The Antihypertensive Drug Telmisartan Protects Oligodendrocytes from Cholesterol Accumulation and Promotes Differentiation by a PPAR-γ-Mediated Mechanism
Abstract
:1. Introduction
2. Results
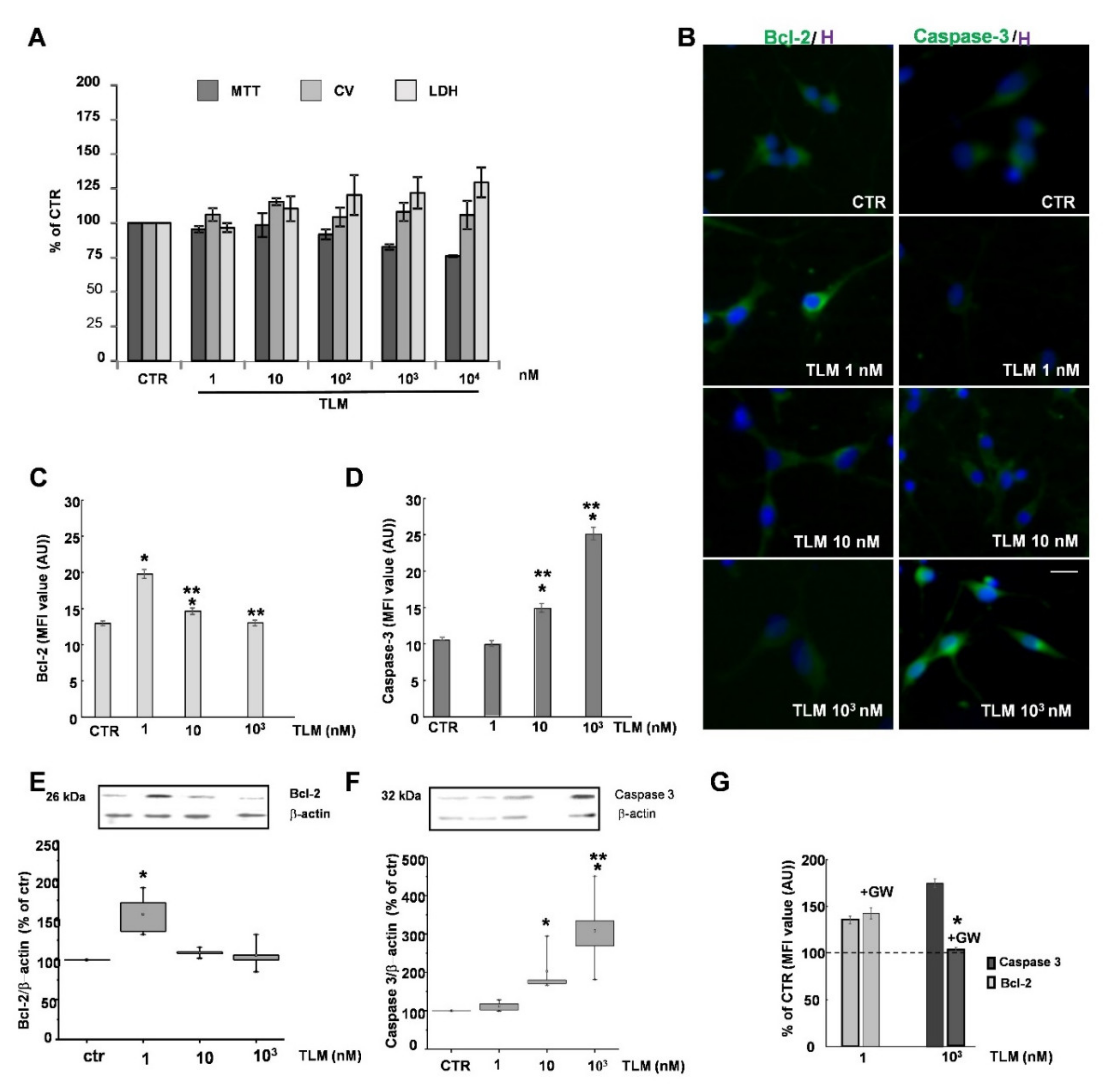
2.1. Effects of TLM on OP Cell Viability in Culture
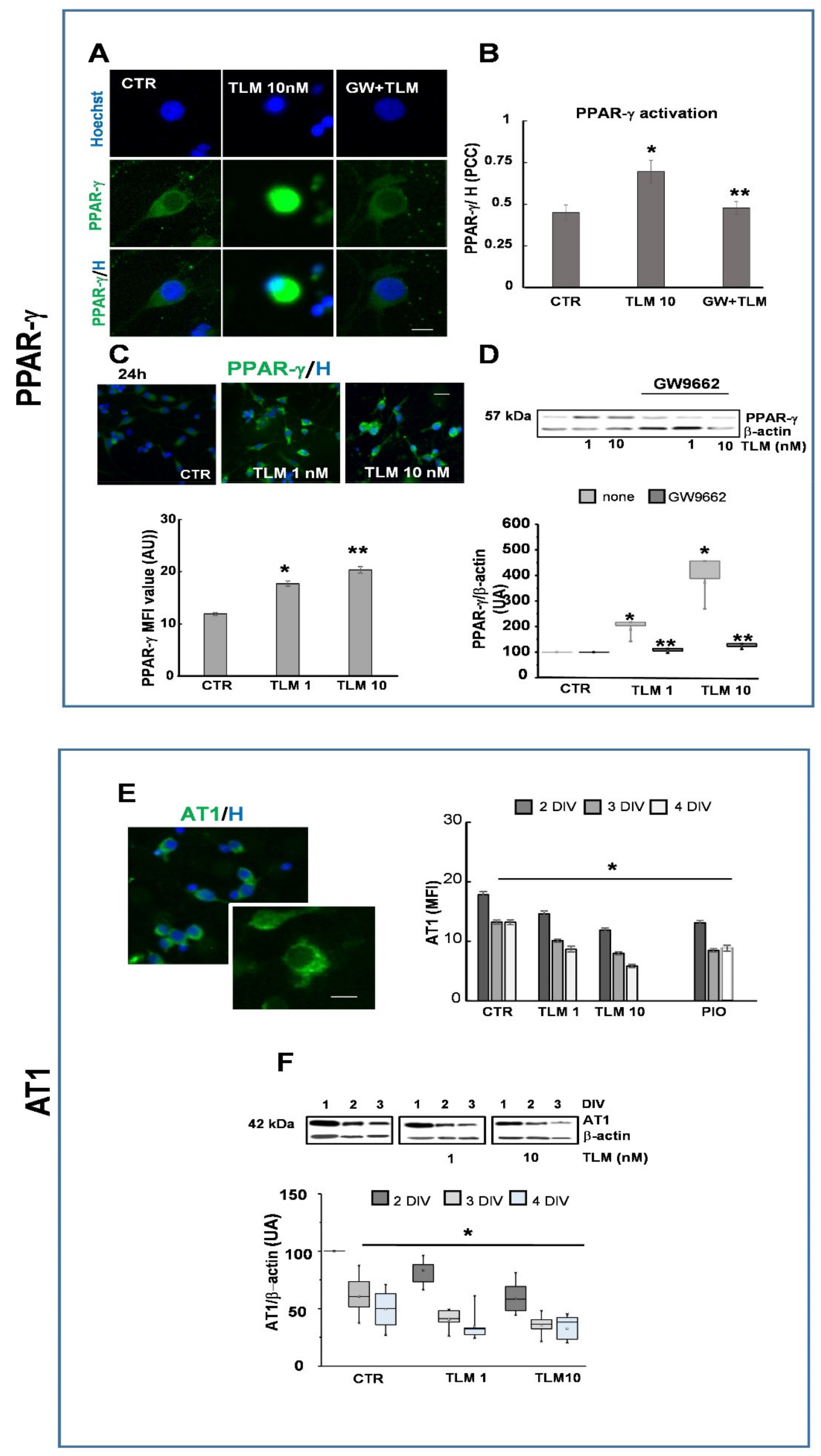
2.2. Telmisartan Activates PPAR-γ and Regulates AT1 Expression by a PPAR-γ-Mediated Mechanism
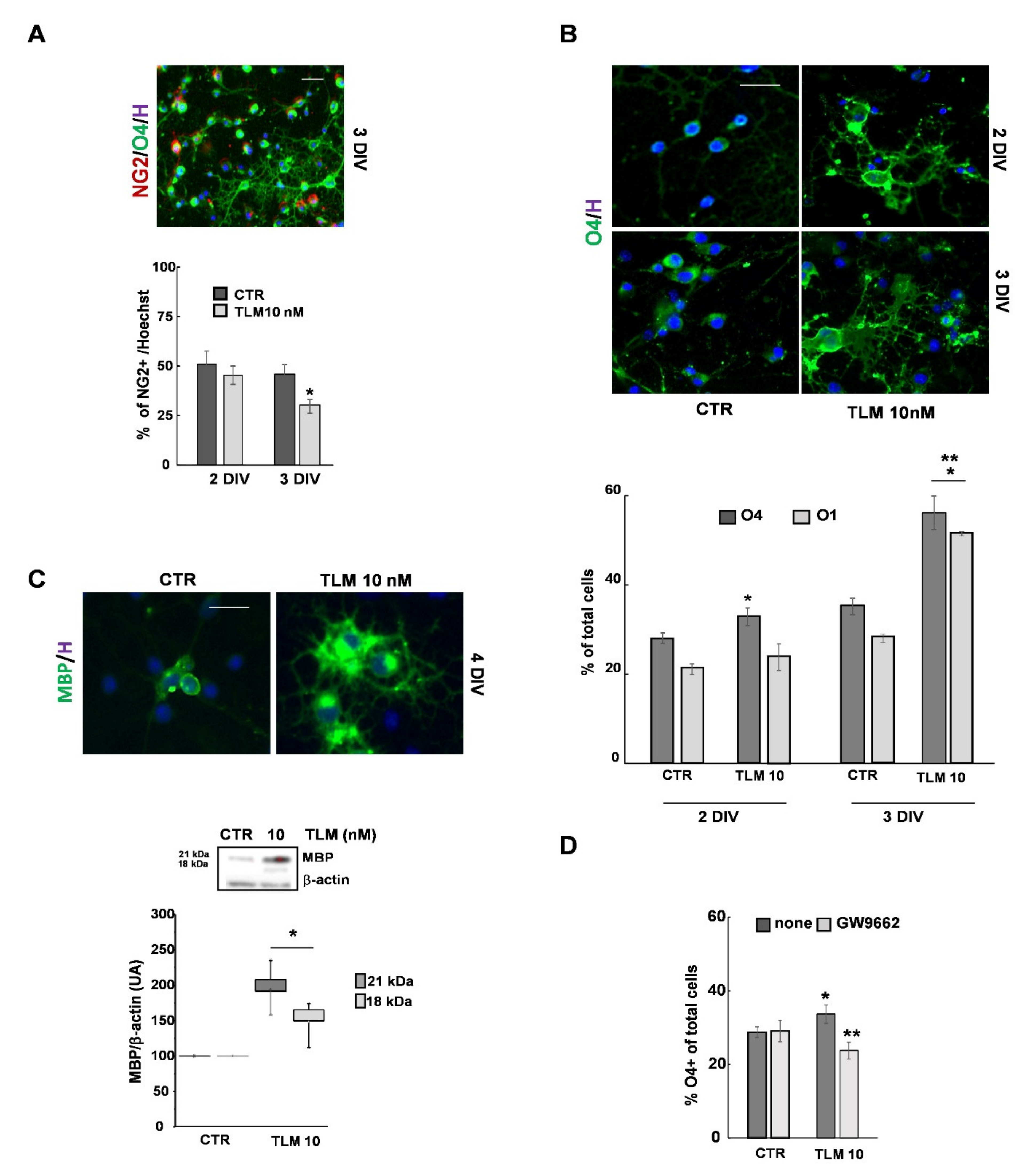
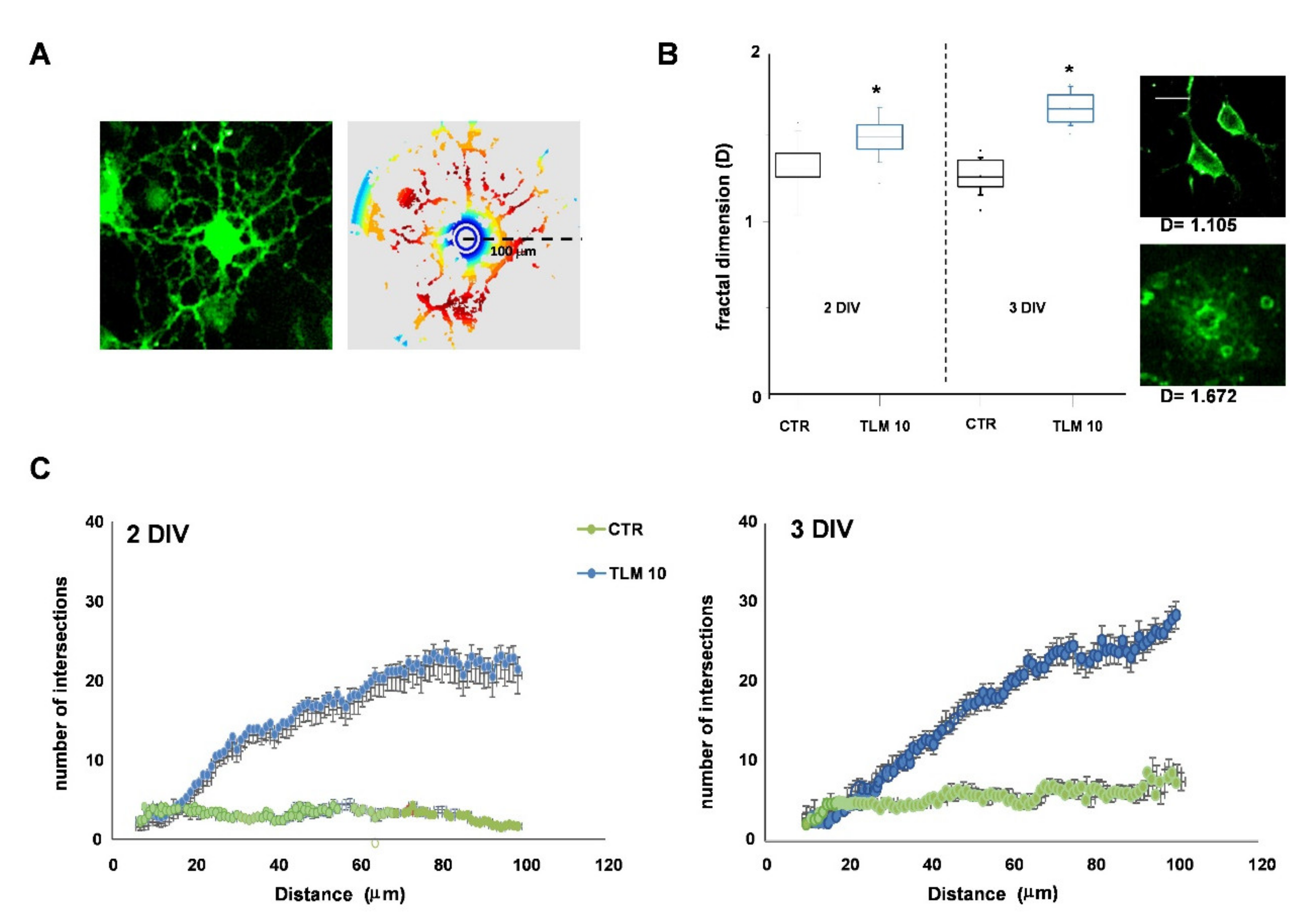
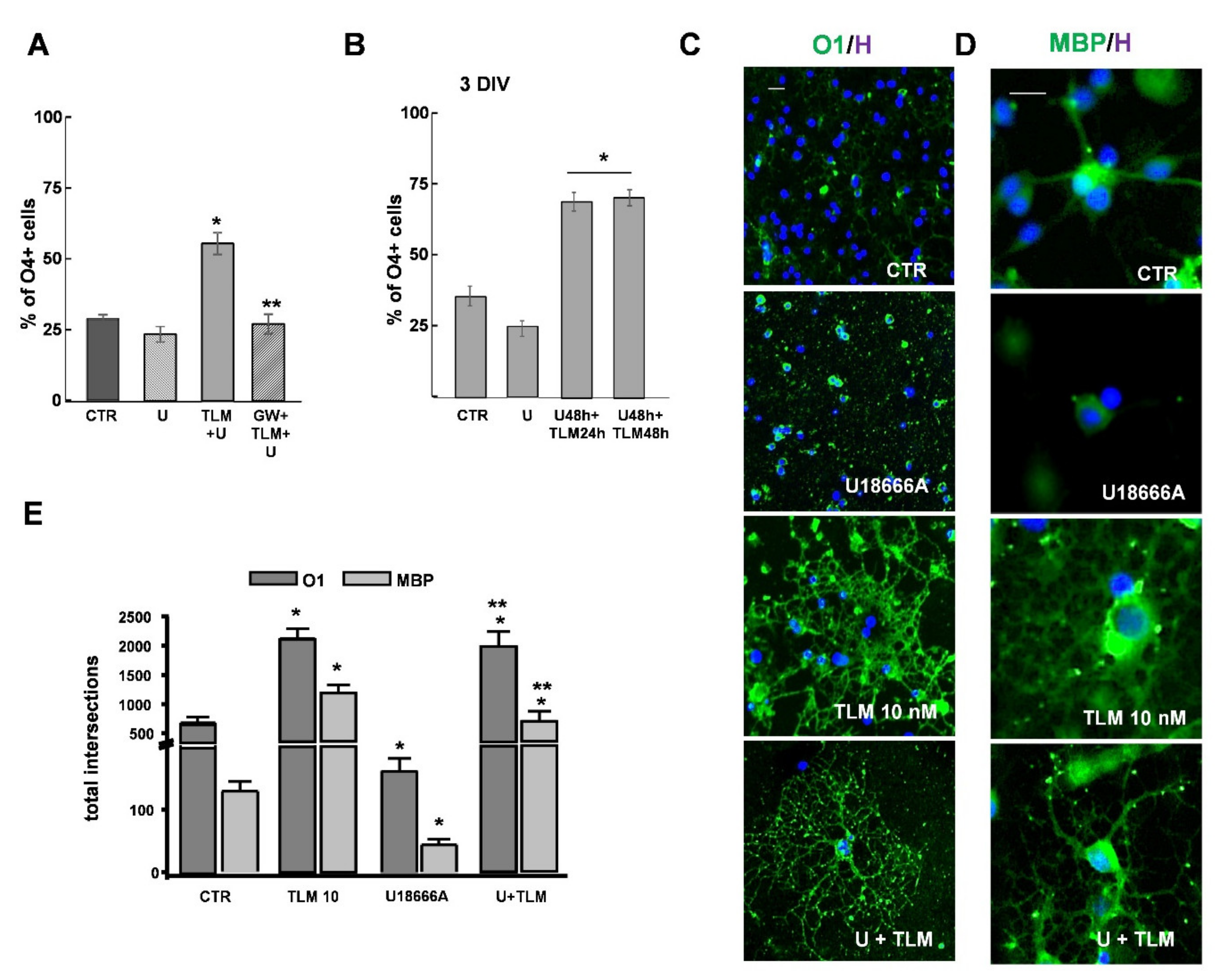
2.3. Telmisartan Promotes the OP Differentiation in a PPAR-γ-Dependent Manner
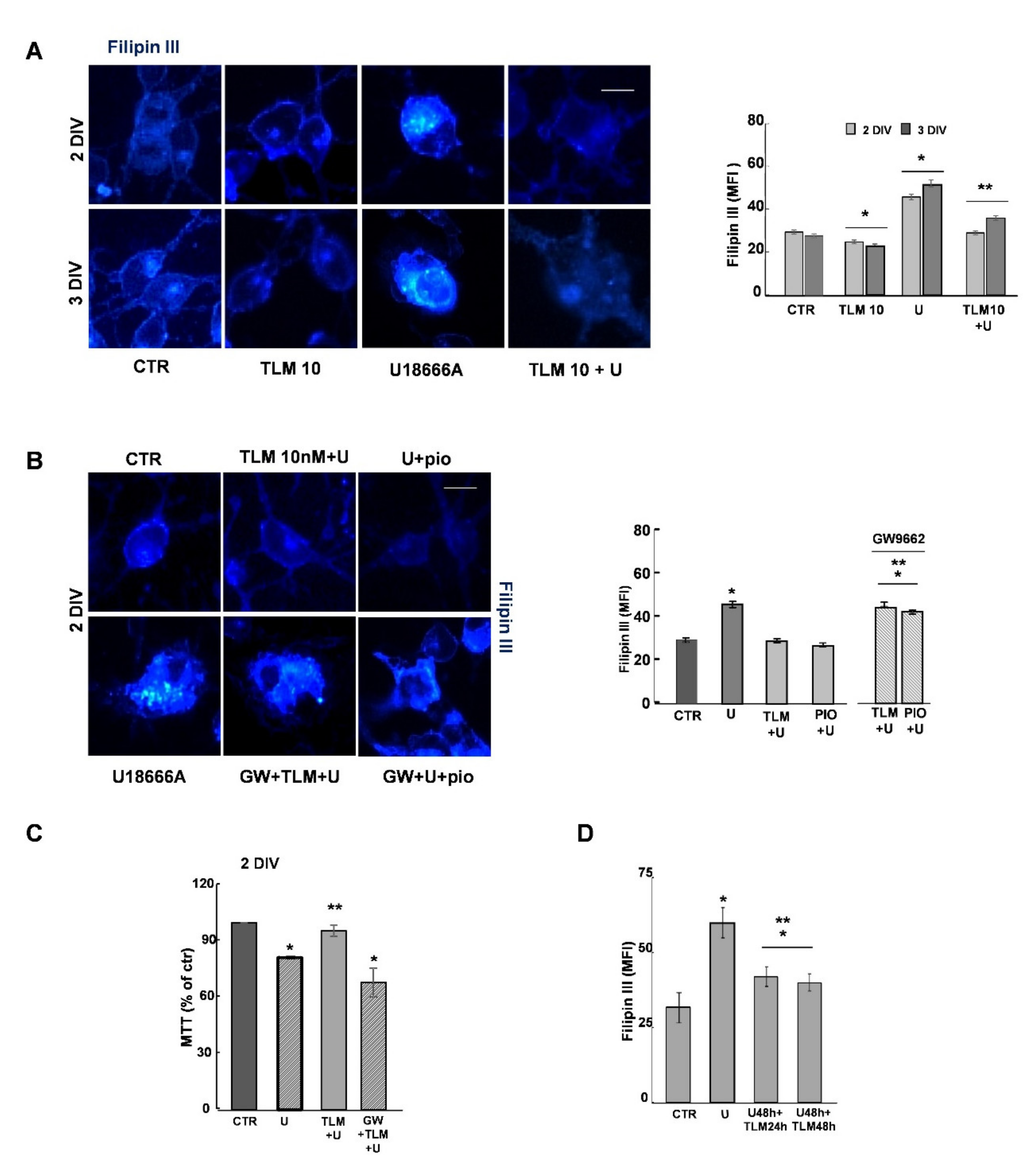
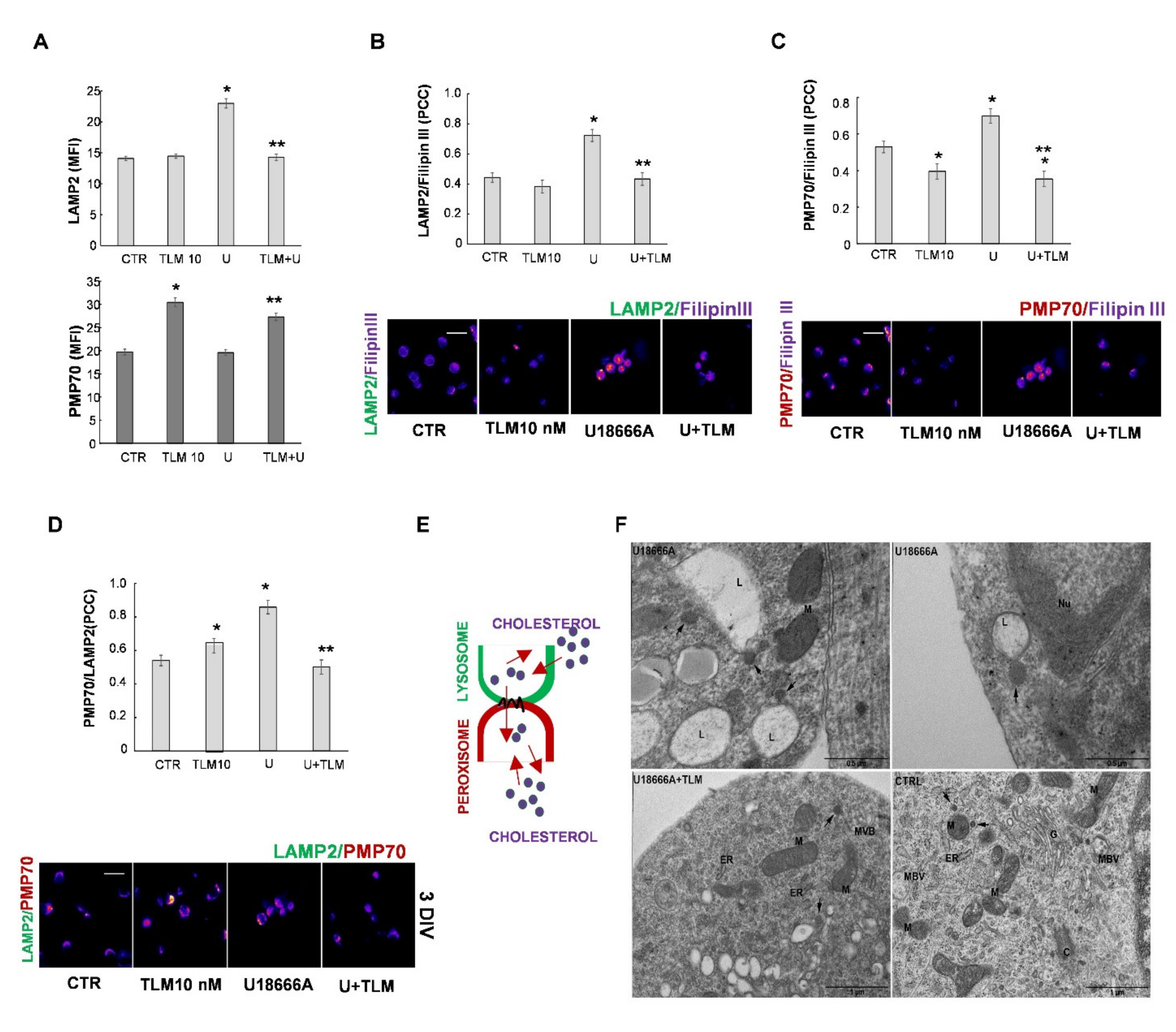
2.4. Effects of Telmisartan in a Cellular Model of Niemann-Pick Type C Disease
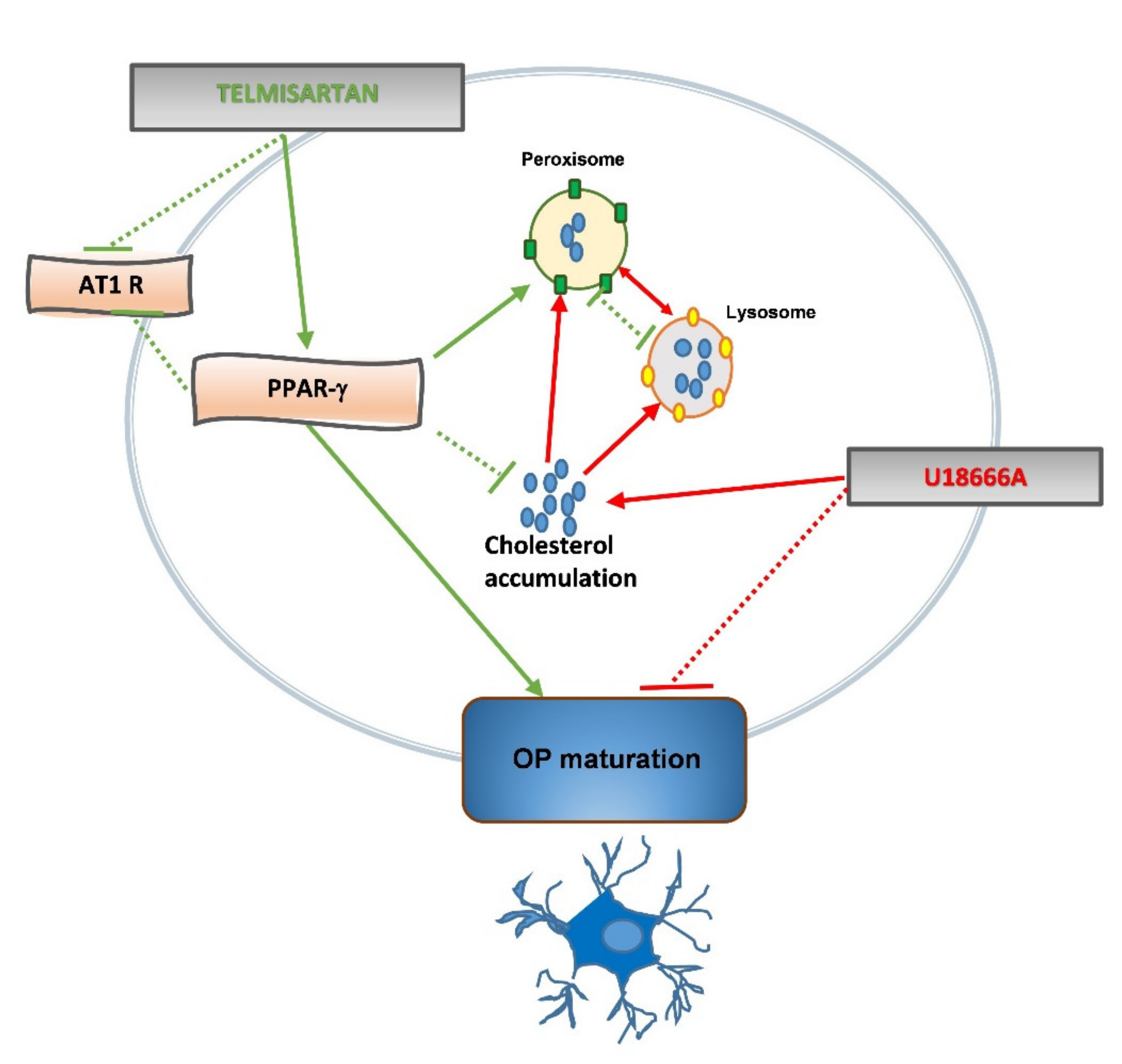
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Cell Viability
4.3. Immunofluorescence
4.4. Image Analysis and Quantification
4.5. Transmission Electron Microscopy (TEM)
4.6. Western Blot Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARB | Angiotensin II receptor blocker |
| AT1 | type 1 angiotensin II receptor |
| BrdU | 5-Bromo-2′-deoxyuridine |
| CNS | Central Neurvus System |
| CTR | Control |
| CV | Crystal violet |
| D | Fractal dimension |
| DIV | Days in vitro |
| IF | Immunofluorescence |
| LAMP2 | Lysosome-associated membrane protein 2 |
| LDH | Lactate dehydrogenase |
| MBP | Myelin basic protein |
| MFI | Mean fluorescence intensity |
| MTT | 3-(4,5-dimethyl thiazol-2-y1)-2,5-diphenyl tetrazolium bromide |
| NG2 | Proteoglycan, Neural/glial antigen 2 |
| NPC | Niemann-Pick type C |
| O1 | galactocerebroside |
| O4 | cell surface sulfatide |
| OL | Oligodendrocyte |
| OP | Oligodendrocyte progenitor |
| PCC | Pearson’s correlation coefficient |
| PCNA | proliferating nuclear antigen |
| PIO | Pioglitazone |
| PMP70 | 70-kDa peroxisomal integral membrane protein |
| PPAR-γ | Peroxisome proliferator-activated receptor gamma |
| RT | Room temperature |
| SPPAR-γ Ms | Selective Modulator of PPAR-γ activity |
| TEM | Transmission Electron Microscopy |
| TLM | Telmisartan |
| TNF-a | Tumor necrosis factor-alpha |
| WB | Western blot |
References
- Villapol, S.; Saavedra, J.M. Neuroprotective Effects of Angiotensin Receptor Blockers. Am. J. Hypertens. 2015, 28, 289–299. [Google Scholar] [CrossRef] [Green Version]
- Schupp, M.; Clemenz, M.; Gineste, R.; Witt, H.; Janke, J.; Helleboid, S.; Hennuyer, N.; Ruiz, P.; Unger, T.; Staels, B.; et al. Molecular Characterization of New Selective Peroxisome Proliferator-Activated Receptor Modulators With Angiotensin Receptor Blocking Activity. Diabetes 2005, 54, 3442–3452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shindo, T.; Takasaki, K.; Uchida, K.; Onimura, R.; Kubota, K.; Uchida, N.; Irie, K.; Katsurabayashi, S.; Mishima, K.; Nishimura, R.; et al. Ameliorative Effects of Telmisartan on the Inflammatory Response and Impaired Spatial Memory in a Rat Model of Alzheimer’s Disease Incorporating Additional Cerebrovascular Disease Factors. Biol. Pharm. Bull. 2012, 35, 2141–2147. [Google Scholar] [CrossRef] [Green Version]
- Kolli, V.; Stechschulte, L.A.; Dowling, A.R.; Rahman, S.; Czernik, P.J.; Lecka-Czernik, B. Partial Agonist, Telmisartan, Maintains PPARγ Serine 112 Phosphorylation, and Does Not Affect Osteoblast Differentiation and Bone Mass. PLoS ONE 2014, 9, e96323. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Pang, T.; Hafko, R.; Benicky, J.; Sanchez-Lemus, E.; Saavedra, J.M. Telmisartan Ameliorates Glutamate-Induced Neurotoxicity: Roles of AT1 Receptor Blockade and PPARγ Activation. Neuropharmacology 2014, 79, 249–261. [Google Scholar] [CrossRef] [Green Version]
- Imayama, I.; Ichiki, T.; Inanaga, K.; Ohtsubo, H.; Fukuyama, K.; Ono, H.; Hashiguchi, Y.; Sunagawa, K. Telmisartan Downregulates Angiotensin II Type 1 Receptor through Activation of Peroxisome Proliferator-Activated Receptor γ. Cardiovasc. Res. 2006, 72, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, W.; Liu, Y.; Wang, Y.; Zhang, J.; Li, M.; Bu, M. Effect of Telmisartan on Preventing Learning and Memory Deficits Via Peroxisome Proliferator-Activated Receptor-γ in Vascular Dementia Spontaneously Hypertensive Rats. J. Stroke Cerebrovasc. Dis. 2018, 27, 277–285. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, T.; Li, C.; Guo, J.; Xu, B.; Xue, L. Telmisartan Attenuates Human Glioblastoma Cells Proliferation and Oncogenicity by Inducing the Lipid Oxidation. Asia-Pac. J. Clin. Oncol. 2021. [Google Scholar] [CrossRef]
- Bernardo, A.; Minghetti, L. PPAR-γ; Agonists as Regulators of Microglial Activation and Brain Inflammation. Curr. Pharm. Des. 2006, 12, 93–109. [Google Scholar] [CrossRef]
- Heneka, M.T.; Landreth, G.E.; Hüll, M. Drug Insight: Effects Mediated by Peroxisome Proliferator-Activated Receptor-γ in CNS Disorders. Nat. Rev. Neurol. 2007, 3, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Fakan, B.; Szalardy, L.; Vecsei, L. Exploiting the Therapeutic Potential of Endogenous Immunomodulatory Systems in Multiple Sclerosis—Special Focus on the Peroxisome Proliferator-Activated Receptors (PPARs) and the Kynurenines. IJMS 2019, 20, 426. [Google Scholar] [CrossRef] [Green Version]
- Villapol, S. Roles of Peroxisome Proliferator-Activated Receptor Gamma on Brain and Peripheral Inflammation. Cell Mol. Neurobiol. 2018, 38, 121–132. [Google Scholar] [CrossRef]
- Tong, Q.; Wu, L.; Jiang, T.; Ou, Z.; Zhang, Y.; Zhu, D. Inhibition of Endoplasmic Reticulum Stress-Activated IRE1α-TRAF2-Caspase-12 Apoptotic Pathway Is Involved in the Neuroprotective Effects of Telmisartan in the Rotenone Rat Model of Parkinson’s Disease. Eur. J. Pharmacol. 2016, 776, 106–115. [Google Scholar] [CrossRef]
- Khallaf, W.A.I.; Messiha, B.A.S.; Abo-Youssef, A.M.H.; El-Sayed, N.S. Protective Effects of Telmisartan and Tempol on Lipopolysaccharide-Induced Cognitive Impairment, Neuroinflammation, and Amyloidogenesis: Possible Role of Brain-Derived Neurotrophic Factor. Can. J. Physiol. Pharmacol. 2017, 95, 850–860. [Google Scholar] [CrossRef]
- Abo-Youssef, A.M.; Khallaf, W.A.; Khattab, M.M.; Messiha, B.A.S. The Anti-Alzheimer Effect of Telmisartan in a Hyperglycemic Ovariectomized Rat Model; Role of Central Angiotensin and Estrogen Receptors. Food Chem. Toxicol. 2020, 142, 111441. [Google Scholar] [CrossRef]
- Abd El Aziz, A.E.; Sayed, R.H.; Sallam, N.A.; El Sayed, N.S. Neuroprotective Effects of Telmisartan and Nifedipine Against Cuprizone-Induced Demyelination and Behavioral Dysfunction in Mice: Roles of NF-ΚB and Nrf2. Inflammation 2021. [Google Scholar] [CrossRef]
- Pang, T.; Sun, L.; Wang, T.; Jiang, Z.; Liao, H.; Zhang, L. Telmisartan Protects Central Neurons against Nutrient Deprivation-Induced Apoptosis in Vitro through Activation of PPARγ and the Akt/GSK-3β Pathway. Acta Pharm. Sin. 2014, 35, 727–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elkahloun, A.G.; Rodriguez, Y.; Alaiyed, S.; Wenzel, E.; Saavedra, J.M. Telmisartan Protects a Microglia Cell Line from LPS Injury Beyond AT1 Receptor Blockade or PPARγ Activation. Mol. Neurobiol. 2019, 56, 3193–3210. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, A.; De Simone, R.; De Nuccio, C.; Visentin, S.; Minghetti, L. The Nuclear Receptor Peroxisome Proliferator-Activated Receptor-γ Promotes Oligodendrocyte Differentiation through Mechanisms Involving Mitochondria and Oscillatory Ca2+ Waves. Biol. Chem. 2013, 394, 1607–1614. [Google Scholar] [CrossRef]
- De Nuccio, C.; Bernardo, A.; De Simone, R.; Mancuso, E.; Magnaghi, V.; Visentin, S.; Minghetti, L. Peroxisome Proliferator-Activated Receptor γ Agonists Accelerate Oligodendrocyte Maturation and Influence Mitochondrial Functions and Oscillatory Ca 2+ Waves. J. Neuropathol. Exp. Neurol. 2011, 70, 900–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Nuccio, C.; Bernardo, A.; Cruciani, C.; De Simone, R.; Visentin, S.; Minghetti, L. Peroxisome Proliferator Activated Receptor-γ Agonists Protect Oligodendrocyte Progenitors against Tumor Necrosis Factor-Alpha-Induced Damage: Effects on Mitochondrial Functions and Differentiation. Exp. Neurol. 2015, 271, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Tauheed, A.M.; Ayo, J.O.; Kawu, M.U. Regulation of Oligodendrocyte Differentiation: Insights and Approaches for the Management of Neurodegenerative Disease. Pathophysiology 2016, 23, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Berry, K.; Wang, J.; Lu, Q.R. Epigenetic Regulation of Oligodendrocyte Myelination in Developmental Disorders and Neurodegenerative Diseases. F1000Research 2020, 9, 105. [Google Scholar] [CrossRef] [Green Version]
- Fogarty, D.J.; Matute, C. Angiotensin Receptor-like Immunoreactivity in Adult Brain White Matter Astrocytes and Oligodendrocytes. Glia 2001, 35, 131–146. [Google Scholar] [CrossRef]
- Bernardo, A.; Bianchi, D.; Magnaghi, V.; Minghetti, L. Peroxisome Proliferator-Activated Receptor-γ Agonists Promote Differentiation and Antioxidant Defenses of Oligodendrocyte Progenitor Cells. J. Neuropathol. Exp. Neurol. 2009, 68, 797–808. [Google Scholar] [CrossRef]
- Behar, T.N. Analysis of Fractal Dimension of O2A Glial Cells Differentiating in Vitro. Methods 2001, 24, 331–339. [Google Scholar] [CrossRef]
- Fernández, E.; Jelinek, H.F. Use of Fractal Theory in Neuroscience: Methods, Advantages, and Potential Problems. Methods 2001, 24, 309–321. [Google Scholar] [CrossRef]
- Ferreira, T.A.; Blackman, A.V.; Oyrer, J.; Jayabal, S.; Chung, A.J.; Watt, A.J.; Sjöström, P.J.; van Meyel, D.J. Neuronal Morphometry Directly from Bitmap Images. Nat. Methods 2014, 11, 982–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valero, J.; Weruaga, E.; Murias, A.R.; Recio, J.S.; Alonso, J.R. Proliferation Markers in the Adult Rodent Brain: Bromodeoxyuridine and Proliferating Cell Nuclear Antigen. Brain Res. Protoc. 2005, 15, 127–134. [Google Scholar] [CrossRef]
- Zheng, Y.; Begum, S.; Zhang, C.; Fleming, K.; Masumura, C.; Zhang, M.; Smith, P.; Darlington, C. Increased BrdU Incorporation Reflecting DNA Repair, Neuronal de-Differentiation or Possible Neurogenesis in the Adult Cochlear Nucleus Following Bilateral Cochlear Lesions in the Rat. Exp. Brain Res. 2011, 210, 477–487. [Google Scholar] [CrossRef]
- Cenedella, R.J. Cholesterol Synthesis Inhibitor U18666A and the Role of Sterol Metabolism and Trafficking in Numerous Pathophysiological Processes. Lipids 2009, 44, 477–487. [Google Scholar] [CrossRef]
- De Nuccio, C.; Bernardo, A.; Ferrante, A.; Pepponi, R.; Martire, A.; Falchi, M.; Visentin, S.; Popoli, P.; Minghetti, L. Adenosine A2A Receptor Stimulation Restores Cell Functions and Differentiation in Niemann-Pick Type C-like Oligodendrocytes. Sci. Rep. 2019, 9, 9782. [Google Scholar] [CrossRef]
- Chu, B.-B.; Liao, Y.-C.; Qi, W.; Xie, C.; Du, X.; Wang, J.; Yang, H.; Miao, H.-H.; Li, B.-L.; Song, B.-L. Cholesterol Transport through Lysosome-Peroxisome Membrane Contacts. Cell 2015, 161, 291–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schedin, S.; Sindelar, P.J.; Pentchev, P.; Brunk, U.; Dallner, G. Peroxisomal Impairment in Niemann-Pick Type C Disease. J. Biol. Chem. 1997, 272, 6245–6251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez, M.C.; Balboa, E.; Alvarez, A.R.; Zanlungo, S. Oxidative Stress: A Pathogenic Mechanism for Niemann-Pick Type C Disease. Oxidative Med. Cell. Longev. 2012, 2012, 205713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanier, M.T. Complex Lipid Trafficking in Niemann-Pick Disease Type C. J. Inherit. Metab Dis. 2015, 38, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.L.; Krus, K.L.; Lieberman, A.P. Lysosome and Endoplasmic Reticulum Quality Control Pathways in Niemann-Pick Type C Disease. Brain Res. 2016, 1649, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Kurtz, T.W. Treating the Metabolic Syndrome: Telmisartan as a Peroxisome Proliferator-Activated Receptor-Gamma Activator. Acta Diabetol. 2005, 42, s9–s16. [Google Scholar] [CrossRef]
- Vitale, C.; Mercuro, G.; Castiglioni, C.; Cornoldi, A.; Tulli, A.; Fini, M.; Volterrani, M.; Rosano, G.M.C. Metabolic Effect of Telmisartan and Losartan in Hypertensive Patients with Metabolic Syndrome. Cardiovasc. Diabetol. 2005, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Shiota, A.; Shimabukuro, M.; Fukuda, D.; Soeki, T.; Sato, H.; Uematsu, E.; Hirata, Y.; Kurobe, H.; Maeda, N.; Sakaue, H.; et al. Telmisartan Ameliorates Insulin Sensitivity by Activating the AMPK/SIRT1 Pathway in Skeletal Muscle of Obese Db/Db Mice. Cardiovasc. Diabetol. 2012, 11, 139. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-F.; Li, J.; Ma, C.; Huang, C.; Li, Z.-Q. Telmisartan Ameliorates Aβ Oligomer-Induced Inflammation via PPARγ/PTEN Pathway in BV2 Microglial Cells. Biochem. Pharmacol. 2020, 171, 113674. [Google Scholar] [CrossRef]
- Omote, Y.; Deguchi, K.; Kurata, T.; Yamashita, T.; Sato, K.; Hishikawa, N.; Abe, K. Telmisartan Promotes Potential Glucose Homeostasis in Stroke-Resistant Spontaneously Hypertensive Rats via Peroxisome Proliferator-Activated Receptor. γ Activation. Curr. Neurovasc. Res. 2015, 12, 91–97. [Google Scholar] [CrossRef]
- Goebel, M.; Clemenz, M.; Staels, B.; Unger, T.; Kintscher, U.; Gust, R. Characterization of New PPARγ Agonists: Analysis of Telmisartan’s Structural Components. ChemMedChem 2009, 4, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, A.; Minghetti, L. Regulation of Glial Cell Functions by PPAR- Natural and Synthetic Agonists. PPAR Res. 2008, 2008, 864140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vance, J.E. Dysregulation of Cholesterol Balance in the Brain: Contribution to Neurodegenerative Diseases. Dis. Models Mech. 2012, 5, 746–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.; Lieberman, A.P. Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin. PLoS Genet. 2013, 9, e1003462. [Google Scholar] [CrossRef] [Green Version]
- Maxfield, F.R.; Wüstner, D. Analysis of Cholesterol Trafficking with Fluorescent Probes. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 108, pp. 367–393. ISBN 978-0-12-386487-1. [Google Scholar]
- Sitarska, D.; Ługowska, A. Laboratory Diagnosis of the Niemann-Pick Type C Disease: An Inherited Neurodegenerative Disorder of Cholesterol Metabolism. Metab. Brain Dis. 2019, 34, 1253–1260. [Google Scholar] [CrossRef] [Green Version]
- Nunes, M.J.; Moutinho, M.; Gama, M.J.; Rodrigues, C.M.P.; Rodrigues, E. Histone Deacetylase Inhibition Decreases Cholesterol Levels in Neuronal Cells by Modulating Key Genes in Cholesterol Synthesis, Uptake and Efflux. PLoS ONE 2013, 8, e53394. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Strunk, B.S.; Weisman, L.S. Close Encounters of the Lysosome-Peroxisome Kind. Cell 2015, 161, 197–198. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Luo, J.; Hu, A.; Xiao, T.; Li, M.; Kong, Z.; Jiang, L.; Zhou, Z.; Liao, Y.; Xie, C.; et al. Cholesterol Transport through the Peroxisome-ER Membrane Contacts Tethered by PI(4,5)P2 and Extended Synaptotagmins. Sci. China Life Sci. 2019, 62, 1117–1135. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, A.; Giammarco, M.L.; De Nuccio, C.; Ajmone-Cat, M.A.; Visentin, S.; De Simone, R.; Minghetti, L. Docosahexaenoic Acid Promotes Oligodendrocyte Differentiation via PPAR-γ Signalling and Prevents Tumor Necrosis Factor-α-Dependent Maturational Arrest. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2017, 1862, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, A.; Plumitallo, C.; De Nuccio, C.; Visentin, S.; Minghetti, L. Curcumin Promotes Oligodendrocyte Differentiation and Their Protection against TNF-α through the Activation of the Nuclear Receptor PPAR-γ. Sci. Rep. 2021, 11, 4952. [Google Scholar] [CrossRef]
- Kaitsuka, T.; Hakim, F. Response of Pluripotent Stem Cells to Environmental Stress and Its Application for Directed Differentiation. Biology 2021, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Hu, L.; Li, G. SH-SY5Y Human Neuroblastoma Cell Line: In Vitro Cell Model of Dopaminergic Neurons in Parkinson’s Disease. Chin. Med. J. 2010, 123, 1086–1092. [Google Scholar] [PubMed]
- Adler, J.; Parmryd, I. Quantifying Colocalization by Correlation: The Pearson Correlation Coefficient Is Superior to the Mander’s Overlap Coefficient. Cytometry 2010, 77A, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, T.A.; Iacono, L.L.; Gross, C.T. Serotonin Receptor 1A Modulates Actin Dynamics and Restricts Dendritic Growth in Hippocampal Neurons: Htr1a and Dendritic Growth. Eur. J. Neurosci. 2010, 32, 18–26. [Google Scholar] [CrossRef]
- Zhang, F.; Lavan, B.E.; Gregoire, F.M. Selective Modulators of PPAR-γ Activity: Molecular Aspects Related to Obesity and Side-Effects. PPAR Res. 2007, 2007, 032696. [Google Scholar] [CrossRef] [Green Version]
- Villarroel-Vicente, C.; Gutiérrez-Palomo, S.; Ferri, J.; Cortes, D.; Cabedo, N. Natural Products and Analogs as Preventive Agents for Metabolic Syndrome via Peroxisome Proliferator-Activated Receptors: An Overview. Eur. J. Med. Chem. 2021, 221, 113535. [Google Scholar] [CrossRef]
| % O4+ Cells | % O1+ Cells | |||
|---|---|---|---|---|
| 2 DIV | ||||
| none | U18666A | none | U18666A | |
| CTR | 28.7 ± 1 | 24.9 ± 2 * | 22.4 ± 2 | 16.8 ± 2 * |
| TLM 10 nM | 33.6 ± 3 * | 58.3 ± 8 */**/*** | 24.7 ± 4 | 55.4 ± 4 */**/*** |
| 3 DIV | ||||
| CTR | 35.3 ± 3 | 27.1 ± 4 * | 28.9 ± 2 | 22.7 ± 2 * |
| TLM 10 nM | 56.4 ± 6 * | 70.2 ± 4 */**/*** | 53.5 ± 1 * | 68.4 ± 4 */**/*** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardo, A.; Malara, M.; Bertuccini, L.; De Nuccio, C.; Visentin, S.; Minghetti, L. The Antihypertensive Drug Telmisartan Protects Oligodendrocytes from Cholesterol Accumulation and Promotes Differentiation by a PPAR-γ-Mediated Mechanism. Int. J. Mol. Sci. 2021, 22, 9434. https://doi.org/10.3390/ijms22179434
Bernardo A, Malara M, Bertuccini L, De Nuccio C, Visentin S, Minghetti L. The Antihypertensive Drug Telmisartan Protects Oligodendrocytes from Cholesterol Accumulation and Promotes Differentiation by a PPAR-γ-Mediated Mechanism. International Journal of Molecular Sciences. 2021; 22(17):9434. https://doi.org/10.3390/ijms22179434
Chicago/Turabian StyleBernardo, Antonietta, Mariagiovanna Malara, Lucia Bertuccini, Chiara De Nuccio, Sergio Visentin, and Luisa Minghetti. 2021. "The Antihypertensive Drug Telmisartan Protects Oligodendrocytes from Cholesterol Accumulation and Promotes Differentiation by a PPAR-γ-Mediated Mechanism" International Journal of Molecular Sciences 22, no. 17: 9434. https://doi.org/10.3390/ijms22179434
APA StyleBernardo, A., Malara, M., Bertuccini, L., De Nuccio, C., Visentin, S., & Minghetti, L. (2021). The Antihypertensive Drug Telmisartan Protects Oligodendrocytes from Cholesterol Accumulation and Promotes Differentiation by a PPAR-γ-Mediated Mechanism. International Journal of Molecular Sciences, 22(17), 9434. https://doi.org/10.3390/ijms22179434






