Structural Features of a Full-Length Ubiquitin Ligase Responsible for the Formation of Patches at the Plasma Membrane
Abstract
1. Introduction
2. Results
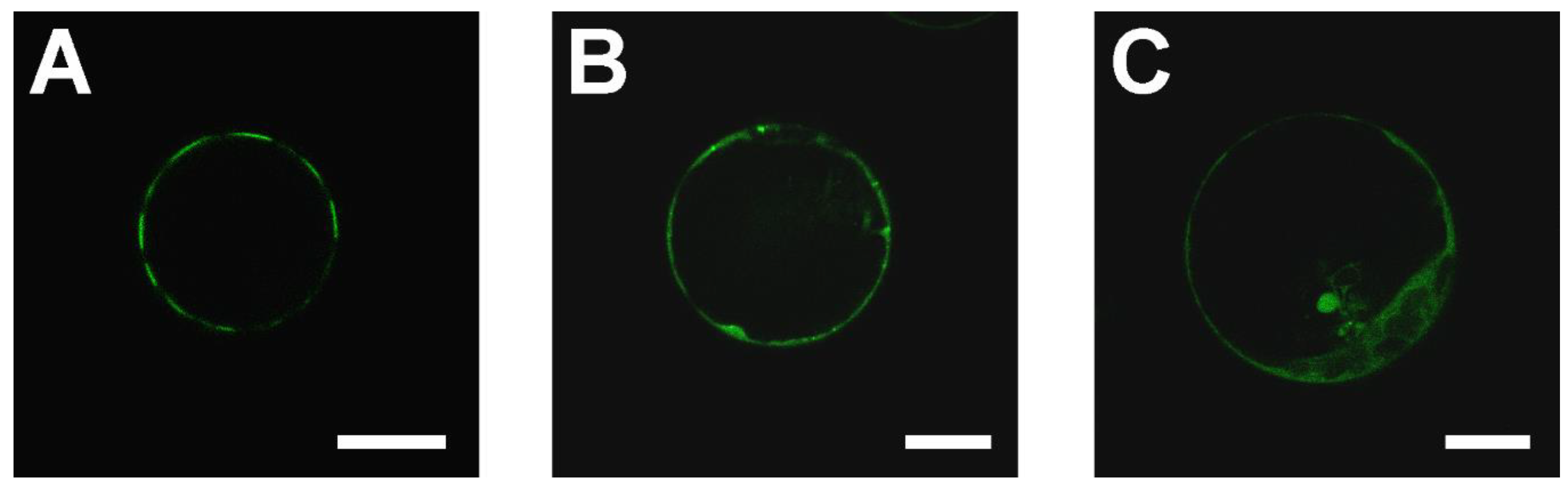
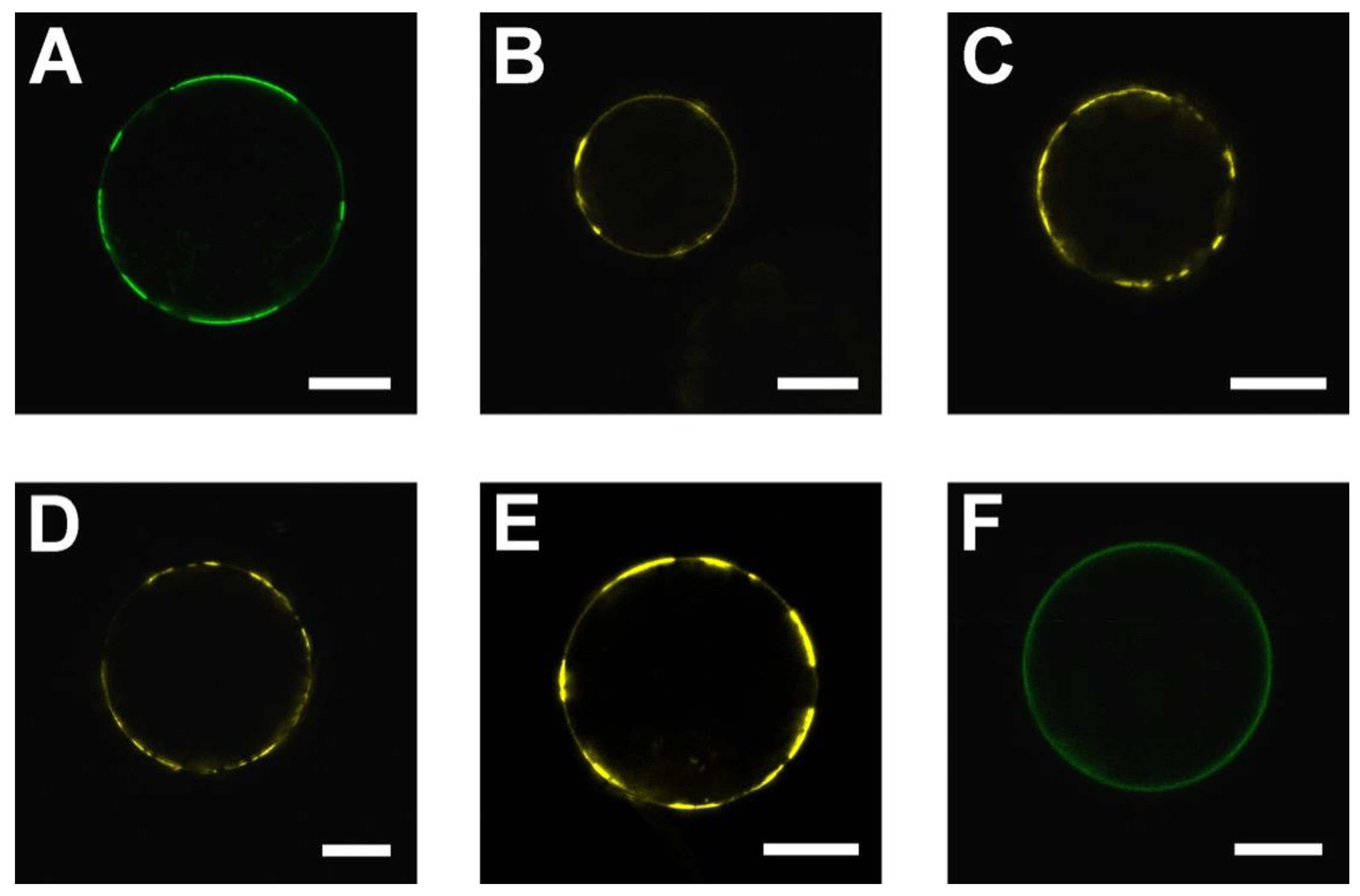
2.1. SAUL1 Domain for the Formation of Patches at the PM
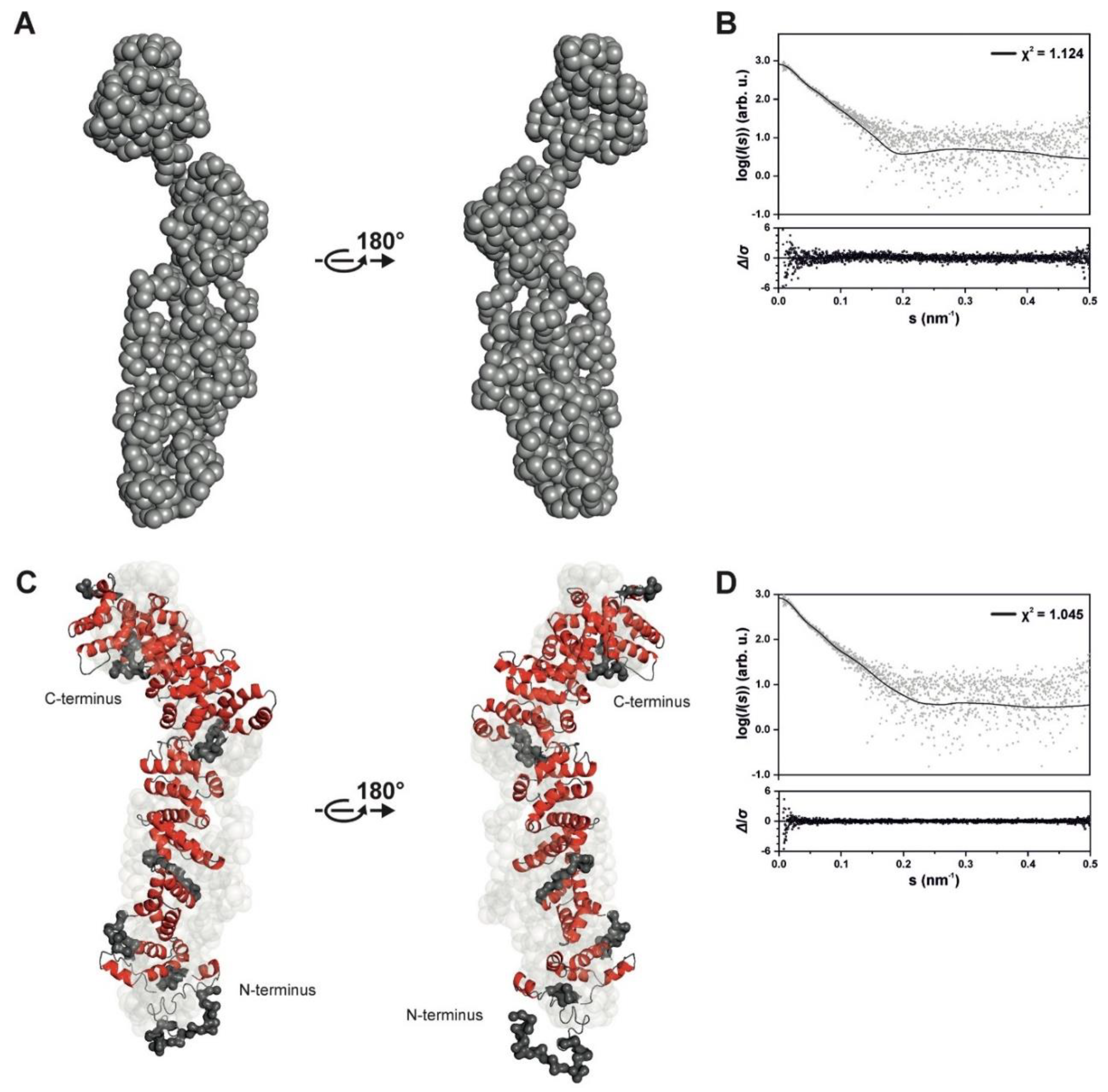
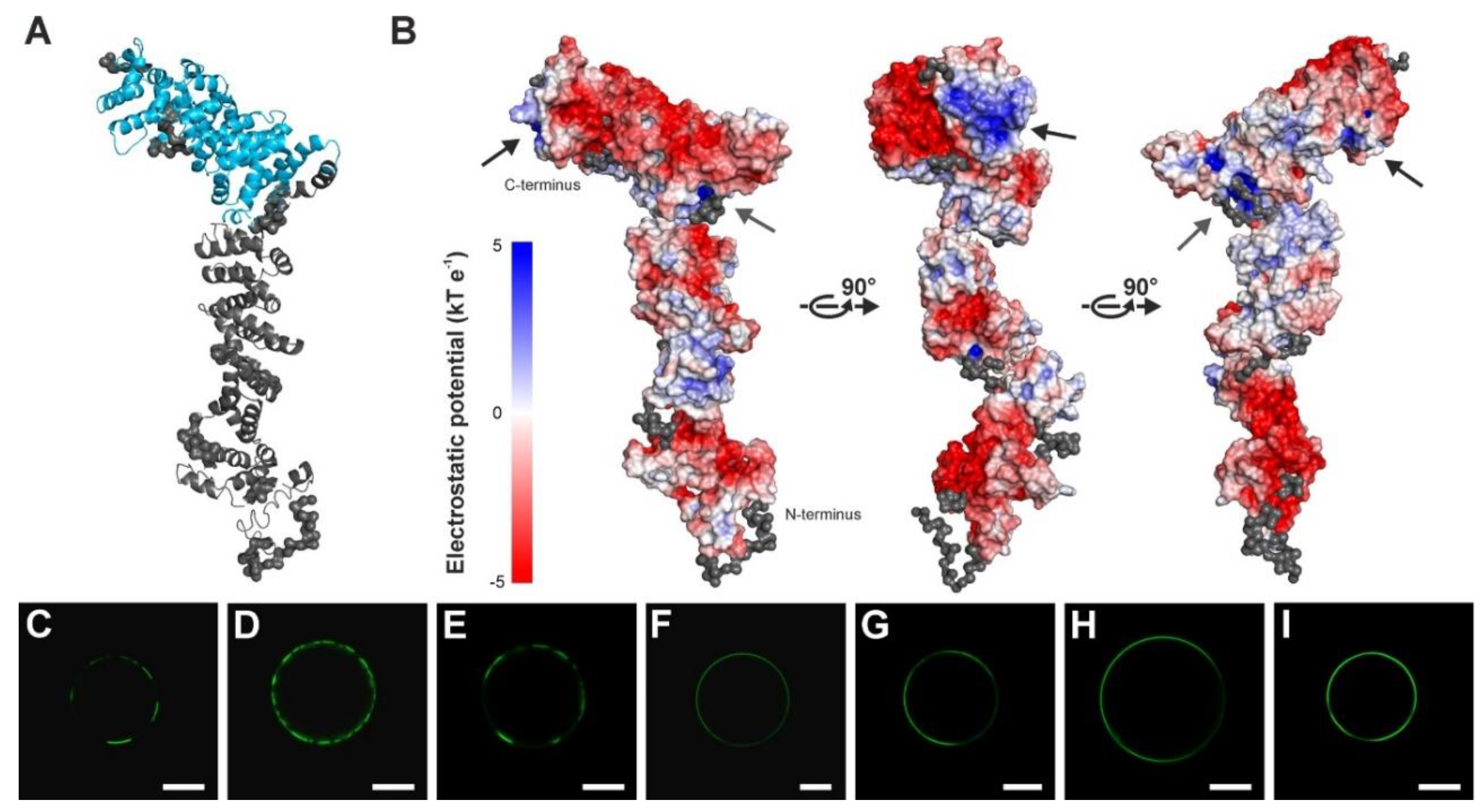
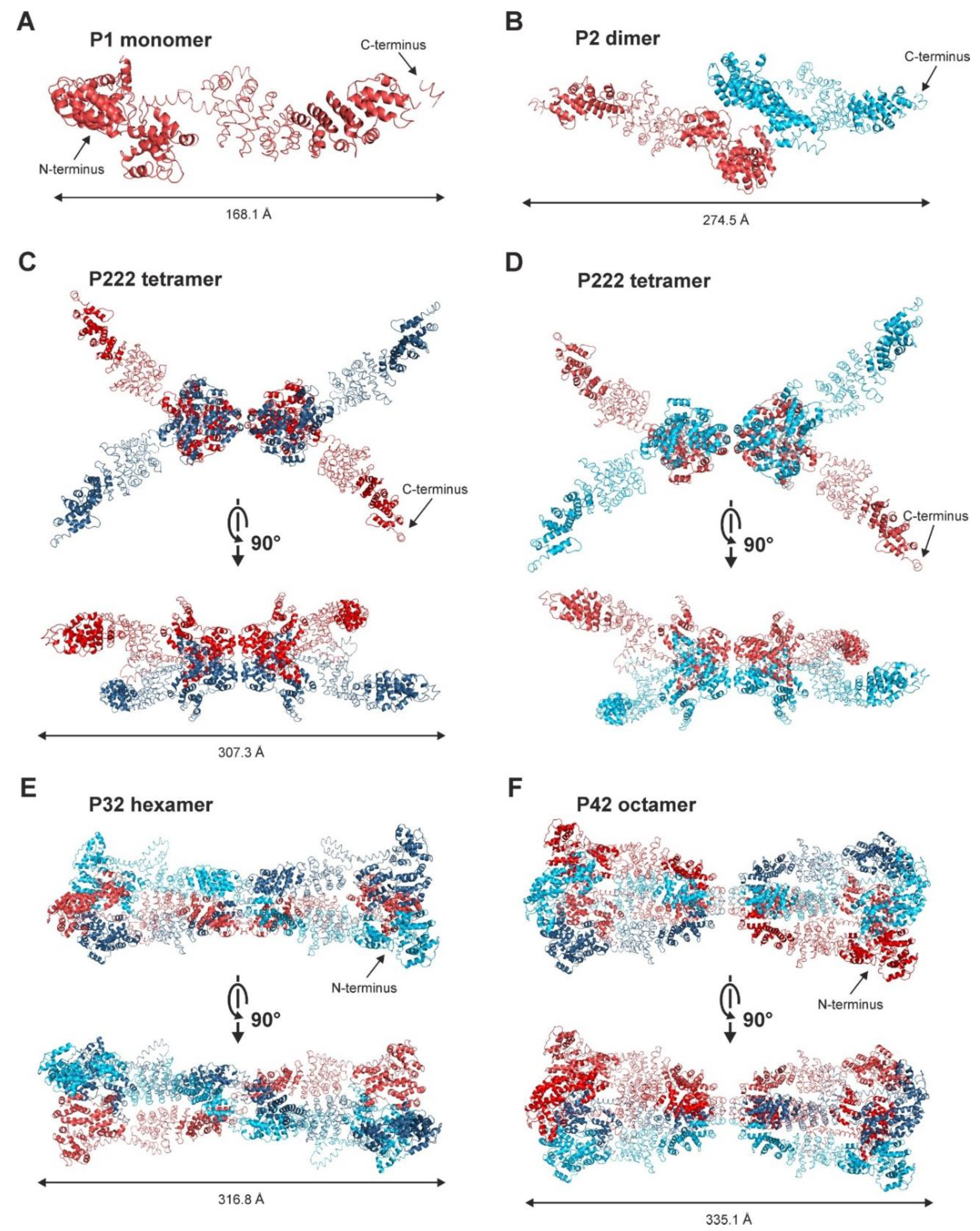
2.2. Structure of SAUL1 Monomers
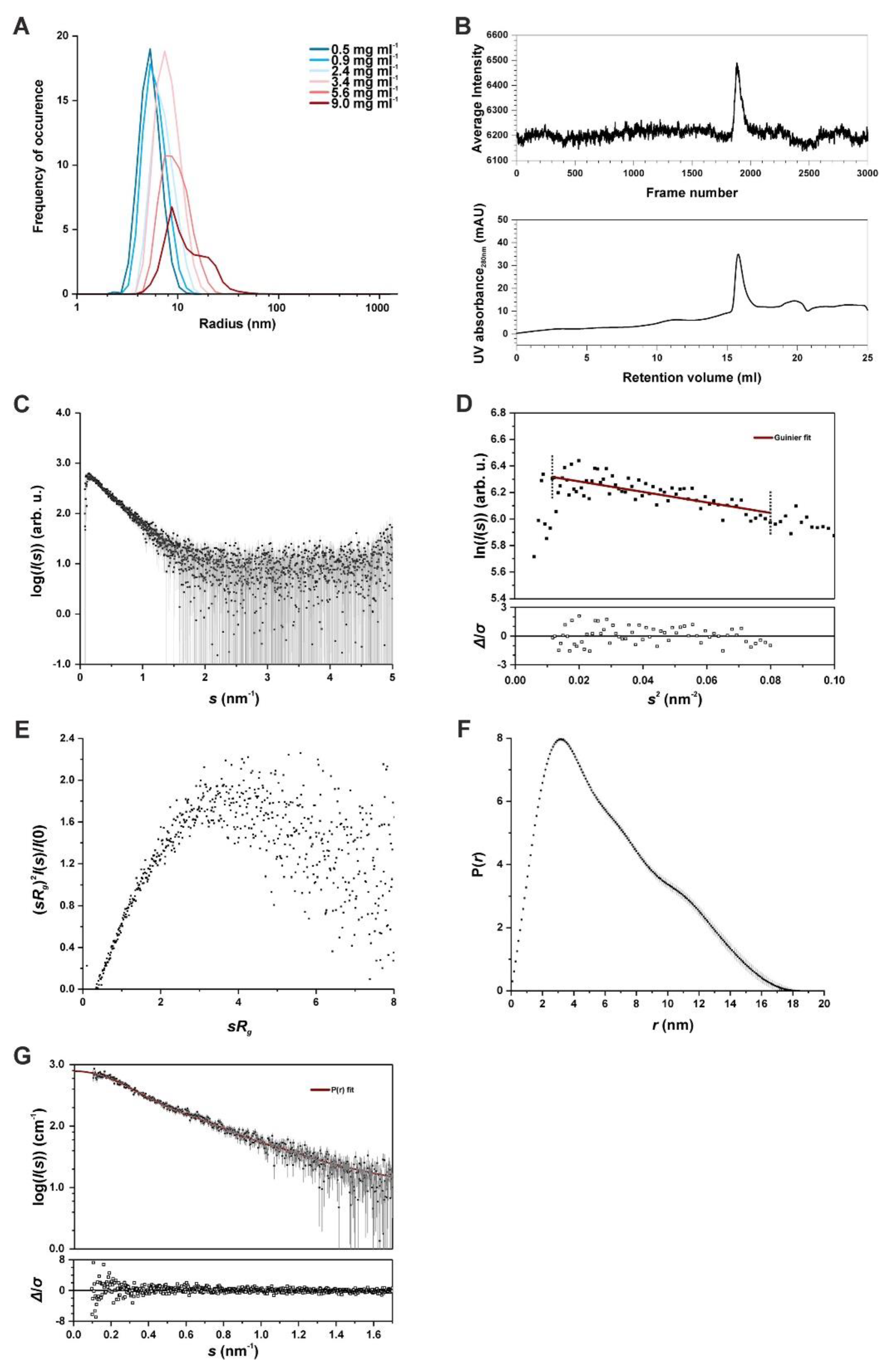
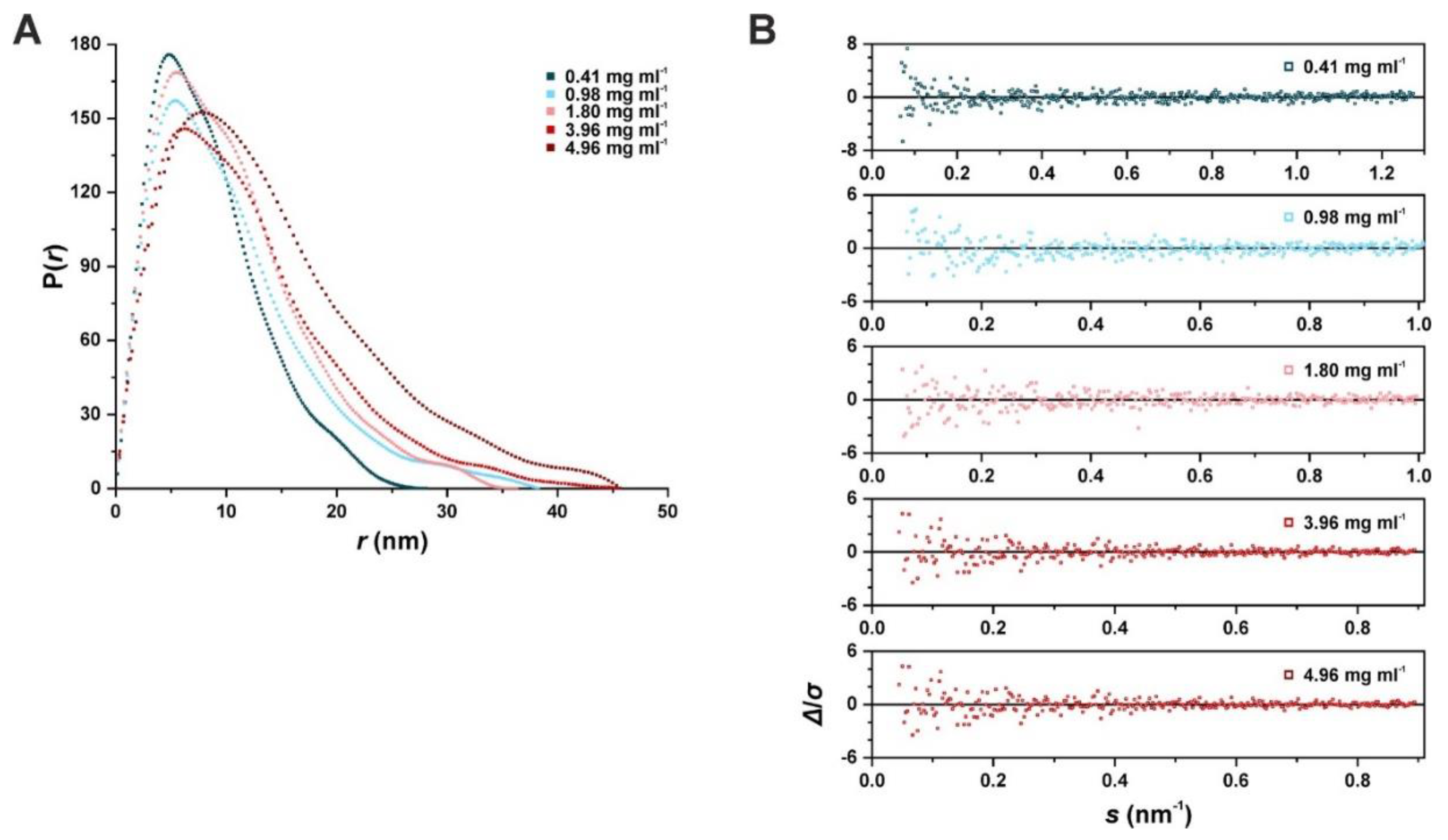
2.3. Structural Analysis of SAUL1 as a Polydisperse System
2.4. Relocalization of SAUL1 upon Masking or Deleting Its N-Terminus
3. Discussion
4. Materials and Methods
4.1. Construction of Plasmids
4.2. Site-Directed Mutagenesis
4.3. Protoplast Transformation
4.4. Confocal Laser Scanning Microscopy
4.5. Expression and Purification of SAUL1
4.6. Dynamic Light Scattering (DLS)
4.7. Circular Dichroism (CD) Spectroscopy
4.8. Small-Angle X-ray Scattering (SAXS) and Inline SEC-SAXS
4.9. Data Processing
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PUB-ARM | Plant U-box armadillo repeat |
| Ub | Ubiquitin |
| SAUL1 | SENESCENCE ASSOCIATED UBIQUITIN E3 LIGASE1 |
| MVBs | Multi-vesicular bodies |
| SAXS | Small-angle X-ray scattering |
| PRR | Pattern recognition receptors |
| PAMP | Pathogen-associated molecular pattern |
| PTI | PAMP-triggered immunity |
| ETI | Effector-triggered immunity |
| SOC3 | Suppressors of chs1-2, 3 |
| PM | Plasma membrane |
| TP | Tonoplast |
| SEC | Size exclusion chromatography |
| DLS | Dynamic light scattering |
| EOM | Ensemble optimization method |
| ARM | Armadillo |
| UBC | Ubiquitin-conjugating |
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef]
- Galan, J.E.; Lara-Tejero, M.; Marlovits, T.C.; Wagner, S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 2014, 68, 415–438. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 2015, 23, 14–22. [Google Scholar] [CrossRef]
- Li, X.; Kapos, P.; Zhang, Y. NLRs in plants. Curr. Opin. Immunol. 2015, 32, 114–121. [Google Scholar] [CrossRef]
- Azevedo, C.; Santos-Rosa, M.J.; Shirasu, K. The U-box protein family in plants. Trends Plant Sci. 2001, 6, 354–358. [Google Scholar] [CrossRef]
- Zhou, B.; Zeng, L. Conventional and unconventional ubiquitination in plant immunity. Mol. Plant Pathol. 2017, 18, 1313–1330. [Google Scholar] [CrossRef]
- Trujillo, M. News from the PUB: Plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 2018, 69, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S.; Yada, M.; Matsumoto, M.; Ishida, N.; Nakayama, K.I. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001, 276, 33111–33120. [Google Scholar] [CrossRef]
- Pruneda, J.N.; Littlefield, P.J.; Soss, S.E.; Nordquist, K.A.; Chazin, W.J.; Brzovic, P.S.; Klevit, R.E. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 2012, 47, 933–942. [Google Scholar] [CrossRef]
- Huber, A.H.; Nelson, W.J.; Weis, W.I. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 1997, 90, 871–882. [Google Scholar] [CrossRef]
- Coates, J.C. Armadillo repeat proteins: Beyond the animal kingdom. Trends Cell Biol. 2003, 13, 463–471. [Google Scholar] [CrossRef]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef]
- Liao, D.; Cao, Y.; Sun, X.; Espinoza, C.; Nguyen, C.T.; Liang, Y.; Stacey, G. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 2017, 214, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, W.; Ning, Y.; Shirsekar, G.; Cai, Y.; Wang, X.; Dai, L.; Wang, Z.; Liu, W.; Wang, G.-L. The U-Box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiol. 2012, 160, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Grubb, L.E.; Wang, J.; Liang, X.; Li, L.; Gao, C.; Ma, M.; Feng, F.; Li, M.; Li, L.; et al. A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 2018, 69, 493–504. [Google Scholar] [CrossRef]
- Trujillo, M.; Ichimura, K.; Casais, C.; Shirasu, K. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 2008, 18, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Anderson, R.G.; Ichimura, K.; Pecenkova, T.; Reuter, P.; Zarsky, V.; McDowell, J.M.; Shirasu, K.; Trujillo, M. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 2012, 24, 4703–4716. [Google Scholar] [CrossRef]
- Gonzalez-Lamothe, R.; Tsitsigiannis, D.I.; Ludwig, A.A.; Panicot, M.; Shirasu, K.; Jones, J.D. The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 2006, 18, 1067–1083. [Google Scholar] [CrossRef]
- Yang, C.-W.; Gonzalez-Lamothe, R.; Ewan, R.A.; Rowland, O.; Yoshioka, H.; Shenton, M.; Ye, H.; O’Donnell, E.; Jones, J.D.; Sadanandom, A. The E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 2006, 18, 1084–1098. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Yamaguchi, K.; Sakamoto, K.; Yoshimura, S.; Inoue, K.; Tsuge, S.; Kojima, C.; Kawasaki, T. Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat. Commun. 2014, 5, 5430. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qu, B.; Dou, S.; Li, L.; Yin, D.; Pang, Z.; Zhou, Z.; Tian, M.; Liu, G.; Xie, Q.; et al. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 2015, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Han, P.-L.; Dong, Y.-H.; Gu, K.-D.; Yu, J.-Q.; Hu, D.-G.; Hao, Y.-J. The apple U-box E3 ubiquitin ligase MdPUB29 contributes to activate plant immune response to the fungal pathogen Botryosphaeria dothidea. Planta 2019, 249, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Kotur, T.; Liang, W.; Vogelmann, K.; Kleine, T.; Leister, D.; Brieske, C.; Yang, S.; Lüdke, D.; Wiermer, M.; et al. E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol. 2017, 215, 1516–1532. [Google Scholar] [CrossRef]
- Raab, S.; Drechsel, G.; Zarepour, M.; Hartung, W.; Koshiba, T.; Bittner, F.; Hoth, S. Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J. 2009, 59, 39–51. [Google Scholar] [CrossRef]
- Disch, E.M.; Tong, M.; Kotur, T.; Koch, G.; Wolf, C.A.; Li, X.; Hoth, S. Membrane-associated ubiquitin ligase SAUL1 suppresses temperature- and humidity-dependent autoimmunity in Arabidopsis. Mol. Plant Microbe Interact. 2016, 29, 69–80. [Google Scholar] [CrossRef]
- Vogelmann, K.; Subert, C.; Danzberger, N.; Drechsel, G.; Bergler, J.; Kotur, T.; Burmester, T.; Hoth, S. Plasma membrane-association of SAUL1-type plant U-box armadillo repeat proteins is conserved in land plants. Front. Plant Sci. 2014, 5, 37. [Google Scholar] [CrossRef]
- Liang, W.; van Wersch, S.; Tong, M.; Li, X. TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol. 2019, 221, 2054–2066. [Google Scholar] [CrossRef]
- Drechsel, G.; Bergler, J.; Wippel, K.; Sauer, N.; Vogelmann, K.; Hoth, S. C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. J. Exp. Bot. 2011, 62, 775–785. [Google Scholar] [CrossRef]
- Tao, K.; Waletich, J.R.; Wise, H.; Arredondo, F.; Tyler, B.M. Tethering of multi-vesicular bodies and the tonoplast to the plasma membrane in plants. Front. Plant Sci. 2019, 10, 636. [Google Scholar] [CrossRef]
- Andersen, P.; Kragelund, B.B.; Olsen, A.N.; Larsen, F.H.; Chua, N.H.; Poulsen, F.M.; Skriver, K. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 2004, 279, 40053–40061. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Windheim, M.; Roe, S.M.; Peggie, M.; Cohen, P.; Prodromou, C.; Pearl, L.H. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 2005, 20, 525–538. [Google Scholar] [CrossRef]
- Kooi, C.W.V.; Ohi, M.D.; Rosenberg, J.A.; Oldham, M.L.; Newcomer, M.E.; Gould, K.L.; Chazin, W.J. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry 2006, 45, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Tu, D.; Li, W.; Ye, Y.; Brunger, A.T. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. USA 2007, 104, 15599–15606. [Google Scholar] [CrossRef] [PubMed]
- Graewert, M.A.; Franke, D.; Jeffries, C.M.; Blanchet, C.E.; Ruskule, D.; Kuhle, K.; Flieger, A.; Schafer, B.; Tartsch, B.; Meijers, R.; et al. Automated pipeline for purification, biophysical and x-ray analysis of biomacromolecular solutions. Sci. Rep. 2015, 5, 10734. [Google Scholar] [CrossRef]
- Mudgil, Y.; Shiu, S.H.; Stone, S.L.; Salt, J.N.; Goring, D.R. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004, 134, 59–66. [Google Scholar] [CrossRef]
- Tewari, R.; Bailes, E.; Bunting, K.A.; Coates, J.C. Armadillo-repeat protein functions: Questions for little creatures. Trends Cell Biol. 2010, 20, 470–481. [Google Scholar] [CrossRef]
- Ritco-Vonsovici, M.; Ababou, A.; Horton, M. Molecular plasticity of beta-catenin: New insights from single-molecule measurements and MD simulation. Protein Sci. 2007, 16, 1984–1998. [Google Scholar] [CrossRef]
- Zhang, Z.; Lin, K.; Gao, L.; Chen, L.; Shi, X.; Wu, G. Crystal structure of the armadillo repeat domain of adenomatous polyposis coli which reveals its inherent flexibility. Biochem. Biophys. Res. Commun. 2011, 412, 732–736. [Google Scholar] [CrossRef]
- Antignani, V.; Klocko, A.L.; Bak, G.; Chandrasekaran, S.D.; Dunivin, T.; Nielsen, E. Recruitment of PLANT U-BOX13 and the PI4Kbeta1/beta2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell 2015, 27, 243–261. [Google Scholar] [CrossRef]
- Yee, D.; Goring, D.R. The diversity of plant U-box E3 ubiquitin ligases: From upstream activators to downstream target substrates. J. Exp. Bot. 2009, 60, 1109–1121. [Google Scholar] [CrossRef]
- Furlan, G.; Nakagami, H.; Eschen-Lippold, L.; Jiang, X.; Majovsky, P.; Kowarschik, K.; Hoehenwarter, W.; Lee, J.; Trujillo, M. Changes in PUB22 ubiquitination modes triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 dampen the immune response. Plant Cell 2017, 29, 726–745. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Zhao, P.; Seo, J.S.; Mitsuda, N.; Deng, S.; Chua, N.H. PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 2015, 27, 2016–2031. [Google Scholar] [CrossRef] [PubMed]
- Zarsky, V.; Kulich, I.; Fendrych, M.; Pecenkova, T. Exocyst complexes multiple functions in plant cells secretory pathways. Curr. Opin. Plant Biol. 2013, 16, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.H.; Ahn, M.Y.; Park, K.Y.; Kim, E.Y.; Kim, W.T. The N-Terminal UND Motif of the Arabidopsis U-Box E3 Ligase PUB18 is critical for the negative regulation of ABA-mediated stomatal movement and determines its ubiquitination specificity for exocyst subunit Exo70B1. Plant Cell 2016, 28, 2952–2973. [Google Scholar] [CrossRef]
- An, Q.; van Bel, A.J.; Hückelhoven, R. Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2007, 2, 4–7. [Google Scholar] [CrossRef]
- Hanson, P.I.; Cashikar, A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 2012, 28, 337–362. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Samuel, M.; Bleackley, M.; Anderson, M.; Mathivanan, S. Extracellular vesicles including exosomes in cross kingdom regulation: A viewpoint from plant-fungal interactions. Front. Plant Sci. 2015, 6, 766. [Google Scholar] [CrossRef] [PubMed]
- Rutter, B.D.; Innes, R.W. Extracellular vesicles as key mediators of plant-microbe interactions. Curr. Opin. Plant Biol. 2018, 44, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, P.; Rutter, B.D.; Karimi, H.Z.; Podicheti, R.; Meyers, B.C.; Innes, R.W. Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-Nucleotide “Tiny” RNAs. Plant Cell 2019, 31, 315–324. [Google Scholar] [CrossRef]
- Provencher, S.W. Contin: A general purpose constrained regularization program for inverting noisy linear algebraic and itegral equations. Comput. Phys. Commun. 1982, 27, 229–242. [Google Scholar] [CrossRef]
- Reed, J.; Reed, T.A. A set of constructed type spectra for the practical estimation of peptide secondary structure from circular dichroism. Anal. Biochem. 1997, 254, 36–40. [Google Scholar] [CrossRef]
- Mylonas, E.; Svergun, D.I. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scatterig. J. Appl. Crsystallogr. 2007, 40, 245–249. [Google Scholar] [CrossRef]
- Panjkovich, A.; Svergun, D.I. Deciphering conformational transitions of proteins by small angle X-ray scattering and normal mode analysis. Phys. Chem. Chem. Phys. 2016, 18, 5707–5719. [Google Scholar] [CrossRef]
- Petoukhov, M.V.; Konarev, P.V.; Kikhney, A.G.; Svergun, D.I. ATSAS 2.1—Towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 2007, 40, 223–228. [Google Scholar] [CrossRef]
- Franke, D.; Petoukhov, M.V.; Konarev, P.V.; Panjkovich, A.; Tuukkanen, A.; Mertens, H.D.T.; Kikhney, A.G.; Hajizadeh, N.R.; Franklin, J.M.; Jeffries, C.M.; et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50, 1212–1225. [Google Scholar] [CrossRef]
- Flory, P.J. Principles of Polymer Chemistry; Cornell University Press: Ithaca, NY, USA, 1953. [Google Scholar]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knop, J.; Lienemann, T.; El-Kilani, H.; Falke, S.; Krings, C.; Sindalovskaya, M.; Bergler, J.; Betzel, C.; Hoth, S. Structural Features of a Full-Length Ubiquitin Ligase Responsible for the Formation of Patches at the Plasma Membrane. Int. J. Mol. Sci. 2021, 22, 9455. https://doi.org/10.3390/ijms22179455
Knop J, Lienemann T, El-Kilani H, Falke S, Krings C, Sindalovskaya M, Bergler J, Betzel C, Hoth S. Structural Features of a Full-Length Ubiquitin Ligase Responsible for the Formation of Patches at the Plasma Membrane. International Journal of Molecular Sciences. 2021; 22(17):9455. https://doi.org/10.3390/ijms22179455
Chicago/Turabian StyleKnop, Jan, Tim Lienemann, Haifa El-Kilani, Sven Falke, Catharina Krings, Maria Sindalovskaya, Johannes Bergler, Christian Betzel, and Stefan Hoth. 2021. "Structural Features of a Full-Length Ubiquitin Ligase Responsible for the Formation of Patches at the Plasma Membrane" International Journal of Molecular Sciences 22, no. 17: 9455. https://doi.org/10.3390/ijms22179455
APA StyleKnop, J., Lienemann, T., El-Kilani, H., Falke, S., Krings, C., Sindalovskaya, M., Bergler, J., Betzel, C., & Hoth, S. (2021). Structural Features of a Full-Length Ubiquitin Ligase Responsible for the Formation of Patches at the Plasma Membrane. International Journal of Molecular Sciences, 22(17), 9455. https://doi.org/10.3390/ijms22179455






