Phase Separation and Mechanical Forces in Regulating Asymmetric Cell Division of Neural Stem Cells
Abstract
:1. Introduction
2. ACD of Drosophila Neuroblasts
2.1. Asymmetric Protein Localization during ACD
2.2. Generation of Distinct Sibling Cells
3. LLPS and Asymmetric Protein Localization during ACD of NBs
3.1. LLPS in Cells
3.2. LLPS-Mediated Basal Localization of Numb and Pon in Dividing NBs
3.3. LLPS-Mediated Apical Localization of the PAR Complexes in Dividing NBs
3.4. LLPS-Mediated Mitotic Implantation of Pros in Dividing GMCs
4. Mechanical Forces Regulating ACD
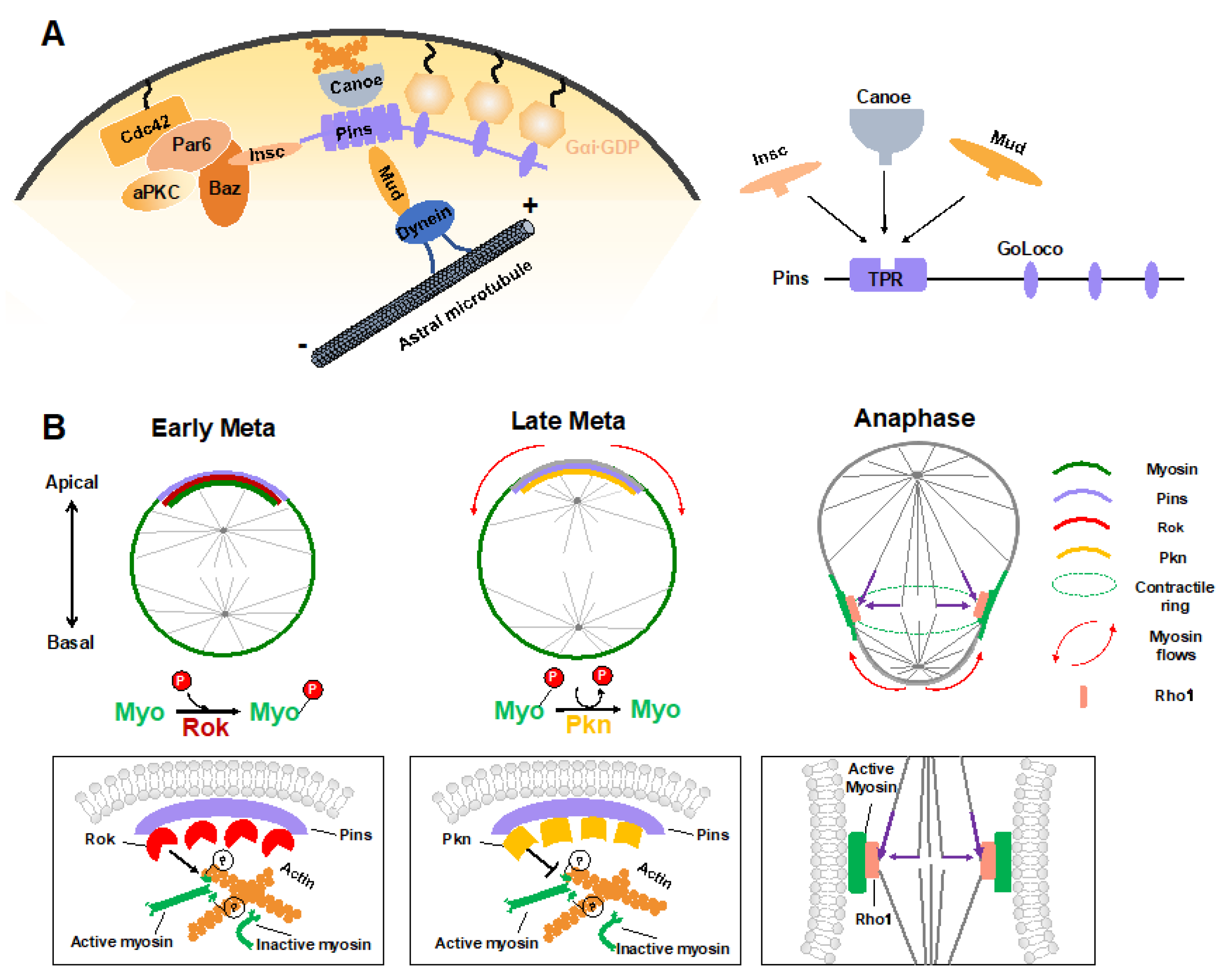
4.1. Polarity Cue-Regulated Spindle Orientation
4.2. Myosin Flows Regulated by Polarity and Spindle Cues
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Knoblich, J.A. Mechanisms of Asymmetric Stem Cell Division. Cell 2008, 132, 583–597. [Google Scholar] [CrossRef] [Green Version]
- Gönczy, P. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 2008, 9, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Knoblich, J.A. Asymmetric cell division: Recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 2010, 11, 849–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, W.; Zhang, M. Protein Complex Assemblies in Epithelial Cell Polarity and Asymmetric Cell Division. J. Mol. Biol. 2018, 430, 3504–3520. [Google Scholar] [CrossRef] [PubMed]
- Knoblich, J.A.; Jan, Y.N. Asymmetric segregation of Numb and Prospero during cell division. Nat. Cell Biol. 1995, 377, 624–627. [Google Scholar] [CrossRef]
- Betschinger, J.; Mechtler, K.; Knoblich, J.A. Asymmetric Segregation of the Tumor Suppressor Brat Regulates Self-Renewal in Drosophila Neural Stem Cells. Cell 2006, 124, 1241–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeshima-Kataoka, H.; Skeath, J.B.; Nabeshima, Y.-I.; Doe, C.Q.; Matsuzaki, F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nat. Cell Biol. 1997, 390, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Wodarz, A.; Ramrath, A.; Kuchinke, U.; Knust, E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nat. Cell Biol. 1999, 402, 544–547. [Google Scholar] [CrossRef]
- Slack, C.; Overton, P.; Tuxworth, R.; Chia, W. Asymmetric localisation of Miranda and its cargo proteins during neuroblast division requires the anaphase-promoting complex/cyclosome. Development 2007, 134, 3781–3787. [Google Scholar] [CrossRef] [Green Version]
- Godard, B.; Heisenberg, C.-P. Cell division and tissue mechanics. Curr. Opin. Cell Biol. 2019, 60, 114–120. [Google Scholar] [CrossRef]
- Delgado, M.K.; Cabernard, C. Mechanical regulation of cell size, fate, and behavior during asymmetric cell division. Curr. Opin. Cell Biol. 2020, 67, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Tsang, B.; Pritišanac, I.; Scherer, S.W.; Moses, A.M.; Forman-Kay, J.D. Phase Separation as a Missing Mechanism for Interpretation of Disease Mutations. Cell 2020, 183, 1742–1756. [Google Scholar] [CrossRef]
- Alberti, S.; Hyman, A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 196–213. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Roden, C.; Gladfelter, A.S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021, 22, 183–195. [Google Scholar] [CrossRef]
- Shan, Z.; Tu, Y.; Yang, Y.; Liu, Z.; Zeng, M.; Xu, H.; Long, J.; Zhang, M.; Cai, Y.; Wen, W. Basal condensation of Numb and Pon complex via phase transition during Drosophila neuroblast asymmetric division. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Yang, Y.; Gu, A.; Xu, J.; Mao, Y.; Lu, H.; Hu, W.; Lei, Q.-Y.; Li, Z.; Zhang, M.; et al. Par complex cluster formation mediated by phase separation. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Wen, W. Phase Separation in Asymmetric Cell Division. Biochemistry 2020, 59, 47–56. [Google Scholar] [CrossRef]
- Venkei, Z.G.; Yamashita, Y.M. Emerging mechanisms of asymmetric stem cell division. J. Cell Biol. 2018, 217, 3785–3795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schober, M.; Schaefer, M.; Knoblich, J.A. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nat. Cell Biol. 1999, 402, 548–551. [Google Scholar] [CrossRef]
- Zhu, J.; Wen, W.; Zheng, Z.; Shang, Y.; Wei, Z.; Xiao, Z.; Pan, Z.; Du, Q.; Wang, W.; Zhang, M. LGN/mInsc and LGN/NuMA Complex Structures Suggest Distinct Functions in Asymmetric Cell Division for the Par3/mInsc/LGN and Gαi/LGN/NuMA Pathways. Mol. Cell 2011, 43, 418–431. [Google Scholar] [CrossRef] [Green Version]
- Atwood, S.; Prehoda, K.E. aPKC Phosphorylates Miranda to Polarize Fate Determinants during Neuroblast Asymmetric Cell Division. Curr. Biol. 2009, 19, 723–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirtz-Peitz, F.; Nishimura, T.; Knoblich, J.A. Linking Cell Cycle to Asymmetric Division: Aurora-A Phosphorylates the Par Complex to Regulate Numb Localization. Cell 2008, 135, 161–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.A.; Lau, K.M.; Rahmani, Z.; Dho, S.E.; Brothers, G.; She, Y.-M.; Berry, D.M.; Bonneil, E.; Thibault, P.; Schweisguth, F.; et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 2007, 26, 468–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramat, A.; Hannaford, M.; Januschke, J. Maintenance of Miranda Localization in Drosophila Neuroblasts Involves Interaction with the Cognate mRNA. Curr. Biol. 2017, 27, 2101–2111.e5. [Google Scholar] [CrossRef] [Green Version]
- Jia, M.; Shan, Z.; Yang, Y.; Liu, C.; Li, J.; Luo, Z.-G.; Zhang, M.; Cai, Y.; Wen, W.; Wang, W. The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division. Nat. Commun. 2015, 6, 8381. [Google Scholar] [CrossRef] [Green Version]
- Broadus, J.; Fuerstenberg, S.; Doe, C.Q. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nat. Cell Biol. 1998, 391, 792–795. [Google Scholar] [CrossRef]
- Connell, M.; Cabernard, C.; Ricketson, D.; Doe, C.Q.; Prehoda, K.E. Asymmetric cortical extension shifts cleavage furrow position in Drosophila neuroblasts. Mol. Biol. Cell 2011, 22, 4220–4226. [Google Scholar] [CrossRef] [PubMed]
- Cabernard, C.; Prehoda, K.E.; Doe, C.Q. A spindle-independent cleavage furrow positioning pathway. Nat. Cell Biol. 2010, 467, 91–94. [Google Scholar] [CrossRef] [Green Version]
- Noatynska, A.; Gotta, M.; Meraldi, P. Mitotic spindle (DIS)orientation and DISease: Cause or consequence? J. Cell Biol. 2012, 199, 1025–1035. [Google Scholar] [CrossRef] [Green Version]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; van den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef] [Green Version]
- Hyman, A.A.; Weber, C.A.; Julicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.G.; Zhang, H. Phase Separation in Membrane Biology: The Interplay between Membrane-Bound Organelles and Membraneless Condensates. Dev. Cell 2020, 55, 30–44. [Google Scholar] [CrossRef]
- Quiroz, F.G.; Fiore, V.F.; Levorse, J.; Polak, L.; Wong, E.; Pasolli, H.A.; Fuchs, E. Liquid-liquid phase separation drives skin barrier formation. Science 2020, 367, eaax9554. [Google Scholar] [CrossRef]
- Ong, J.Y.; Torres, J.Z. Phase Separation in Cell Division. Mol. Cell 2020, 80, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cai, Q.; Feng, Z.; Zhang, M. Liquid-Liquid Phase Separation in Neuronal Development and Synaptic Signaling. Dev. Cell 2020, 55, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ji, X.; Li, P.; Liu, C.; Lou, J.; Wang, Z.; Wen, W.; Xiao, Y.; Zhang, M.; Zhu, X. Liquid-liquid phase separation in biology: Mechanisms, physiological functions and human diseases. Sci. China Life Sci. 2020, 63, 953–985. [Google Scholar] [CrossRef]
- Jain, A.; Vale, A.J.R.D. RNA phase transitions in repeat expansion disorders. Nat. Cell Biol. 2017, 546, 243–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.; et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Zhang, H. Phase Separation, Transition, and Autophagic Degradation of Proteins in Development and Pathogenesis. Trends Cell Biol. 2019, 29, 417–427. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Posey, A.E.; Holehouse, A.S.; Pappu, R.V. Phase Separation of Intrinsically Disordered Proteins. Methods Enzymol. 2018, 611, 1–30. [Google Scholar] [CrossRef]
- Banani, S.F.; Rice, A.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.; Shang, Y.; Araki, Y.; Guo, T.; Huganir, R.L.; Zhang, M. Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 2016, 166, 1163–1175.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Banjade, S.; Cheng, H.-C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nat. Cell Biol. 2012, 483, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Nott, T.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, B.; Ackerman, L.; Jan, L.; Jan, Y.-N. Modes of Protein Movement that Lead to the Asymmetric Localization of Partner of Numb during Drosophila Neuroblast Division. Mol. Cell 1999, 4, 883–891. [Google Scholar] [CrossRef]
- Patel, S.S.; Belmont, B.; Sante, J.M.; Rexach, M.F. Natively Unfolded Nucleoporins Gate Protein Diffusion across the Nuclear Pore Complex. Cell 2007, 129, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Kono, K.; Yoshiura, S.; Fujita, I.; Okada, Y.; Shitamukai, A.; Shibata, T.; Matsuzaki, F. Reconstruction of Par-dependent polarity in apolar cells reveals a dynamic process of cortical polarization. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- Wilson, M.I.; Gill, D.J.; Perisic, O.; Quinn, M.; Williams, R.L. PB1 Domain-Mediated Heterodimerization in NADPH Oxidase and Signaling Complexes of Atypical Protein Kinase C with Par6 and p62. Mol. Cell 2003, 12, 39–50. [Google Scholar] [CrossRef]
- Soriano, E.V.; Ivanova, M.E.; Fletcher, G.; Riou, P.; Knowles, P.P.; Barnouin, K.; Purkiss, A.; Kostelecky, B.; Saiu, P.; Linch, M.; et al. aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization. Dev. Cell 2016, 38, 384–398. [Google Scholar] [CrossRef] [Green Version]
- Holly, R.W.; Jones, K.; Prehoda, K.E. A Conserved PDZ-Binding Motif in aPKC Interacts with Par-3 and Mediates Cortical Polarity. Curr. Biol. 2020, 30, 893–898.e5. [Google Scholar] [CrossRef]
- De Sá, E.M.; Mirouse, V.; Johnston, D.S. aPKC Phosphorylation of Bazooka Defines the Apical/Lateral Border in Drosophila Epithelial Cells. Cell 2010, 141, 509–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.-C.; Low, T.Y.F.; Nishimura, Y.; Gole, L.; Yukako, N.; Motegi, F. Cortical forces and CDC-42 control clustering of PAR proteins for Caenorhabditis elegans embryonic polarization. Nat. Cell Biol. 2017, 19, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.; Peglion, F.; Martin, J.; Hubatsch, L.; Reich, J.; Hirani, N.; Gubieda, A.G.; Roffey, J.; Fernandes, A.R.; Johnston, D.S.; et al. aPKC Cycles between Functionally Distinct PAR Protein Assemblies to Drive Cell Polarity. Dev. Cell 2017, 42, 400–415.e9. [Google Scholar] [CrossRef] [Green Version]
- Oon, C.H.; Prehoda, K.E. Asymmetric recruitment and actin-dependent cortical flows drive the neuroblast polarity cycle. eLife 2019, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-Y.; Robinson, K.J.; Doe, C.Q. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nat. Cell Biol. 2005, 439, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, F.; Ohshiro, T.; Ikeshima-Kataoka, H.; Izumi, H. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development 1998, 125, 4089–4098. [Google Scholar] [CrossRef]
- Liu, X.; Shen, J.; Xie, L.; Wei, Z.; Wong, C.; Li, Y.; Zheng, X.; Li, P.; Song, Y. Mitotic Implantation of the Transcription Factor Prospero via Phase Separation Drives Terminal Neuronal Differentiation. Dev. Cell 2020, 52, 277–293.e8. [Google Scholar] [CrossRef] [PubMed]
- Speicher, S.; Fischer, A.; Knoblich, J.; Carmena, A. The PDZ Protein Canoe Regulates the Asymmetric Division of Drosophila Neuroblasts and Muscle Progenitors. Curr. Biol. 2008, 18, 831–837. [Google Scholar] [CrossRef]
- Wee, B.; Johnston, C.A.; Prehoda, K.E.; Doe, C.Q. Canoe binds RanGTP to promote PinsTPR/Mud-mediated spindle orientation. J. Cell Biol. 2011, 195, 369–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.; Acharya, B.; Peyret, G.; Fardin, M.-A.; Mège, R.-M.; Ladoux, B.; Yap, A.; Fanning, A.S.; Peifer, M. Remodeling the zonula adherens in response to tension and the role of afadin in this response. J. Cell Biol. 2016, 213, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yang, Z.; Hiremath, C.; Zimmerman, S.E.; Long, B.; Brakeman, P.R.; Mostov, K.E.; Bryant, D.M.; Luby-Phelps, K.; Marciano, D.K. Developing renal tubules orient cell division via Afadin to position the tubule lumen. Development 2017, 144, 3511–3520. [Google Scholar] [CrossRef] [Green Version]
- Keder, A.; Rives-Quinto, N.; Aerne, B.L.; Franco, M.; Tapon, N.; Carmena, A. The Hippo Pathway Core Cassette Regulates Asymmetric Cell Division. Curr. Biol. 2015, 25, 2739–2750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewey, E.; Sanchez, D.; Johnston, C.A. Warts Phosphorylates Mud to Promote Pins-Mediated Mitotic Spindle Orientation in Drosophila, Independent of Yorkie. Curr. Biol. 2015, 25, 2751–2762. [Google Scholar] [CrossRef] [Green Version]
- Carminati, M.; Gallini, S.; Pirovano, L.; Alfieri, A.; Bisi, S.; Mapelli, M. Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation. Nat. Struct. Mol. Biol. 2016, 23, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Gloerich, M.; Bianchini, J.M.; Siemers, K.A.; Cohen, D.J.; Nelson, W.J. Cell division orientation is coupled to cell–cell adhesion by the E-cadherin/LGN complex. Nat. Commun. 2017, 8, 13996. [Google Scholar] [CrossRef] [PubMed]
- Finegan, T.M.; Bergstralh, D.T. Division orientation: Disentangling shape and mechanical forces. Cell Cycle 2019, 18, 1187–1198. [Google Scholar] [CrossRef]
- Tsankova, A.; Pham, T.; Garcia, D.S.; Otte, F.; Cabernard, C. Cell Polarity Regulates Biased Myosin Activity and Dynamics during Asymmetric Cell Division via Drosophila Rho Kinase and Protein Kinase N. Dev. Cell 2017, 42, 143–155.e5. [Google Scholar] [CrossRef] [Green Version]
- Ou, G.; Stuurman, N.; D’Ambrosio, M.; Vale, R.D. Polarized Myosin Produces Unequal-Size Daughters during Asymmetric Cell Division. Science 2010, 330, 677–680. [Google Scholar] [CrossRef] [Green Version]
- Roubinet, C.; Tsankova, A.; Pham, T.; Monnard, A.; Caussinus, E.; Affolter, M.; Cabernard, C. Spatio-temporally separated cortical flows and spindle geometry establish physical asymmetry in fly neural stem cells. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Mayer, M.; Depken, M.; Bois, J.; Julicher, F.; Grill, S.W. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nat. Cell Biol. 2010, 467, 617–621. [Google Scholar] [CrossRef]
- Pham, T.T.; Monnard, A.; Helenius, J.; Lund, E.; Lee, N.; Müller, D.J.; Cabernard, C. Spatiotemporally Controlled Myosin Relocalization and Internal Pressure Generate Sibling Cell Size Asymmetry. iScience 2019, 13, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Wen, W. Phase Separation in Cell Polarity. Biochemistry 2021, 60, 2677–2684. [Google Scholar] [CrossRef]
- Langdon, E.M.; Qiu, Y.; Niaki, A.G.; McLaughlin, G.A.; Weidmann, C.A.; Gerbich, T.M.; Smith, J.A.; Crutchley, J.M.; Termini, C.M.; Weeks, K.M.; et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 2018, 360, 922–927. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Zheng, G.; Xie, W.; Mayr, C. In vivo reconstitution finds multivalent RNA–RNA interactions as drivers of mesh-like condensates. eLife 2021, 10, e64252. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, X.; Wasser, M.; Cai, Y.; Chia, W. Inscuteable and Staufen Mediate Asymmetric Localization and Segregation of prospero RNA during Drosophila Neuroblast Cell Divisions. Cell 1997, 90, 437–447. [Google Scholar] [CrossRef] [Green Version]
- Weirich, K.; Banerjee, S.; Dasbiswas, K.; Witten, T.A.; Vaikuntanathan, S.; Gardel, M.L. Liquid behavior of cross-linked actin bundles. Proc. Natl. Acad. Sci. USA 2017, 114, 2131–2136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Wang, S.; Huang, Y.; He, X.; Cui, H.; Zhu, X.; Zheng, Y. Phase Transition of Spindle-Associated Protein Regulate Spindle Apparatus Assembly. Cell 2015, 163, 108–122. [Google Scholar] [CrossRef] [Green Version]
- Gan, L.; Cookson, M.R.; Petrucelli, L.; La Spada, A.R. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 2018, 21, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Vega, A.; Braun, M.; Scharrel, L.; Jahnel, M.; Wegmann, S.; Hyman, B.T.; Alberti, S.; Diez, S.; Hyman, A.A. Local Nucleation of Microtubule Bundles through Tubulin Concentration into a Condensed Tau Phase. Cell Rep. 2017, 20, 2304–2312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegmann, S.; Eftekharzadeh, B.; Tepper, K.; Zoltowska, K.M.; Bennett, R.E.; Dujardin, S.; Laskowski, P.R.; MacKenzie, D.; Kamath, T.; Commins, C.; et al. Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 2018, 37, e98049. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wei, H.; Wen, W. Phase Separation and Mechanical Forces in Regulating Asymmetric Cell Division of Neural Stem Cells. Int. J. Mol. Sci. 2021, 22, 10267. https://doi.org/10.3390/ijms221910267
Zhang Y, Wei H, Wen W. Phase Separation and Mechanical Forces in Regulating Asymmetric Cell Division of Neural Stem Cells. International Journal of Molecular Sciences. 2021; 22(19):10267. https://doi.org/10.3390/ijms221910267
Chicago/Turabian StyleZhang, Yiqing, Heyang Wei, and Wenyu Wen. 2021. "Phase Separation and Mechanical Forces in Regulating Asymmetric Cell Division of Neural Stem Cells" International Journal of Molecular Sciences 22, no. 19: 10267. https://doi.org/10.3390/ijms221910267
APA StyleZhang, Y., Wei, H., & Wen, W. (2021). Phase Separation and Mechanical Forces in Regulating Asymmetric Cell Division of Neural Stem Cells. International Journal of Molecular Sciences, 22(19), 10267. https://doi.org/10.3390/ijms221910267





