Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer
Abstract
:1. Introduction
2. Results
2.1. Patients’ Characteristics
2.2. Radiomics Signature Building
2.3. Supervised Learning Classification
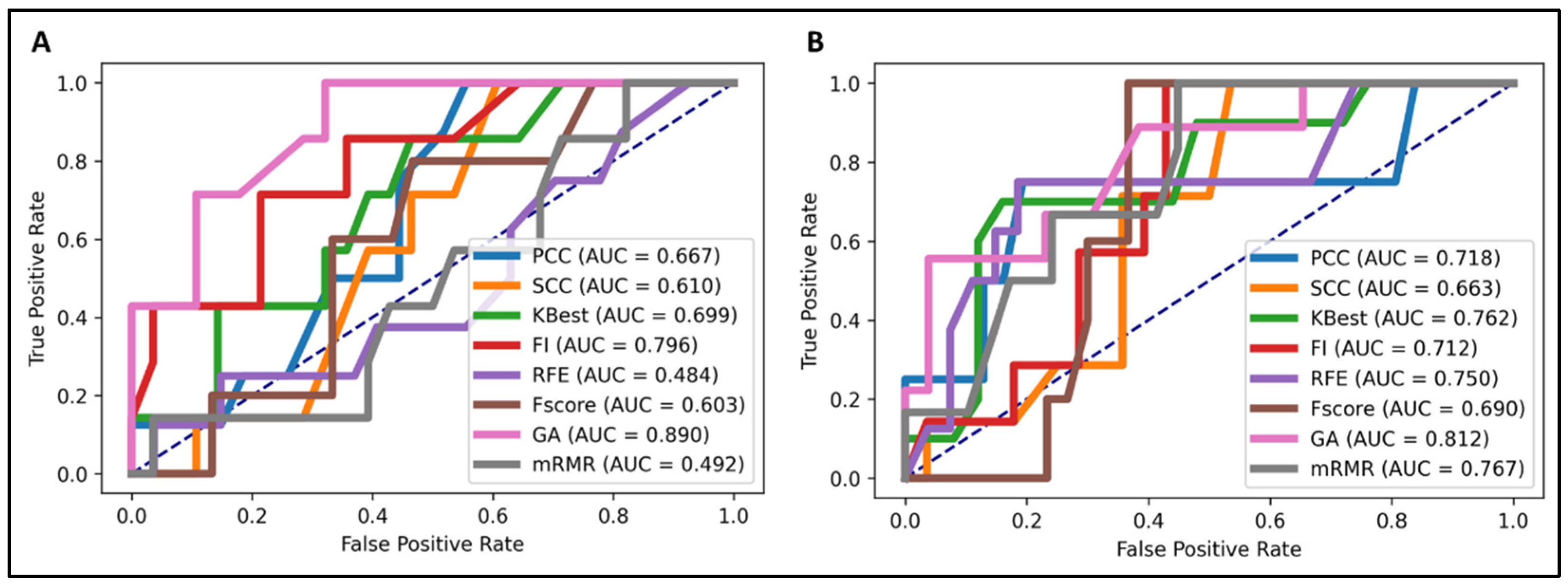
2.4. Validation of Models
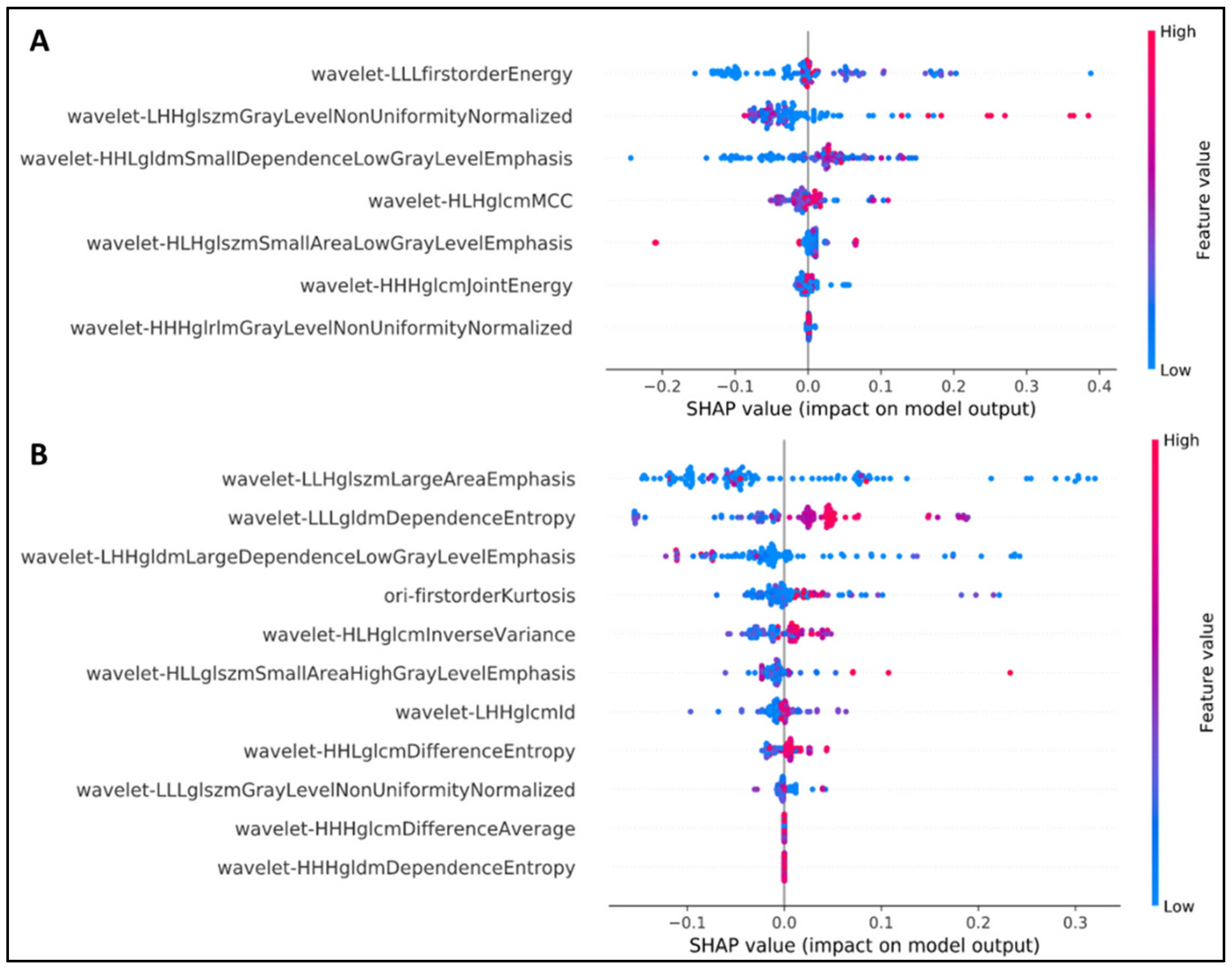
2.5. Explanation of Feature Selection Using SHAP
2.6. Comparison with Previous Radiomics-Based EGFR and KRAS Prediction Models
3. Discussion
4. Materials and Methods
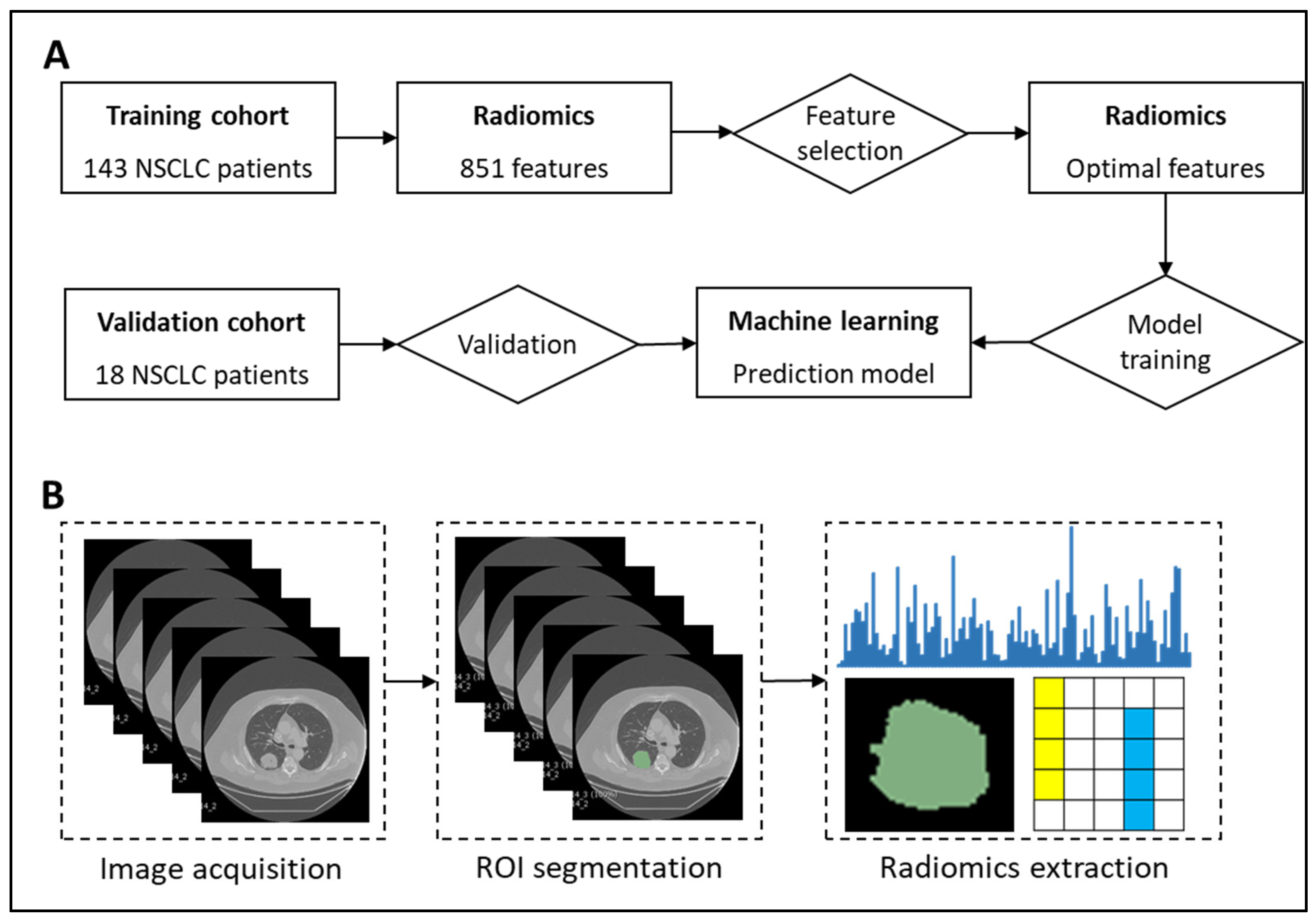
4.1. NSCLC Patient Cohort
4.2. Segmentation of Lung Tumors
4.3. Radiomics Feature Extraction
4.4. Feature Selection
4.4.1. Univariate Selection
4.4.2. RFE
4.4.3. Feature Importance
4.4.4. Filter Methods
4.4.5. F-Score
4.4.6. GA
4.4.7. Minimum Redundancy Feature Selection
4.4.8. KBest Algorithm
4.5. Machine Learning
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non–Small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1623–1640. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. 2020 CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Riely, G.J.; Marks, J.; Pao, W. KRAS mutations in non–small cell lung cancer. Proc. Am. Thorac. Soc. 2009, 6, 201–205. [Google Scholar] [CrossRef]
- Boch, C.; Kollmeier, J.; Roth, A.; Stephan-Falkenau, S.; Misch, D.; Grüning, W.; Bauer, T.T.; Mairinger, T. The frequency of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC): Routine screening data for central Europe from a cohort study. BMJ Open 2013, 3, e002560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heeke, S.; Hofman, V.; Benzaquen, J.; Otto, J.; Tanga, V.; Zahaf, K.; Allegra, M.; Long-Mira, E.; Lassalle, S.; Marquette, C.H.; et al. Detection of EGFR Mutations From Plasma of NSCLC Patients Using an Automatic Cartridge-Based PCR System. Front. Pharm. 2021, 12, 657743. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Miller, V.; Zakowski, M.; Doherty, J.; Politi, K.; Sarkaria, I.; Singh, B.; Heelan, R.; Rusch, V.; Fulton, L. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA 2004, 101, 13306–13311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pao, W.; Wang, T.Y.; Riely, G.J.; Miller, V.A.; Pan, Q.; Ladanyi, M.; Zakowski, M.F.; Heelan, R.T.; Kris, M.G.; Varmus, H.E. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005, 2, e17. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.; Liu, S.V.; Subramaniam, D.S.; Giaccone, G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat. Rev. Clin. Oncol. 2015, 12, 511–526. [Google Scholar] [CrossRef]
- Li, Z.; Shu, J.; Yang, B.; Zhang, Z.; Huang, J.; Chen, Y. Emerging non-invasive detection methodologies for lung cancer. Oncol. Lett. 2020, 19, 3389–3399. [Google Scholar] [CrossRef]
- Farooq, M.; Herman, J.G. Noninvasive diagnostics for early detection of lung cancer: Challenges and potential with a focus on changes in DNA methylation. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2416–2422. [Google Scholar] [CrossRef]
- Tang, D.; Shen, Y.; Wang, M.; Yang, R.; Wang, Z.; Sui, A.; Jiao, W.; Wang, Y. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur. J. Cancer Prev. 2013, 22, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toloza, E.M.; Harpole, L.; McCrory, D.C. Noninvasive staging of non-small cell lung cancer: A review of the current evidence. Chest 2003, 123, 137S–146S. [Google Scholar] [CrossRef]
- Xu, Y.; Hosny, A.; Zeleznik, R.; Parmar, C.; Coroller, T.; Franco, I.; Mak, R.H.; Aerts, H.J. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin. Cancer Res. 2019, 25, 3266–3275. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, S.; Di Dong, J.W.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; Tian, J. The applications of radiomics in precision diagnosis and treatment of oncology: Opportunities and challenges. Theranostics 2019, 9, 1303. [Google Scholar] [CrossRef]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 2015, 5, 1–11. [Google Scholar]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H.J. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef] [Green Version]
- Gevaert, O.; Echegaray, S.; Khuong, A.; Hoang, C.D.; Shrager, J.B.; Jensen, K.C.; Berry, G.J.; Guo, H.H.; Lau, C.; Plevritis, S.K. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Rizzo, S.; Raimondi, S.; de Jong, E.E.; van Elmpt, W.; De Piano, F.; Petrella, F.; Bagnardi, V.; Jochems, A.; Bellomi, M.; Dingemans, A.M. Genomics of non-small cell lung cancer (NSCLC): Association between CT-based imaging features and EGFR and K-RAS mutations in 122 patients—An external validation. Eur. J. Radiol. 2019, 110, 148–155. [Google Scholar] [CrossRef]
- Shiri, I.; Maleki, H.; Hajianfar, G.; Abdollahi, H.; Ashrafinia, S.; Hatt, M.; Zaidi, H.; Oveisi, M.; Rahmim, A. Next-generation radiogenomics sequencing for prediction of EGFR and KRAS mutation status in NSCLC patients using multimodal imaging and machine learning algorithms. Mol. Imaging Biol. 2020, 22, 1132–1148. [Google Scholar] [CrossRef] [Green Version]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar]
- Bakr, S.; Gevaert, O.; Echegaray, S.; Ayers, K.; Zhou, M.; Shafiq, M.; Zheng, H.; Benson, J.A.; Zhang, W.; Leung, A.N.C.; et al. A radiogenomic dataset of non-small cell lung cancer. Sci. Data 2018, 5, 180202. [Google Scholar] [CrossRef] [Green Version]
- Mirjalili, S. Genetic algorithm. In Evolutionary Algorithms and Neural Networks; Springer: Berlin, Germany, 2019; pp. 43–55. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Lundberg, S.; Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, G.; Pereira, T.; Dias, C.; Freitas, C.; Hespanhol, V.; Costa, J.L.; Cunha, A.; Oliveira, H.P. Identifying relationships between imaging phenotypes and lung cancer-related mutation status: EGFR and KRAS. Sci. Rep. 2020, 10, 3625. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, X.; Zhao, Y.; Zhang, J.; Zhang, Z.; Wang, J.; Wang, Y.; Dai, M.; Han, J. Value of pre-therapy 18 F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Motoda, H. Computational Methods of Feature Selection; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuzé, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018, 78, 4786–4789. [Google Scholar] [CrossRef] [Green Version]
- Moreno, S.; Bonfante, M.; Zurek, E.; Cherezov, D.; Goldgof, D.; Hall, L.; Schabath, M. A Radiogenomics Ensemble to Predict EGFR and KRAS Mutations in NSCLC. Tomography 2021, 7, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Jiang, L.; Zhang, J.; Shi, Y.; Gray, J.E.; Tunali, I.; Gao, C.; Sun, Y.; Tian, J.; Zhao, X. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Yang, J.; Ni, B.; Bi, D.; Sun, Y.; Xu, M.; Zhu, X.; Li, C.; Jin, L.; Gao, P. Toward automatic prediction of EGFR mutation status in pulmonary adenocarcinoma with 3D deep learning. Cancer Med. 2019, 8, 3532–3543. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Hou, L.; Yang, W.; Han, J.; Wang, J.; Qiang, Y.; Zhao, J.; Hou, J.; Song, K.; Ma, Y.; et al. Multi-channel multi-task deep learning for predicting EGFR and KRAS mutations of non-small cell lung cancer on CT images. Quant. Imaging Med. Surg. 2021, 11, 2354–2375. [Google Scholar] [CrossRef]
- Li, K.; Yang, M.; Liang, N.; Li, S. Determining EGFR-TKI sensitivity of G719X and other uncommon EGFR mutations in non-small cell lung cancer: Perplexity and solution (Review). Oncol. Rep. 2017, 37, 1347–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.-W.; Lei, L.; Wang, W.-X.; Lin, L.; Zhu, Y.-C.; Wang, H.; Miao, L.-Y.; Wang, L.-P.; Zhuang, W.; Fang, M.-Y.; et al. Molecular Characteristics and Clinical Outcomes of EGFR Exon 19 C-Helix Deletion in Non–Small Cell Lung Cancer and Response to EGFR TKIs. Transl. Oncol. 2020, 13, 100791. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.L.; Akdogan, M.U.; Altindag, C.; Alkim, E. ePAD: An image annotation and analysis platform for quantitative imaging. Tomography 2019, 5, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Subho, M.R.H.; Chowdhury, M.R.; Chaki, D.; Islam, S.; Rahman, M.M. A Univariate Feature Selection Approach for Finding Key Factors of Restaurant Business. In 2019 IEEE Region 10 Symposium (TENSYMP); IEEE: Piscataway Township, NJ, USA, 2019; pp. 605–610. [Google Scholar]
- Zeng, X.; Chen, Y.-W.; Tao, C. Feature selection using recursive feature elimination for handwritten digit recognition. In Proceedings of the 2009 Fifth International Conference on Intelligent Information Hiding and Multimedia Signal Processing, Kyoto, Japan, 12–14 September 2009; IEEE: Piscataway Township, NJ, USA, 2009; pp. 1205–1208. [Google Scholar]
- Geng, X.; Liu, T.-Y.; Qin, T.; Li, H. Feature selection for ranking. In Proceedings of the 30th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Amsterdam, The Netherlands, 23–27 July 2007; Association for Computing Machinery: Amsterdam, The Netherlands, 2007; pp. 407–414. [Google Scholar]
- Sánchez-Marono, N.; Alonso-Betanzos, A.; Tombilla-Sanromán, M. Filter methods for feature selection–A comparative study. In Proceedings of the International Conference on Intelligent Data Engineering and Automated Learning, Birmingham, UK, 16–19 December 2007; Springer: Berlin, Germany, 2007; pp. 178–187. [Google Scholar]
- Chen, Y.-W.; Lin, C.-J. Combining SVMs with Various Feature Selection Strategies. In Feature Extraction: Foundations and Applications; Guyon, I., Nikravesh, M., Gunn, S., Zadeh, L.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 315–324. [Google Scholar]
- Radovic, M.; Ghalwash, M.; Filipovic, N.; Obradovic, Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinform. 2017, 18, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine learning for medical imaging. Radiographics 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Hung, T.N.K.; Do, D.T.; Lam, L.H.T.; Dang, L.H.; Huynh, T.-T. Radiomics-based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Comput. Biol. Med. 2021, 132, 104320. [Google Scholar] [CrossRef]
- Zupan, B.; Demšar, J.; Kattan, M.W.; Beck, J.R.; Bratko, I. Machine learning for survival analysis: A case study on recurrence of prostate cancer. Artif. Intell. Med. 2000, 20, 59–75. [Google Scholar] [CrossRef] [Green Version]
- Mattonen, S.A.; Davidzon, G.A.; Benson, J.; Leung, A.N.C.; Vasanawala, M.; Horng, G.; Shrager, J.B.; Napel, S.; Nair, V.S. Bone Marrow and Tumor Radiomics at (18)F-FDG PET/CT: Impact on Outcome Prediction in Non-Small Cell Lung Cancer. Radiology 2019, 293, 451–459. [Google Scholar] [CrossRef]
- Le, N.Q.; Do, D.T.; Chiu, F.-Y.; Yapp, E.K.; Yeh, H.-Y.; Chen, C.-Y. XGBoost Improves Classification of MGMT Promoter Methylation Status in IDH1 Wildtype Glioblastoma. J. Pers. Med. 2020, 10, 128. [Google Scholar] [CrossRef] [PubMed]
| Training (n = 143) | Validation (n = 18) | p Value | |
|---|---|---|---|
| Age (mean ± SD, years) | 69.2 ± 8.84 | 66.9 ± 13.85 | 0.334 |
| Sex | 0.123 | ||
| Male | 107 | 4 | |
| Female | 36 | 14 | |
| Smoking status | 0.069 | ||
| Current | 30 | ||
| Former | 91 | 9 | |
| Nonsmoker | 22 | 9 | |
| Histological type | 0.5 | ||
| Adenocarcinoma | 111 | 18 | |
| NSCLC NOS 1 | 3 | 0 | |
| Squamous cell carcinoma | 29 | 0 | |
| EGFR mutation | 0.074 | ||
| Mutant | 23 | 6 | |
| Wild-type | 93 | 12 | |
| KRAS mutation | 0.074 | ||
| Mutant | 27 | 3 | |
| Wild-type | 87 | 15 | |
| Recurrence | 0.123 | ||
| Yes | 40 | 3 | |
| No | 103 | 15 |
| Original | SMOTE | ||||||
|---|---|---|---|---|---|---|---|
| Sens | Spec | Acc | Sens | Spec | Acc | ||
| EGFR | LR | 4.3 | 100 | 81 | 43.5 | 78.5 | 71.6 |
| kNN | 34.8 | 92.5 | 81 | 60.9 | 67.7 | 66.4 | |
| RF | 21.7 | 97.8 | 82.8 | 52.2 | 84.9 | 78.4 | |
| XGBoost | 43.5 | 94.6 | 84.5 | 65.2 | 88.2 | 83.6 | |
| KRAS | LR | 11.1 | 98.9 | 78.1 | 48.1 | 73.6 | 67.5 |
| kNN | 18.5 | 98.9 | 79.8 | 55.6 | 67.8 | 64.9 | |
| RF | 33.3 | 96.6 | 81.6 | 51.9 | 75.9 | 70.2 | |
| XGBoost | 55.6 | 89.3 | 77.2 | 55.6 | 95.4 | 86 | |
| Sens | Spec | Acc | AUC | ||
|---|---|---|---|---|---|
| EGFR | Pinheiro et al. | - | - | - | 0.7458 |
| Shiri et al. | - | - | - | 0.78 | |
| Zhang et al. | 91.7 | 70.3 | 80.8 | 0.87 | |
| Ours | 65.2 | 88.2 | 83.6 | 0.89 | |
| KRAS | Pinheiro et al. | 11.1 | 98.9 | 78.1 | 0.5035 |
| Shiri et al. | 18.5 | 98.9 | 79.8 | 0.83 | |
| Ours | 55.6 | 95.4 | 86 | 0.812 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, N.Q.K.; Kha, Q.H.; Nguyen, V.H.; Chen, Y.-C.; Cheng, S.-J.; Chen, C.-Y. Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 9254. https://doi.org/10.3390/ijms22179254
Le NQK, Kha QH, Nguyen VH, Chen Y-C, Cheng S-J, Chen C-Y. Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer. International Journal of Molecular Sciences. 2021; 22(17):9254. https://doi.org/10.3390/ijms22179254
Chicago/Turabian StyleLe, Nguyen Quoc Khanh, Quang Hien Kha, Van Hiep Nguyen, Yung-Chieh Chen, Sho-Jen Cheng, and Cheng-Yu Chen. 2021. "Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer" International Journal of Molecular Sciences 22, no. 17: 9254. https://doi.org/10.3390/ijms22179254
APA StyleLe, N. Q. K., Kha, Q. H., Nguyen, V. H., Chen, Y.-C., Cheng, S.-J., & Chen, C.-Y. (2021). Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer. International Journal of Molecular Sciences, 22(17), 9254. https://doi.org/10.3390/ijms22179254







