, I.; Akar, M.E.; Dora, B.; Akar, Y.; Taskin, O.; Ozer, H.O. Effect of the menstrual cycle on standard achromatic and blue-on-yellow visual field analysis of women with migraine. Can. J. Ophthalmol. 2005, 40, 51–57. [Google Scholar] [CrossRef]
- Green, K.; Phillips, C.I.; Cheeks, L.; Slagle, T. Aqueous humor flow rate and intraocular pressure during and after pregnancy. Ophthalmic Res. 1988, 20, 353–357. [Google Scholar] [CrossRef]
- Centofanti, M.; Migliardi, R.; Bonini, S.; Manni, G.; Bucci, M.G.; Pesavento, C.B.; Amin, C.S.; Harris, A. Pulsatile ocular blood flow during pregnancy. Eur. J. Ophthalmol. 2002, 12, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Bahadir Kilavuzoglu, A.E.; Cosar, C.B.; Bildirici, I.; Cetin, O.; Ozbasli, E. Estrogen- and Progesterone-Induced Variation in Corneal Parameters According to Hormonal Status. Eye Contact Lens 2018, 44, S179–S184. [Google Scholar] [CrossRef]
- Sekhar, G.C.; Nagarajan, R. Ocular toxicity of tamoxifen. Indian J. Ophthalmol. 1995, 43, 23–26. [Google Scholar]
- Centofanti, M.; Zarfati, D.; Manni, G.L.; Bonini, S.; Migliardi, R.; Oddone, F.; Harris, A.; Bucci, M.G. The influence of oestrogen on the pulsatile ocular blood flow. Acta Ophthalmol. Scand. Suppl. 2000, 232, 38–39. [Google Scholar] [CrossRef]
- Kosior-Jarecka, E.; Sagan, M.; Wróbel-Dudzińska, D.; Łukasik, U.; Aung, T.; Khor, C.C.; Kocki, J.; Żarnowski, T. Estrogen receptor gene polymorphisms and their influence on clinical status of Caucasian patients with primary open angle glaucoma. Ophthalmic Genet. 2019, 40, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Jansson, I.; Stoilov, I.; Sarfarazi, M.; Schenkman, J.B. Effect of two mutations of human CYP1B1, G61E and R469W, on stability and endogenous steroid substrate metabolism. Pharmacogenetics 2001, 11, 793–801. [Google Scholar] [CrossRef]
- Pasquale, L.R.; Loomis, S.J.; Weinreb, R.N.; Kang, J.H.; Yaspan, B.L.; Bailey, J.C.; Gaasterland, D.; Gaasterland, T.; Lee, R.K.; Scott, W.K.; et al. Estrogen pathway polymorphisms in relation to primary open angle glaucoma: An analysis accounting for gender from the United States. Mol. Vis. 2013, 19, 1471–1481. [Google Scholar]
- Chen, X.; Liu, Y.; Zhang, Y.; Kam, W.R.; Pasquale, L.R.; Sullivan, D.A. Impact of aromatase absence on murine intraocular pressure and retinal ganglion cells. Sci. Rep. 2018, 8, 3280. [Google Scholar] [CrossRef]
- Kobayashi, K.; Kobayashi, H.; Ueda, M.; Honda, Y. Estrogen receptor expression in bovine and rat retinas. Investig. Ophthalmol. Vis. Sci. 1998, 39, 2105–2110. [Google Scholar]
- Kumar, D.M.; Perez, E.; Cai, Z.Y.; Aoun, P.; Brun-Zinkernagel, A.M.; Covey, D.F.; Simpkins, J.W.; Agarwal, N. Role of nonfeminizing estrogen analogues in neuroprotection of rat retinal ganglion cells against glutamate-induced cytotoxicity. Free Radic. Biol. Med. 2005, 38, 1152–1163. [Google Scholar] [CrossRef]
- Russell-Randall, K.R.; Dortch-Carnes, J. Kappa opioid receptor localization and coupling to nitric oxide production in cells of the anterior chamber. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5233–5239. [Google Scholar] [CrossRef]
- Bucolo, C.; Campana, G.; Di Toro, R.; Cacciaguerra, S.; Spampinato, S. Sigma1 recognition sites in rabbit iris-ciliary body: Topical sigma1-site agonists lower intraocular pressure. J. Pharmacol. Exp. Ther. 1999, 289, 1362–1369. [Google Scholar]
- Bucolo, C.; Drago, F. Effects of neurosteroids on ischemia-reperfusion injury in the rat retina: Role of sigma1 recognition sites. Eur. J. Pharmacol. 2004, 498, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Delafontaine, P.; Song, Y.H.; Li, Y. Expression, regulation, and function of IGF-1, IGF-1R, and IGF-1 binding proteins in blood vessels. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Kanda, S.; Miyata, Y.; Kanetake, H. T-cell factor-4-dependent up-regulation of fibronectin is involved in fibroblast growth factor-2-induced tube formation by endothelial cells. J. Cell Biochem. 2005, 94, 835–847. [Google Scholar] [CrossRef]
- Hofmann, J.J.; Luisa Iruela-Arispe, M. Notch expression patterns in the retina: An eye on receptor-ligand distribution during angiogenesis. Gene Expr Patterns 2007, 7, 461–470. [Google Scholar] [CrossRef]
- Kume, T. Novel insights into the differential functions of Notch ligands in vascular formation. J. Angiogenes Res. 2009, 1, 8. [Google Scholar] [CrossRef]
- Xie, L.; Frank, P.G.; Lisanti, M.P.; Sowa, G. Endothelial cells isolated from caveolin-2 knockout mice display higher proliferation rate and cell cycle progression relative to their wild-type counterparts. Am. J. Physiol. Cell Physiol. 2010, 298, C693–C701. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kume, T. Ligand-dependent Notch signaling in vascular formation. Adv. Exp. Med. Biol. 2012, 727, 210–222. [Google Scholar]
- Napp, L.C.; Augustynik, M.; Paesler, F.; Krishnasamy, K.; Woiterski, J.; Limbourg, A.; Bauersachs, J.; Drexler, H.; Le Noble, F.; Limbourg, F.P. Extrinsic Notch ligand Delta-like 1 regulates tip cell selection and vascular branching morphogenesis. Circ. Res. 2012, 110, 530–535. [Google Scholar] [CrossRef]
- Panneerselvam, M.; Patel, H.H.; Roth, D.M. Caveolins and heart diseases. Adv. Exp. Med. Biol. 2012, 729, 145–156. [Google Scholar]
- Melincovici, C.S.; Boşca, A.B.; Şuşman, S.; Mărginean, M.; Mihu, C.; Istrate, M.; Moldovan, I.M.; Roman, A.L.; Mihu, C.M. Vascular endothelial growth factor (VEGF)—Key factor in normal and pathological angiogenesis. Rom. J. Morphol. Embryol. 2018, 59, 455–467. [Google Scholar] [PubMed]
- Sáinz-Jaspeado, M.; Claesson-Welsh, L. Cytokines regulating lymphangiogenesis. Curr. Opin. Immunol. 2018, 53, 58–63. [Google Scholar] [CrossRef]
- Arnal, J.F.; Fontaine, C.; Billon-Galés, A.; Favre, J.; Laurell, H.; Lenfant, F.; Gourdy, P. Estrogen receptors and endothelium. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1506–1512. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, E.; Constantin, B.; Drabkin, H.; Roche, J. Semaphorin SEMA3F localization in malignant human lung and cell lines: A suggested role in cell adhesion and cell migration. Am. J. Pathol. 2000, 156, 939–950. [Google Scholar] [CrossRef]
- Scimone, C.; Donato, L.; Alibrandi, S.; Vadalà, M.; Giglia, G.; Sidoti, A.; D’Angelo, R. N-retinylidene-N-retinylethanolamine adduct induces expression of chronic inflammation cytokines in retinal pigment epithelium cells. Exp. Eye Res. 2021, 209, 108641. [Google Scholar] [CrossRef] [PubMed]
- Donato, L.; Abdalla, E.M.; Scimone, C.; Alibrandi, S.; Rinaldi, C.; Nabil, K.M.; D’Angelo, R.; Sidoti, A. Impairments of Photoreceptor Outer Segments Renewal and Phototransduction Due to a Peripherin Rare Haplotype Variant: Insights from Molecular Modeling. Int. J. Mol. Sci. 2021, 22, 3484. [Google Scholar] [CrossRef]
- Murata, A.; Okuyama, K.; Sakano, S.; Kajiki, M.; Hirata, T.; Yagita, H.; Zúñiga-Pflücker, J.C.; Miyake, K.; Akashi-Takamura, S.; Moriwaki, S.; et al. A Notch ligand, Delta-like 1 functions as an adhesion molecule for mast cells. J. Immunol. 2010, 185, 3905–3912. [Google Scholar] [CrossRef]
- Aga, M.; Bradley, J.M.; Wanchu, R.; Yang, Y.F.; Acott, T.S.; Keller, K.E. Differential effects of caveolin-1 and -2 knockdown on aqueous outflow and altered extracellular matrix turnover in caveolin-silenced trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5497–5509. [Google Scholar] [CrossRef]
- Zhang, J.P.; Li, N.; Bai, W.Z.; Qiu, X.C.; Ma, B.A.; Zhou, Y.; Fan, Q.Y.; Shan, L.Q. Notch ligand Delta-like 1 promotes the metastasis of melanoma by enhancing tumor adhesion. Braz. J. Med. Biol. Res. 2014, 47, 299–306. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Sinderen, M.; Oyanedel, J.; Menkhorst, E.; Cuman, C.; Rainczuk, K.; Winship, A.; Salamonsen, L.; Edgell, T.; Dimitriadis, E. Soluble Delta-like ligand 1 alters human endometrial epithelial cell adhesive capacity. Reprod. Fertil. Dev. 2017, 29, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Yang, Y.; Sun, P.; Tian, Y.; Gao, F.; Wang, C.; Zong, T.; Li, M.; Zhang, Y.; Yu, T.; et al. βII spectrin (SPTBN1): Biological function and clinical potential in cancer and other diseases. Int. J. Biol. Sci. 2021, 17, 32–49. [Google Scholar] [CrossRef]
- Wiggs, J.L.; Kang, J.H.; Yaspan, B.L.; Mirel, D.B.; Laurie, C.; Crenshaw, A.; Brodeur, W.; Gogarten, S.; Olson, L.M.; Abdrabou, W.; et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum. Mol. Genet. 2011, 20, 4707–4713. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Reh, T.A. Relationship between Delta-like and proneural bHLH genes during chick retinal development. Dev. Dyn. 2008, 237, 1565–1580. [Google Scholar] [CrossRef]
- Stone, E.M.; Lotery, A.J.; Munier, F.L.; Héon, E.; Piguet, B.; Guymer, R.H.; Vandenburgh, K.; Cousin, P.; Nishimura, D.; Swiderski, R.E.; et al. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat. Genet. 1999, 22, 199–202. [Google Scholar] [CrossRef]
- Cheng, L.; Chen, C.; Guo, W.; Liu, K.; Zhao, Q.; Lu, P.; Yu, F.; Xu, X. EFEMP1 Overexpression Contributes to Neovascularization in Age-Related Macular Degeneration. Front. Pharmacol. 2020, 11, 547436. [Google Scholar] [CrossRef]
- Gauthier, A.C.; Wiggs, J.L. Childhood glaucoma genes and phenotypes: Focus on FOXC1 mutations causing anterior segment dysgenesis and hearing loss. Exp. Eye Res. 2020, 190, 107893. [Google Scholar] [CrossRef]
- Khaled, M.L.; Bykhovskaya, Y.; Yablonski, S.E.R.; Li, H.; Drewry, M.D.; Aboobakar, I.F.; Estes, A.; Gao, X.R.; Stamer, W.D.; Xu, H.; et al. Differential Expression of Coding and Long Noncoding RNAs in Keratoconus-Affected Corneas. Investig. Ophthalmol. Vis. Sci. 2018, 59, 2717–2728. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Stamer, W.D.; Seftor, R.E.; Williams, S.K.; Samaha, H.A.; Snyder, R.W. Isolation and culture of human trabecular meshwork cells by extracellular matrix digestion. Curr. Eye Res. 1995, 14, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Keller, K.E.; Bhattacharya, S.K.; Borrás, T.; Brunner, T.M.; Chansangpetch, S.; Clark, A.F.; Dismuke, W.M.; Du, Y.; Elliott, M.H.; Ethier, C.R.; et al. Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp. Eye Res. 2018, 171, 164–173. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
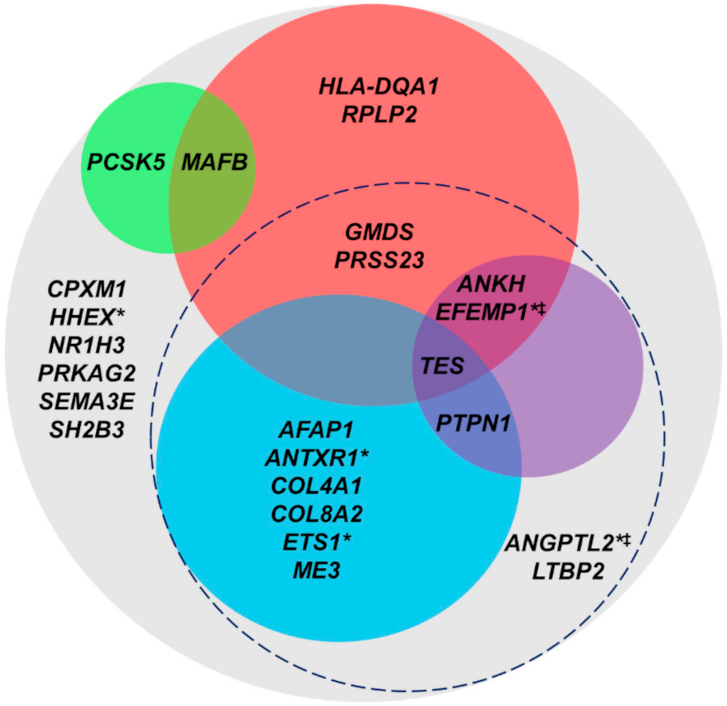
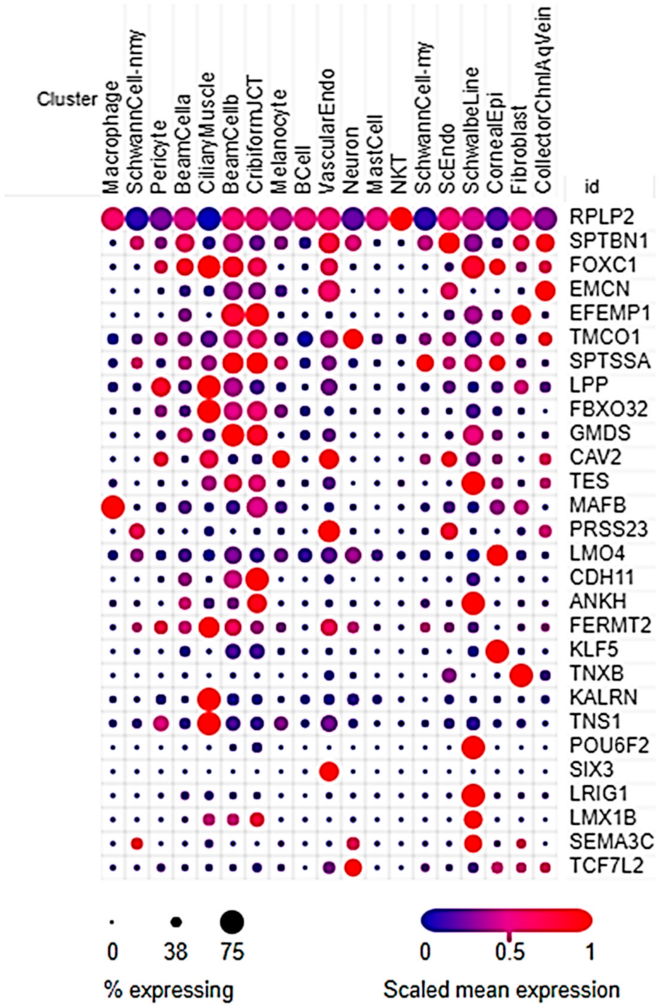

| Gene | Description | Fold Change | p |
|---|---|---|---|
| ANKH | ANKH inorganic pyrophosphate transport regulator | 2.69 | 3.16 × 10-7 |
| ANTXR1 | ANTXR cell adhesion molecule 1 | 1.79 | 6.46 × 10-3 |
| COL8A2 | Collagen type VIII α2 chain | 2.66 | 1.32 × 10-4 |
| EFEMP1 | EGF containing fibulin extracellular matrix protein 1 | 1.82 | 4.43 × 10-3 |
| GMDS | GDP-mannose 4,6-dehydratase | 1.93 | 3.14 × 10-3 |
| HLA-DQA1 | Major histocompatibility complex, class II, DQ α1 | 2.13 | 3.10 × 10-3 |
| LTBP2 | Latent TGF-β binding protein 2 | 2.25 | 1.65 × 10-3 |
| MAFB | MAF BZIP transcription factor B | 1.54 | 1.22 × 10-2 |
| RPLP2 | Ribosomal protein lateral stalk subunit P2 | 1.58 | 1.32 × 10-1 |
| SEMA3E | Semaphorin 3E | 1.94 | 1.26 × 10-5 |
| Gene | Description | Fold Change | p |
|---|---|---|---|
| AFAP1 | Actin filament associated protein 1 | 1.56 | 1.35 × 10−1 |
| ANGPTL2 | Angiopoietin like 2 | −3.33 | 6.73 × 10−2 |
| ANTXR1 | ANTXR cell adhesion molecule 1 | −1.67 | 4.77 × 10−2 |
| COL4A1 | Collagen type IV α1 chain | 2.84 | 1.43 × 10−1 |
| CPXM1 | Carboxypeptidase X, M14 family member 1 | 1.57 | 2.18 × 10−2 |
| ETS1 | ETS proto-oncogene 1, transcription factor | 1.61 | 1.28 × 10−1 |
| HHEX | Hematopoietically expressed homeobox | −1.62 | 1.81 × 10−2 |
| LTBP2 | Latent TGF-β binding protein 2 | −1.63 | 1.56 × 10−1 |
| MAFB | MAF BZIP transcription factor B | −1.79 | 2.82 × 10−1 |
| ME3 | Malic enzyme 3 | −1.59 | 3.23 × 10−3 |
| NR1H3 | Nuclear receptor subfamily 1 group H member 3 | −1.59 | 5.54 × 10−2 |
| PCSK5 | Proprotein convertase subtilisin/kexin type 5 | −2.04 | 3.93 × 10−2 |
| PRKAG2 | Protein kinase AMP-activated non-catalytic subunit γ2 | 1.63 | 1.70 × 10−2 |
| PRSS23 | Serine protease 23 | −1.65 | 1.60 × 10−1 |
| PTPN1 | Protein tyrosine phosphatase non-receptor type 1 | 1.63 | 1.63 × 10−2 |
| SH2B3 | SH2B adaptor protein 3 | 1.58 | 7.07 × 10−3 |
| TES | Testin LIM domain protein | 1.54 | 4.75 × 10−2 |
| Gene | Cell/Tissue Type | RNA-Seq Expression (FPKM) |
|---|---|---|
| AFAP1 | SC | 46.64 |
| ANGPTL2 | SC | 30.90 |
| ANKH | TM | 64.41 |
| ANTXR1 * | TM, SC | 32.78, 47.13 |
| COL4A1 | SC | 17.92 |
| COL8A2 | TM | 34.44 |
| CPXM1 | SC | 0.49 |
| EFEMP1 | TM | 89.70 |
| ETS1 | SC | 15.70 |
| GMDS | TM | 29.40 |
| HHEX | SC | 2.60 |
| HLA-DQA1 | TM | 0.00 |
| LTBP2 * | TM, SC | 104.70, 171.74 |
| MAFB * | TM, SC | 1.37, 1.79 |
| ME3 | SC | 11.09 |
| NR1H3 | SC | 4.45 |
| PCSK5 | SC | 1.97 |
| PRKAG2 | SC | 6.49 |
| PRSS23 | SC | 184.92 |
| PTPN1 | SC | 12.46 |
| RPLP2P1 | TM | 0.00 |
| SEMA3E | TM | 1.51 |
| SH2B3 | SC | 13.63 |
| TES | SC | 64.48 |
| Gene | Trabecular Meshwork | Ciliary Body | Cornea | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PLIER | DABG Average | DABG Range | PLIER | DABG Average | DABG Range | PLIER | DABG Average | DABG Range | |
| AFAP1 | 33.64 | 0.08 | 0–0.58 | 31.53 | 0.07 | 0–0.49 | 30.54 | 0.14 | 0–0.77 |
| ANGPTL2 | 45.11 | 0.16 | 0–0.71 | 30.32 | 0.19 | 0–0.8 | 74.34 | 0.07 | 0–0.42 |
| ANKH | 112.61 | 0.05 | 0–0.76 | 70.15 | 0.05 | 0–0.51 | 88.56 | 0.07 | 0–0.51 |
| ANTXR1 * | 145.19 | 0.06 | 0–0.89 | 90.84 | 0.10 | 0–0.77 | 58.56 | 0.09 | 0–0.84 |
| COL4A1 | 46.71 | 0.08 | 0–0.92 | 42.61 | 0.16 | 0–0.97 | 24.59 | 0.24 | 0–0.98 |
| COL8A2 | 99.82 | 0.12 | 0–0.56 | 86.39 | 0.14 | 0–0.52 | 81.40 | 0.11 | 0–0.57 |
| CPXM1 | 26.48 | 0.11 | 0–0.58 | 24.60 | 0.17 | 0–0.85 | 26.29 | 0.10 | 0–0.32 |
| EFEMP1 | 214.28 | 0.01 | 0–0.11 | 667.78 | 0.03 | 0–0.35 | 165.29 | 0.04 | 0–0.52 |
| ETS1 | 86.82 | 0.07 | 0–0.76 | 41.16 | 0.08 | 0–0.51 | 27.07 | 0.05 | 0–0.35 |
| GMDS | 76.23 | 0.06 | 0–0.20 | 88.91 | 0.19 | 0–0.78 | 93.19 | 0.06 | 0–0.16 |
| HHEX | 92.41 | 0.14 | 0–0.64 | 63.12 | 0.20 | 0–0.54 | 95.97 | 0.23 | 0–0.95 |
| HLA-DQA1 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| LTBP2 * | 79.44 | 0.06 | 0–0.66 | 69.94 | 0.08 | 0–0.77 | 62.10 | 0.08 | 0–0.53 |
| MAFB * | 54.38 | 0.19 | 0–0.93 | 62.34 | 0.17 | 0–0.82 | 77.52 | 0.09 | 0–0.50 |
| ME3 | 29.17 | 0.13 | 0–0.86 | 36.24 | 0.17 | 0–0.52 | 34.63 | 0.11 | 0–0.52 |
| NR1H3 | 32.84 | 0.18 | 0–0.72 | 32.88 | 0.23 | 0–1.00 | 33.11 | 0.16 | 0–0.57 |
| PCSK5 | 37.87 | 0.07 | 0–0.33 | 31.31 | 0.17 | 0–0.81 | 234.57 | 0.01 | 0–0.18 |
| PRKAG2 | 65.93 | 0.08 | 0–0.68 | 57.75 | 0.14 | 0–0.78 | 51.41 | 0.13 | 0–0.72 |
| PRSS23 | 76.78 | 0.06 | 0–0.34 | 155.24 | 0.02 | 0–0.15 | 121.46 | 0.04 | 0–0.28 |
| PTPN1 | 84.10 | 0.02 | 0–0.22 | 61.15 | 0.06 | 0–0.62 | 78.01 | 0.07 | 0–0.37 |
| RPLP2P1 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| SEMA3E | 130.67 | 0.01 | 0–0.20 | 28.40 | 0.10 | 0–0.82 | 35.46 | 0.07 | 0–0.83 |
| SH2B3 | 40.00 | 0.15 | 0–0.99 | 29.77 | 0.15 | 0–0.68 | 37.72 | 0.12 | 0–0.5 |
| TES | 290.39 | 0.01 | 0–0.11 | 67.06 | 0.07 | 0–0.27 | 193.21 | 0.01 | 0–0.08 |
| Gene | Trabecular Meshwork | Ciliary Body | Cornea |
|---|---|---|---|
| AFAP1 | 3.10 | 4.71 | 2.46 |
| ANGPTL2 | 18.84 | 15.75 | 8.69 |
| ANKH | 50.88 | 32.83 | 21.43 |
| ANTXR1 * | 67.92 | 5.40 | 10.42 |
| COL4A1 | 9.15 | 8.06 | 0.73 |
| COL8A2 | 118.54 | 33.40 | 28.26 |
| CPXM1 | 6.63 | 9.52 | 0.22 |
| EFEMP1 | 142.27 | 153.01 | 77.16 |
| ETS1 | 16.18 | 1.85 | 3.56 |
| GMDS | 81.59 | 55.42 | 89.55 |
| HHEX | 2.25 | 0.53 | 0.48 |
| HLA-DQA1 | 31.13 | 4.11 | 9.36 |
| LTBP2 * | 54.69 | 142.61 | 3.93 |
| MAFB * | 13.35 | 1.45 | 8.08 |
| ME3 | 7.26 | 15.74 | 5.42 |
| NR1H3 | 8.60 | 19.88 | 7.02 |
| PCSK5 | 5.04 | 2.71 | 21.57 |
| PRKAG2 | 16.79 | 2.30 | 16.08 |
| PRSS23 | 23.22 | 8.20 | 16.50 |
| PTPN1 | 16.16 | 34.22 | 17.24 |
| RPLP2P1 | 0.00 | 0.00 | 0.00 |
| SEMA3E | 8.27 | 0.16 | 1.93 |
| SH2B3 | 4.54 | 2.79 | 1.28 |
| TES | 56.81 | 0.81 | 30.17 |
| SNP | Gene | Histone Modification | Enhancer or Promoter Region | Significant eQTL |
|---|---|---|---|---|
| rs28649910 | AFAP1 | Yes | Yes | Skin; Whole blood |
| rs11795066 | ANGPTL2 | Yes | No | Cerebellum; Esophageal mucosa; Skin; Testis; Tibial nerve; Thyroid; Whole blood |
| rs368503 | ANKH | Yes | No | Esophageal mucosa |
| rs6546486 | ANTXR1 * | Yes | Yes | None |
| rs112972174 | COL4A1 | Yes | Yes | None |
| rs12123086 | COL8A2 | Yes | Yes | Whole blood |
| rs4672075 | EFEMP1 | Yes | No | Skin; Thyroid |
| rs7924522 | ETS1 | Yes | Yes | None |
| rs722585 | GMDS | Yes | No | None |
| rs74384554 | LTBP2 * | Yes | No | Tibial nerve |
| rs2433414 | ME3 | Yes | Yes | Esophageal muscularis; Lung; Skeletal muscle; Skin; Subcutaneous adipose; Tibial nerve; Transformed fibroblasts |
| rs11606902 | PRSS23 | Yes | No | Adrenal gland; Esophagus; Skeletal muscle |
| rs6095946 | PTPN1 | Yes | Yes | None |
| rs10774624 | SH2B3 | Yes | No | Esophageal mucosa; Skin |
| rs55892100 | TES | Yes | Yes | Adrenal gland; Lung; Pancreas; Subcutaneous adipose; Testis; Tibial artery; Tibial nerve; Thyroid; Whole blood |
| Gene | Description | Fold Change | p |
|---|---|---|---|
| MCPH1 | Microcephalin 1 | 2.18 | 2.4 × 10−2 |
| MAFB | MAF BZIP transcription factor B | −2.12 | 1.9 × 10−2 |
| HGF | Hepatocyte growth factor | −2.52 | 3.1 × 10−2 |
| PCSK5 | Proprotein convertase subtilisin/kexin type 5 | −2.75 | 3.2 × 10−2 |
| lncRNA | Sequence Homology | Fold Change | p |
|---|---|---|---|
| NONHSAT157336.1:208–1549 | Myoferlin | 1.61 × 105 | 3.6 × 10−2 |
| NONHSAT106523.2:1153–1748 | Forkhead box C1 | 2.26 × 103 | 4.5 × 10−2 |
| NONHSAT184643.1:226–631 | Spectrin β, non-erythrocytic 1 | 4.77 | 1.6 × 10−2 |
| NONHSAT156133.1:88–552 | Transcription factor 7 like 2 | −2.42 | 2.5 × 10−2 |
| Donor | 17β-Estradiol Concentration | |
|---|---|---|
| Non-glaucomatous (n = 4) | 0.14 ± 0.05 nM | 37.90 ± 14.80 pg/mL |
| Glaucomatous (n = 4) | 0.13 ± 0.03 nM | 34.46 ± 7.08 pg/mL |
| Bovine (n = 5) | 0.22 ± 0.03 nM | 59.70 ± 8.58 pg/mL |
| Porcine (n = 4) | 0.55 ± 0.62 nM | 149.44 ± 168.21 pg/mL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Youngblood, H.A.; Parker, E.; Cai, J.; Perkumas, K.; Yu, H.; Sun, J.; Smith, S.B.; Bollinger, K.E.; Wiggs, J.L.; Pasquale, L.R.; et al. Identification of Estrogen Signaling in a Prioritization Study of Intraocular Pressure-Associated Genes. Int. J. Mol. Sci. 2021, 22, 10288. https://doi.org/10.3390/ijms221910288
Youngblood HA, Parker E, Cai J, Perkumas K, Yu H, Sun J, Smith SB, Bollinger KE, Wiggs JL, Pasquale LR, et al. Identification of Estrogen Signaling in a Prioritization Study of Intraocular Pressure-Associated Genes. International Journal of Molecular Sciences. 2021; 22(19):10288. https://doi.org/10.3390/ijms221910288
Chicago/Turabian StyleYoungblood, Hannah A., Emily Parker, Jingwen Cai, Kristin Perkumas, Hongfang Yu, Jason Sun, Sylvia B. Smith, Kathryn E. Bollinger, Janey L. Wiggs, Louis R. Pasquale, and et al. 2021. "Identification of Estrogen Signaling in a Prioritization Study of Intraocular Pressure-Associated Genes" International Journal of Molecular Sciences 22, no. 19: 10288. https://doi.org/10.3390/ijms221910288
APA StyleYoungblood, H. A., Parker, E., Cai, J., Perkumas, K., Yu, H., Sun, J., Smith, S. B., Bollinger, K. E., Wiggs, J. L., Pasquale, L. R., Hauser, M. A., Stamer, W. D., & Liu, Y. (2021). Identification of Estrogen Signaling in a Prioritization Study of Intraocular Pressure-Associated Genes. International Journal of Molecular Sciences, 22(19), 10288. https://doi.org/10.3390/ijms221910288






