Abstract
Garlic (Allium sativum L.) is an important vegetable and is cultivated and consumed worldwide for its economic and medicinal values. Garlic cloves, the major reproductive and edible organs, are derived from the axillary meristems. KNOTTED-like homeobox (KNOX) proteins, such as SHOOT MERISTEM-LESS (STM), play important roles in axillary meristem formation and development. However, the KNOX proteins in garlic are still poorly known. Here, 10 AsKNOX genes, scattered on 5 of the 8 chromosomes, were genome-wide identified and characterized based on the newly released garlic genome. The typical conserved domains of KNOX proteins were owned by all these 10 AsKNOX homologs, which were divided into two Classes (Class I and Class II) based on the phylogenetic analysis. Prediction and verification of the subcellular localizations revealed the diverse subcellular localization of these 10 AsKNOX proteins. Cis-element prediction, tissue expression analysis, and expression profilings in responding to exogenous GA3 and 6-BA showed the potential involvement of AsKNOX genes in the gibberellin and cytokinin signaling pathways. Overall, the results of this work provided a better understanding of AsKNOX genes in garlic and laid an important foundation for their further functional studies.
1. Introduction
Garlic, an economically important vegetable, spice, and medicinal crop, originated in central Asia and the Mediterranean and Caucasus zones [1]. Nowadays, as the second most important Allium crop after the bulb onion, garlic is cultivated and consumed worldwide for its remarkable medicinal and nutraceutical properties [2]. Commercial garlics are usually sterile and asexually reproduced with cloves or bulbils, which results in a relatively lower reproduction coefficient. Garlic clove, as the major reproductive and edible organ, contains a growth bud, a thick storage leaf, and a protective leaf, which is derived from the axillary meristem and similar to the tiller in rice or wheat and branch in woody plants [3]. Therefore, promoting the formation and development of axillary meristems in garlic is an effective way to increase clove numbers and further improve the reproduction coefficient. On the contrary, it is possible to obtain one-clove garlics by inhibiting the growth of garlic axillary meristems. Cytokinin (CK) has long been considered to be the key phytohormone to promote lateral bud outgrowth. Recently, researchers have reported that exogenous gibberellin (GA) was a positive regulator in lateral bud development [4], for example, GA3 treatment increased the number of tubers per plant in potato [5], induced tillering in welsh onions [6], and promoted shoot branching in Jatropha carcass [4]. Our previous work also indicated that exogenous GA3 with appropriate concentration, either injected into garlic plants or soaking the seed cloves before sowing, was able to significantly increase the cloves in a single garlic bulb [7,8]. Further histological and physiological analysis revealed that the exogenous GA3 promoted axillary bud formation and outgrowth via increasing the level of cytokinin (zeatin riboside, ZR) and soluble proteins and reducing the content of GA3, IAA, and sugar in the stems of the garlic plant, resulting in increased cloves per bulb [7,8].
Although the regulation of exogenous GA3 in promoting lateral bud formation and growth has been revealed from the aspects of histology and physiology, the related regulating or responding genes are still unknown in garlic. In plants, branching or tillering is achieved by axillary meristems, which are established in the axil of each leaf base. Research into axillary meristems revealed that several transcription factors (TFs) and phytohormones are key regulators that enable axillary meristem formation, in which the expression levels of SHOOT MERISTEM-LESS (STM) played essential roles [9]. STM is a member of the homeodomain (HD) KNOTTED-like homeobox transcription factors (KNOX) that regulate several processes of plant organ development such as meristem development, hormone metabolisms, and lignin biosynthesis [10]. KNOX genes belong to the group of TFs known as the three-amino acid loop extension (TALE) superclass [11] and usually contain 4 typical domains: KNOX1 domain, KNOX2 domain, ELK domain, and Homeobox KN domain [12], although some lack the ELK and Homeobox KN domains. The KNOX proteins, which have been thoroughly investigated, influence plant growth and development in a versatile context-dependent manner [13].
In Arabidopsis, there are eight KNOX homologs which have been divided into two Classes: Class I including KNAT1 (BP), KNAT2, KNAT6, and STM, and Class II containing KNAT3, KNAT4, KNAT5, and KNAT7 [14]. Studies on the functions of Class II KNOX proteins (KNAT3, KNAT4, KNAT5, and KNAT7) are less reported. The KNAT3, KNAT4, and KNAT5 might play distinct roles in root development [15], while the KNAT7 is involved in the fine regulation of secondary wall and lignin synthesis [16]. Additionally, a Class II KNOX protein, MdKNOX19, participating in the abscisic acid (ABA) pathway and affecting seed and fruit development, was recently characterized in apple [17]. For the Class I KNOX proteins, STM is essential for meristem formation and maintenance [13]; KNAT1 works redundantly with STM in the Arabidopsis shoot apical meristem (SAM) under the absence of AS1(ASYMMETRIC LEAVES1) [18] and takes part in controlling inflorescence stem development [19]; KNAT6, similar to KNAT1, contributes to meristem function and inflorescence development [20]; and, finally, KNAT2 regulates flower patterning acting in the inner whorls [21]. Furthermore, the Class I KNOX proteins were evidenced as the general orchestrators of growth-regulator homeostasis at the shoot apex by simultaneously activating cytokinin and repressing GA biosynthesis, thus promoting meristem activity in Arabidopsis [22]. Overall, KNOX proteins as versatile regulators play essential roles in sculpting plant form and its diversity [13].
Despite the identification, characterization, and functions of KNOX proteins that have been widely studied in Arabidopsis [12,22], poplar [23], peach [10], and pear [24], the KNOX homologs in garlic is unknown. Furthermore, whether the garlic AsKNOX proteins have the same function involved in the formation and outgrowth of axillary meristems and further regulate the cloves’ differentiation by interacting with phytohormones of gibberellin and cytokinin is also unclear. Genome-wide identification and characterization of AsKNOX proteins will provide us more information in understanding the potential functions of AsKNOX genes in regulating garlic axillary bud formation and outgrowth. Fortunately, the newly released chromosome-level garlic genome [2] provides a good opportunity to investigate the KNOX gene family members in garlic from a whole-genome perspective. Therefore, the objectives of the present study is to conduct a genome-wide characterization of AsKNOX gene families in the garlic genome and to reveal their expression profilings in responding to exogenous cytokinin and gibberellin. Results of this study not only provide target genes for the study of the KNOX family in garlic but also lay a foundation for the investigation of the molecular regulation of GA-induced garlic axillary meristem formation and further enrich our knowledge of improving garlic propagation efficiency via exogenous phytohormones application.
2. Results
2.1. Identification and Characterization of KNOX Family Members
Genome-wide identification of KNOX gene family members was performed with the newly assembled garlic genome [2], in which the HHM profiles of the conserved KNOX protein domains were firstly used as queries to perform an HMMER search (http://hmmer.org/, accessed on 15 January 2021). Then, the SMART search and Pfam search were performed to confirm the presence of the conserved domains for those potential garlic KNOX proteins. Finally, 10 KNOX genes were identified in the garlic genome. For convenience, these 10 genes were assigned names from AsKNOX1 to AsKNOX10 based on their chromosomal locations (Table 1). These AsKNOX genes encoded predicted peptides ranging from 160 to 354 aa with the isoelectric point (pI) value from 4.05 to 7.29 and the molecular weight (Mw) from 17.25 to 40.21 kDa. All AsKNOX homologs were predicted to be hydrophilic proteins and localized in the nucleus, of which one (AsKNOX1) and two (AsKNOX7 and AsKNOX10) members also showed localizations in chloroplast and cytoplasm, respectively (Table 1). All the related sequences information and detailed positions of these AsKNOX genes are listed in the Supplemental Table S1.

Table 1.
Information of 10 AsKNOX gene homologs in garlic genome.
2.2. Chromosomal Distribution and Gene Structures of AsKNOX Genes
Based on the physical locations in the garlic genome, nine of the ten AsKNOX genes (AsKNOX1 to AsKNOX9) were anchored onto five of the eight chromosomes (Chr1, 2, 4, 6, and 8). However, KNOX gene Asa0G05087.1 was not located on any assembled garlic chromosomes and was named with AsKNOX10. As shown in Figure 1, both Chr1 and Chr6 harbored one AsKNOX gene, while both Chr4 and Chr8 had two AsKNOX genes, and three AsKNOX genes were located on Chr2.
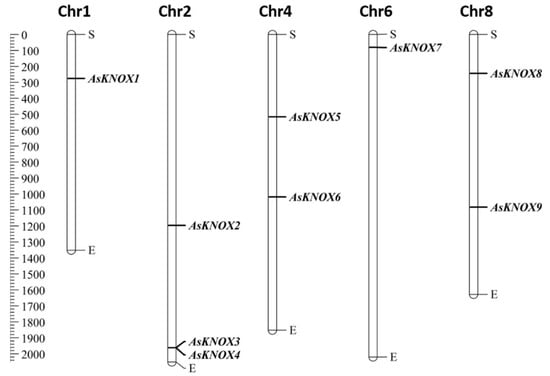
Figure 1.
Distribution of AsKNOX genes on garlic chromosomes. The left scale represents the length of garlic chromosomes with a unit of Megabase pairs (Mb). The letters of S and E indicate the start and end positions of each chromosome.
The Exon-Intron structures of AsKNOX genes were analyzed and plotted with TBtools. As shown in Figure 2, the exon numbers of these AsKNOX genes ranged from one to five, in which AsKNOX2 was the only member that had one exon. Five AsKNOX genes (AsKNOX1, AsKNOX3, AsKNOX5, AsKNOX6, and AsKNOX7) showed with the structures of 5 exons and 4 introns. In addition, all the other four AsKNOX genes (AsKNOX4, AsKNOX8, AsKONX9, and AsKNOX10) had four exons and three introns. Among these AsKNOX genes, AsKNOX9 and AsKNOX10 had similar gene structures with each other, which might suggest that these two genes have similar functions in garlic. Additionally, the gene structure of AsKNOX3 was also somewhat similar to that of AsKNOX4, although the 2nd exon of AsKNOX4 was spliced into two small exons (2nd and 3rd exon) in AsKNOX3.
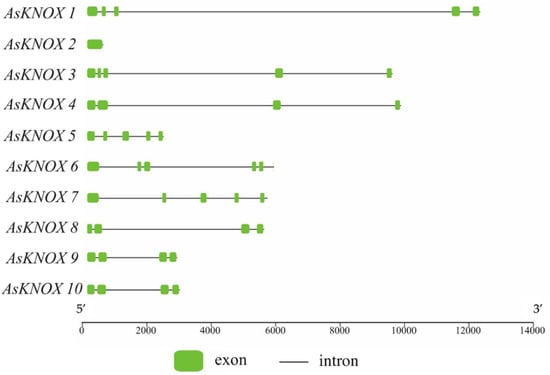
Figure 2.
Exon-Intron structures of AsKNOX genes in garlic genome. The green boxes indicate the exons and the black lines indicate the introns. The bottom scale is represented with base pair (bp).
2.3. Cis-Element and Conserved Motif Analysis of AsKNOX Genes
To further explore the possible regulation mechanism of AsKNOX genes in garlic, the cis-elements in their promoter regions were scanned in the database of PlantCARE. In total, 14 cis-elements were obtained and listed in Figure 3, which included two development-related, four stress responses-related, and seven phytohormone-related cis-elements. The development-related cis-elements (O2-site and GCN4-motif) are relevant to zein metabolism regulation and endosperm expression, respectively. The stress response-related cis-elements included the MYB binding site involved in drought-inducibility (MBS), anaerobic induction element (ARE), low-temperature responsiveness (LTR), and stress response element (STRE). The phytohormone-related cis-elements are involved in several hormone signalings, such as the abscisic acid (ABRE), auxin (AuxRR), gibberellin (GARE and P-box), jasmonic acid (CGTCA-motif and TGACG-motif), and ethylene (ERE). Moreover, the TCA cis-element was also presented in the promoters of AsKNOX genes. These findings might indicate the involvement of AsKNOX genes in the regulation of development and responses to abiotic stresses and phytohormones.
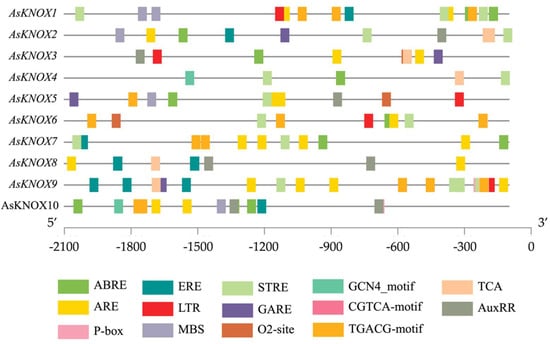
Figure 3.
Putative cis-elements in the promoter regions of AsKNOX genes. Different colored rectangles denote different cis-elements with various biological functions.
Furthermore, to obtain insights into the diversity of conserved motif compositions in AsKNOX proteins, the protein sequences of these 10 AsKNOX genes were assessed using the MEME program. As shown in Figure 4, eight conserved motifs were identified among these 10 AsKNOX proteins, of which the first four motifs (motif 1 to 4) correspond to the 4 typical KNOX domains (Homeobox KN domain, KNOX1 domain, ELK domain, and KNOX2 domain). These 4 typical domains of KNOX protein were owned by all the AsKNOX proteins, except for AsKNOX2, which contains 2 of these 4 typical domains (KNOX1 domain and KNOX2 domain). Additionally, the remaining four motifs (motif 5 to 8) were scattered among nine of the 10 AsKNOX homologs (Figure 4A). Furthermore, based on the stacked motif logos of those four typical domains among AsKNOX proteins, the KNOX2 domain, and Homeobox KN domain showed relatively more conservation than the KNOX1 domain and ELK domain (Figure 4B) among these 10 AsKNOX proteins. To more accurately understand the specific positions of these conserved domains in their protein sequences, the Pfam website was used to compare the sequences and to retrieve the detailed positions of each domain, which are listed in Supplemental Table S1.
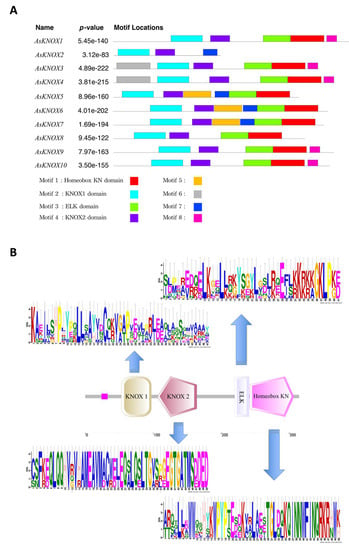
Figure 4.
Conserved motif analysis of garlic AsKNOX homologs. (A) The conserved motifs within the 10 garlic AsKNOX proteins. Motif 1 to 8 were indicated with different colors; (B) The stacked motif logos of the KNOX1 domain, KNOX2 domain, ELK domain, and Homeobox KN domain.
2.4. Phylogenetic Analysis of AsKNOX Homologs
To clarify the evolutionary relationships of 10 AsKNOX proteins and to predict their potential biological functions, a phylogenetic tree was constructed based on the alignment of protein sequences including the KNOX proteins collected from Arabidopsis, tomato, rice, and maize (Figure 5 and Table S2). The neighbor-joining (NJ) phylogenetic tree divided the KNOX proteins into two groups, Class I and Class II, in which Class I includes six AsKNOX members (AsKNOX1, AsKNOX3, AsKNOX4, AsKNOX8, AsKNOX9, and AsKNOX10), while the remaining four AsKNOX homologs (AsKNOX2, AsKNOX5, AsKNOX6, and AsKNOX7) belong to the Class II. According to the classification method reported by Cheng et al. [24], these proteins were further divided into 5 subgroups: Class I includes STM-like, BP-like, and KNAT2/6-like subfamilies; Class II consists of KNAT7-like and KNAT3-5-like subfamilies. Among these 10 AsKNOX proteins, both AsKNOX9 and AsKNOX10 belong to the STM-like subfamily; AsKNOX1 is a member of the BP-like subfamily; AsKNOX3, AsKNOX4, and AsKNOX8 belong to the KNAT2/6-like subfamily; AsKNOX2, AsKNOX6, and AsKNOX7 are classified into the KNAT3-5-like subfamily, while AsKNOX5 belongs to KNAT7-like subfamily.
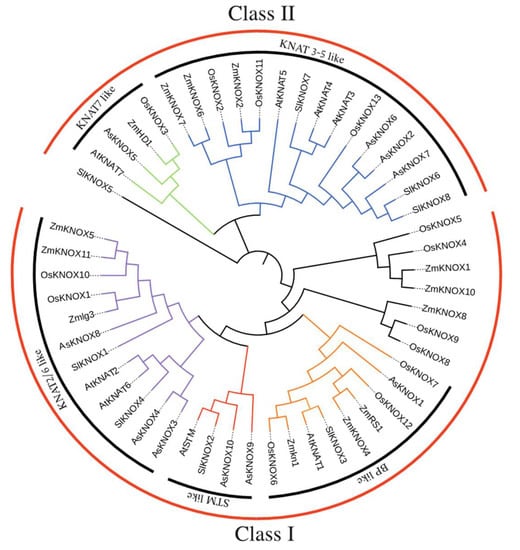
Figure 5.
Interspecific phylogenetic tree of KNOX proteins from various species. A phylogenetic tree was constructed using the neighbor-joining method with bootstrap test (1000 iterations) by MEGA X.
2.5. Expression Patterns of AsKNOX Genes in Different Garlic Tissues
To primarily investigate the functions of AsKNOX genes, their expression patterns were analyzed among six different garlic tissues including root, stem, pseudostem, leaf, scape, and clove. As shown in Figure 6, different expression patterns were found for these 10 AsKNOX genes. AsKNOX1, AsKNOX5, AsKNOX8 and AsKNOX10 were expressed in stem that contains the apical meristem of garlic. Both AsKNOX1 and AsKNOX8 were expressed in the pseudostem and leaf, while AsKNOX8 was barely expressed in root and AsKNOX1 had rarely expression in both scape and clove. AsKNOX2 showed low expression levels among all the investigated garlic tissues, especially for the pseudostem and clove. Compared with other garlic tissues, AsKNOX6 had more rarely expressions in root and leaf. Expressions of AsKNOX4 in the root, leaf, and clove were higher than other tissues, especially for the scape. Similarly, AsKNOX3 also showed very low expression in the scape. AsKNOX5, AsKNOX7, and AsKNOX9 were all highly expressed in garlic scape and clove, and higher expression in garlic stem was also found for AsKNOX5. The expression levels of AsKNOX10 were much higher in garlic stem and leaf, while almost no expressions of AsKNOX10 were shown in garlic root and clove.
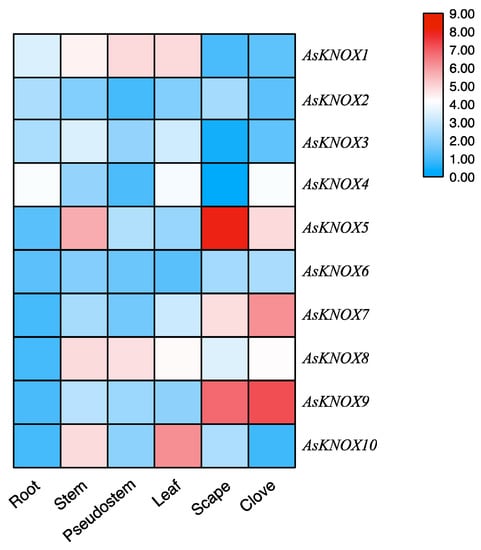
Figure 6.
Expression analysis of AsKNOX genes in different garlic tissues. The color scale from blue to red represents the lower to higher relative expression levels.
2.6. Responses of AsKNOX Genes to Exogenous GA3 and 6-BA Treatments
To reveal the potential responses of AsKNOX genes to exogenous GA3 and 6-BA, the expression patterns of these 10 AsKNOX genes were checked in the garlic stems that were collected at the 1, 3, 5, and 7 DAT (Days after treatment) (Figure 7). These 10 AsKNOX genes had different expression patterns under either GA3 or 6-BA treatments.
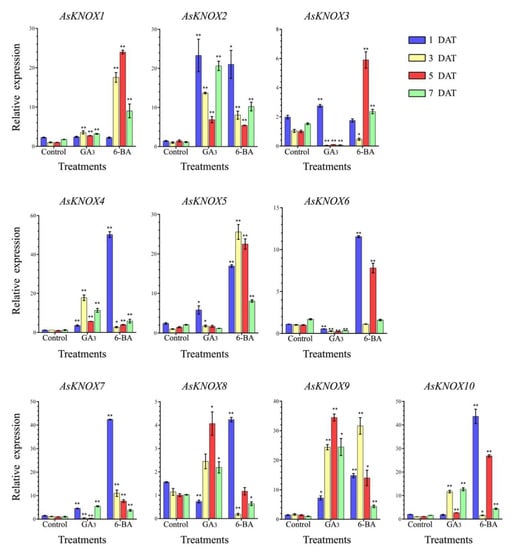
Figure 7.
Expression analysis of the AsKNOX genes under exogenous GA3 and 6-BA treatments. Student’s t-test was used to determine significant differences at the same period between control group and treatment group. Control means the control without any treatment, GA3 means treated with exogenous GA3 solution, 6-BA means treated with exogenous 6-BA solution. Significance level: * p < 0.05. ** p < 0.01.
When treated with GA3, the expressions of AsKNOX1, AsKNOX2, AsKNOX4, and AsKNOX9 showed more significant increases than those of their controls. Compared with the control groups, the expression of AsKNOX3 was up-regulated at the 1 DAT, while it was down-regulated at the rest sample points. The expression of AsKNOX5 was more significantly increased than that of the control at both 1 DAT and 3 DAT. The AsKNOX6 had different expressions which were significantly decreased at all sample points compared with the control groups. AsKNOX7 showed obvious increases at the 1 DAT and 7 DAT, while decreased expressions were obtained at 3 DAT and 5 DAT. Compared with their controls, the expressing of AsKNOX8 was significantly down-regulated at the 1 DAT, while AsKNOX10 had no obvious changes at this sample point. Both the expressions of AsKNOX8 and AsKNOX10 were up-regulated compared with their controls at all other sample points (3, 5, and 7 DAT).
For the 6-BA treatments, the expressions of AsKNOX2, AsKNOX4, AsKNOX5, AsKNOX7, AsKNOX9, and AsKNOX10 showed obvious increases than their controls at all sample points. AsKNOX1 and AsKNOX3 had no obvious changes compared with their controls at the 1 DAT, while the expressions of AsKNOX1 and AsKNOX3 at the 3 DAT were significantly increased and decreased than their controls, respectively. Expressions of AsKNOX6 had no obvious difference compared with its controls for the 3 DAT and 7 DAT samples, while its expressions were significantly increased compared with the controls at 1 DAT and 5 DAT. For the 3 DAT and 7 DAT samples, the expressions of AsKNOX8 were obviously decreased compared with its controls.
2.7. Subcellular Localization of AsKNOX8 and AsKNOX6 Proteins
To preliminarily verify the results of predicted subcellular localization, two family members were randomly selected for verification. The subcellular localization of AsKNOX6 and AsKNOX8 proteins were verified with transient expression of GFP fusion proteins GFP-AsKNOX6 and GFP-AsKNOX8 in tobacco leaf cells. The results of the control experiment showed that the fluorescence signal of GFP could be detected in both the nucleus and cytoplasm of tobacco cells (Figure 8).
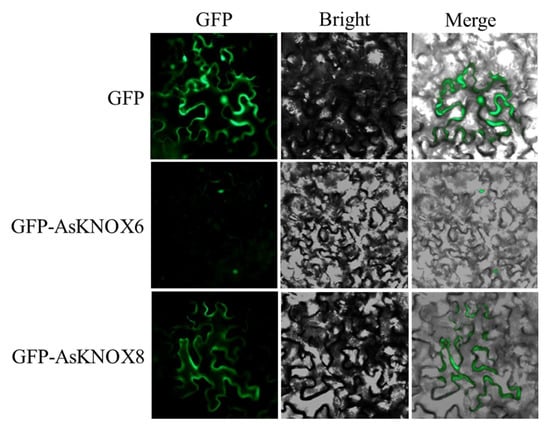
Figure 8.
Subcellular localizations of AsKNOX8 and AsKNOX6 proteins. Both the GFP protein and the fusion proteins GFP-AsKNOX8 and GFP-AsKNOX6 were transiently expressed in the epidermal cells of tobacco leaves. Green fluorescence, bright-field, and merged images were orderly shown from left to right.
The GFP signal of GFP-AsKNOX6 was successfully observed in the nucleus, which was consistent with the predicted results in Table 1. However, the GFP signal of GFP-AsKNOX8 was unexpectedly localized on the cytoplasm.
3. Discussion
The first reported KNOX gene in plants was the ZmKn1 from maize [25], which is involved in the maintenance of the shoot apical meristem and is related with the cell fates switching from indeterminate to determinate [26]. Subsequently, more KNOX genes were identified and characterized in other plant species [13]. The KNOX genes seem to have a limited number in different species, for example, 8 in Arabidopsis, 13 in rice, 12 in maize, 15 in poplar [12], 10 in peach [10], 22 in apple [27], and 18 in pear [24]. Furthermore, the number of KNOX genes among different plant species seems not to correlate with their genome sizes, which was further confirmed in the present study. Although garlic has a genome size of ~16 GB, here only 10 AsKNOX genes were genome-wide identified. These AsKNOX genes are scattered on 5 garlic chromosomes (Chr1, 2, 4, 6, and 8) and subcellular localization prediction of these 10 AsKNOX proteins suggested that all of them could have nuclear localizations. In addition, the results also suggested that AsKNOX1, AsKNOX7, and AsKNOX10 could have locations on the chloroplast or cytoplasm. Intriguingly, verification of subcellular locations of AsKNOX6 and AsKNOX8 showed that AsKNOX6 was consistently localized in the nucleus, while AsKNOX8 was unexpectedly localized on the cytoplasm (Figure 8). The subcellular localization of KNOX proteins outside the nucleus was also found in other plant species, such as the MdKNOX15 in apple located in both the nucleus and plasma membrane [28] and the KNAT1 in Arabidopsis when co-expressed with AtOFP1 or AtOFP5, in which the AtOFP negatively controls the activity of KNOX-BELL protein dimers by changing their localization from the nucleus to the cytoplasmic space [29]. Transcription factors might be retained outside the nucleus for several potential reasons, for examples containing membrane-binding domains, interacting with protein partners that inhibit nuclear import or directly interplaying with membranes, and the posttranslational modifications (phosphorylation, acetylation, glycosylation, ubiquitination, sumoylation, and redox-dependent changes) [30]. Therefore, the cytoplasm localization of AsKNOX8 might indicate that it works with other proteins to play its regulatory function. Another possible reason for the subcellular localization of AsKNOX proteins located outside the nucleus probably was the intercellular transport of KNOX transcription factors. For example, the microinjection studies performed by Lucas et al. (1995) showed that encoded knotted1 (kn1) of maize can be transported to adjacent cells by plasmodesmata [31].
Four typical conserved domains including Homeobox KN domain, KNOX1 domain, ELK domain, and KNOX2 domain were usually contained in KNOX proteins [12]. Similarly, all the AsKNOX homologs in garlic also showed these four typical domains, except for the AsKNOX2 which has not the Homeobox KN domain and ELK domain. Multiple sequence alignment and phylogenetic analyses of these AsKNOX proteins revealed that these 10 AsKNOX, similar to Arabidopsis, can be divided into two classes (Class I and II) and five subfamilies (STM-like, BP-like, KNAT2/6-like, KNAT3-5-like, and KNAT7-like). Among these 10 AsKNOX proteins, both AsKNOX9 and AsKNOX10 belong to the STM-like subfamily; AsKNOX1 is a member of the BP-like subfamily; AsKNOX3, AsKNOX4, and AsKNOX8 belong to the KNAT2/6-like subfamily; AsKNOX2, AsKNOX6, and AsKNOX7 are classified into the KNAT3-5-like subfamily, while AsKNOX5 belongs to KNAT7-like subfamily. In Arabidopsis, the Class I KNOX genes (STM, BP/KNAT1, KNAT2, and KNAT6) play important roles in the maintenance of meristematic potentials and are expressed mainly in the meristematic regions, less in differentiated tissues, and not at all in the mature organs [32]. However, the Class II KNOX genes (KNAT3, KNAT4, KNAT5, and KNAT7) of Arabidopsis, showing functions in root [15] and lateral organ differentiation [14] and secondary cell wall biosynthesis [33], have expressions in both differentiating tissues and mature organs, but not in the meristematic zones [14]. Tissues expressions of these 10 AsKNOX genes indicated that the expressions of AsKONX genes might not strictly follow the tissue expression patterns of Arabidopsis, in which either the members of Class I and II AsKNOX genes, for example, AsKNOX5 and AsKNOX9, showed expressions in mature organs such as the garlic scapes and cloves. AsKNOX1, AsKNOX8, and AsKNOX10 showed relatively higher expressions in the organs derived from the apical meristem tissues such as garlic stems and leaves. Since the stem of garlic is a condensed stem, both the apical and axillary meristems are growing in this part. Therefore, the AsKNOX genes with higher expressions in garlic stems (AsKNOX1, 8, and 10), perhaps have similar functions with the Arabidopsis Class I KNOX genes that play roles in the meristem maintenance. The high expressions of AsKNOX5 and AsKNOX9 in garlic scape might suggest their functions in the development of garlic inflorescences, which might be similar to the KNAT7 and STM in Arabidopsis, that repress the cell wall biosynthesis of inflorescence stems and influence the development of inflorescence architecture, respectively [34,35].
Research work has indicated that KNOX genes regulate the metabolic activities and signaling pathways associated with several phytohormones [36], including cytokinin, gibberellin, auxin, brassinosteroid, and abscisic acid. In Arabidopsis, KNOX function is mediated by cytokinin (CK) which is a growth regulator that promotes cell division and meristem function [22]. KNOX proteins regulate the balance between gibberellin (GA) and CK in the shoot apex to prevent cell differentiation and thus maintain pluripotent cell fate in the shoot apex meristem [37]. Furthermore, both in the natural context of KNOX function in the SAM and upon ectopic KNOX expression in Arabidopsis leaves, reducing activity of the GA growth regulators could promotes meristematic activity [38]. Recently, studies in apple indicated that expressions of KNOX genes are also inducible by exogenous phytohormones [17,27]. For example, MdKNOX19, a Class II KNOX gene regulating apple fruit and seed developments through ABA signaling, was significantly up-regulated when applied ABA to apple leaves, fruits, and seeds [17]. Furthermore, expressions of the fourteen MdKNOX genes that strongly exhibited expressions in the floral bud of apple were also more or less affected by the applications of exogenous phytohormones of 6-BA, GA3, ABA, and SA during the flower induction period [27]. In the present study, phytohormone-associated cis-elements, including auxin, abscisic acid, gibberellin, and ethylene-responsive elements were also shown within the promoters of AsKNOX genes (depicted in Figure 3), which might indicate the involvement of AsKNOX genes in these related phytohormone signaling pathways. Additionally, several abiotic stress responsive elements were also predicted in the promoters of AsKNOX genes, which suggest that AsKNOX genes might also contribute to the resistance of garlic to abiotic stresses.
In Arabidopsis, the KNOX Class I transcription factors have functions in repressing GA synthesis and promoting cytokinin synthesis, respectively, to maintain the meristem identity [13]. Furthermore, lateral organ founder cells with low CK and high GA could lead to the absence of KNOX expression [22,37]. To reveal the potential function of AsKNOX genes responsive to GA and CK, we checked the expression profilings of the identified 10 AsKNOX genes under either exogenous GA3 or 6-BA treatments. Compared with the untreated control groups, expressions of AsKNOX1, AsKNOX5, and AsKNOX7 showed a significant increase under two treatments, while these AsKNOX genes showed much higher expressions in their 6-BA treatments than their GA3 treatments. The expression levels of AsKNOX3, AsKNOX6, and AsKNOX7 under these two treatments showed opposite trends, for example the expression of AsKNOX6 decreased at all sample points when treated with GA3 but significantly increased under the 6-BA treatment. Conversely, AsKNOX2, AsKNOX4, and AsKNOX9 showed identical expression patterns in response to 6-BA and GA3 treatments, and the transcription of these AsKNOXs were strongly induced by both 6-BA and GA3. These results were similar to a previously reported study, for example, the transcription of MdKNOX1/20 was strongly induced by both exogenous 6-BA and exogenous GA3 at 30 and 70 days after full bloom in apple [27]. For both the treatments of exogenous GA3 and 6-BA, AsKNOX genes showed relatively complicated expression patterns at the sampling points of 1, 3, 5, and 7 DAT. This might indicate that each of the AsKNOX genes perhaps has a different regulating function in garlic. Notably, the relative expression levels of AsKNOX9 and AsKNOX10 under the GA3 treatment were significantly higher than their control groups. This might differ from previous studies of the KNOX gene family in other plants such as the Arabidopsis. Despite this, more research still needs to be conducted to reveal the detailed functions of AsKNOXs in garlic either from the perspective of the organ development (such as the differentiation of garlic cloves) or from the side of abiotic stresses responsiveness.
4. Materials and Methods
4.1. Plant Materials and Phytohormone Treatments
The garlic cultivar G024, widely cultivated in the northwest of China, was used in this study. Bulbs of G024 show the following morphological characteristics: fresh weight 30 to 50 g, bulb diameter 3.7 to 5.7 cm, and 10 to 12 cloves arranged in two whorls for each bulb. The seed cloves of G024 were provided by the Vegetable Physiology and Biotechnology Laboratory of Northwest A&F University. At the beginning of September 2019, the healthy and uniform-sized cloves of G024 were selected and planted in the Wuquan Experiment Station of Northwest A&F University in Yangling, China. Throughout the growing period, standard agronomic practices were performed to maintain the garlic plants.
To evaluate the expression patterns of the AsKNOX genes, their expression levels were checked in six kinds of organs or tissues of garlic. The samples of garlic leaf, stem, pseudostem, and root were collected on the 90th day after planting. The stems containing meristems were sampled with a height of 1 cm above the root. The pseudostem samples were cut from the lower part of the leaf, also with a height of 1 cm. The scape samples were collected on the 215th day after planting and the clove samples were collected from the seed cloves. All the samples were separated from garlic plants using scalpels and tweezers. To reveal the potential responses of AsKNOX genes to gibberellin and cytokinin, the exogenous phytohormones GA3 (1 mmol/L) and 6-BA (1 mmol/L) were used to treat garlic plants on the 90th day after planting. The treatment method strictly followed Liu et al. (2019) [7], in which the prepared phytohormones were injected into the stems from the top of garlic plants. Since the research of Liu et al. (2019) [7] also showed no difference in bulb structure between non-injection and injection of water treatments, so the control group of this study was directly performed with non-injection. Garlic stem samples were collected at the 1, 3, 5, and 7 days after treatment (DAT) to check the expression levels of all AsKNOX genes. The stem samples with a height of 1 cm and containing meristems were separated from garlic plants through the same method as before. All the collected tissue samples were immediately frozen with liquid nitrogen and then stored in a −80 °C freezer for further use.
4.2. Identification and Sequence Analysis of AsKNOX Family Members
The newly released garlic genome was used for AsKNOX genes genome-wide identification and it is available at the following website: https://doi.org/10.6084/m9.figshare.12570947.v1 (accessed on 5 November 2020). The typical conserved domain sequences of KNOX in Arabidopsis were downloaded from the website of TAIR (http://www.arabidopsis.org/, accessed on 1 January 2021). In addition, the HMM file built based on KNOX1 (PF03790) domain was used as a query to search the garlic local protein database by HMMER3.1 (E-value = 0.01). Other typical domains of KNOX proteins including KNOX2 (PF03791), ELK(PF03789), and Homeobox KN (PF05920) were further checked for the identified candidate homologs using the SMART (http://smart.embl-heidelberg.de/, accessed on 15 January 2021) and the Pfam (http://pfam.xfam.org/, accessed on 15 January 2021) databases. The position of each domain of AsKNOX proteins was also predicted with Pfam. Only those proteins containing at least two of these four conserved domains were considered as members of garlic AsKNOX. After removing the repeated and redundant sequences, the isoelectric point (pI) and molecular weight (MW) of each AsKNOX protein were predicted by using the ExPASy Proteomics Server (https://www.expasy.org/, accessed on 30 January 2021). Additionally, the subcellular localization of these AsKNOX proteins were predicted using the WoLF PSORT (https://www.genscript.com/wolf-psort.html/, accessed on 30 January 2021). All the related obtained information of these AsKNOX genes are listed in the Supplemental Table S1.
4.3. Chromosome Distribution and Gene Structure Analysis
According to the physical locations of each gene on the draft garlic genome (https://doi.org/10.6084/m9.figshare.12570947.v1, accessed on 5 November 2020), the identified AsKNOX genes were mapped onto the corresponding garlic chromosomes using the Mapchart (https://www.wur.nl/en/show/Mapchart.htm, accessed on 5 February 2021). The detailed positions of these genes are also provided in Table S1. Gene structures of these AsKNOX genes were analyzed using the TBtools software (https://github.com/CJ-Chen/TBtools/%20releases, accessed on 6 February 2021). Conserved motif structures were identified using Multiple EM for Motif Elicitation (MEME, http://meme-suite.org/, accessed on 7 February 2021), in which the parameter of a motif width larger than four and less than 100 was used. The potential cis-regulatory elements were analyzed by scanning the 2.0 kb upstream sequences from the transcription start codon ATG with the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 10 February 2021).
4.4. Phylogenetic Analysis and Multiple Sequence Alignment
Based on the previously reported literature, we collected the KNOX proteins of different species [12,39,40] including rice, maize, tomato, and Arabidopsis. The sequences of these KNOX proteins were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 15 February 2021), in which the detailed sequence accession numbers were listed in Table S2. Multiple sequence alignments of all these obtained KNOX proteins were performed by CLUSTALW (https://www.genome.jp/tools-bin/clustalw/, accessed on 15 February 2021). A phylogenetic tree was constructed using MEGA X, in which the neighbor-joining (NJ) method was used with a 1000-iterations bootstrap test.
4.5. RNA Isolation and Gene Expression Analysis
Total RNA was isolated from different garlic tissues using a Plant RNA Kit (Omega Bio-Tek) following the manufacturer’s instructions. Subsequently, the first-strand cDNA synthesis was performed using the Easy Script One-Step gDNA Removal and cDNA Synthesis Super Mix Kit (Trans Gen Biotech Co., Ltd., Beijing, China). The primers used for quantitative real-time PCR (qPCR) analysis were designed with Primer 3 Input (http://bioinfo.ut.ee/primer3-0.4.0/, accessed on 5 April 2021). Sequence information of all these primers were provided in Table S3. qPCR was conducted using the 2 × Sybr Green qPCR mix (Aidlab Biotechnologies Co., Ltd., China) in the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) with the following procedures: 95 °C for 3 min; 95 °C for 15 s, 60 °C for the 30 s with 40 cycles. Relative quantification was calculated according to the 2−ΔΔCT method described by Livak and Schmittgen [41], in which the garlic ACTIN gene (AsACT) was used as the reference gene [42]. Each PCR assay was run with three independent biological and technological replicates. The expression levels of each KNOX gene among different garlic tissues were displayed with a heatmap using the TBtools software (https://github.com/CJ-Chen/TBtools, accessed on 25 April 2021). The significance tests between the control and the exogenous phytohormones treatments were performed with the t-test analysis.
4.6. Verification of Subcellular Localization
To preliminarily check the predicted subcellular localizations of AsKNOX proteins, two of them (AsKNOX6 and AsKNOX8) were randomly selected for experimental verification. Full-length coding sequences of AsKNOX genes without stop codons were amplified using gene-specific primers and they were ligated into pGreen 0229 vector containing the green fluorescent protein (GFP) to generate GFP-AsKNOX fusion proteins. Sequence information of all these primers were provided in Table S4. The connection mode of AsKNOX genes and vector was shown in Figure 9. The Gene Star Seamless Cloning Kit was used to fuse the AsKNOX gene sequences to the linearized plasmid that was obtained through the double enzyme digestion with Xhol and EcoRV. The recombinant constructs were checked by sanger sequencing and were transfected into the abaxial side of 4- to 6-week-old N. benthamiana leaves by infiltration with strain GV3101 (Agrobacteriumt tumefaciens). The GFP empty vector was served as the positive control. After 48 h co-infiltration, the GFP signals in N. benthamiana leaves were observed using an automatic fluorescence microscope (Olympus, Tokyo, Japan) with 488-nm excitation.
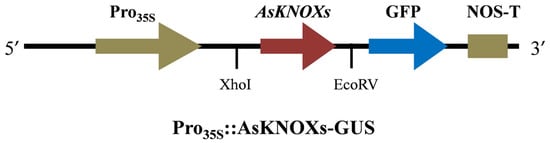
Figure 9.
The connection mode of AsKNOX genes and vector.
5. Conclusions
Ten AsKNOX genes were genome-wide identified and characterized in the garlic genome. Their gene structure, cis-elements, conserved domains, subcellular localizations, phylogenetic relationships, chromosome locations, and expression profiles in different tissues and in responding to exogenous GA3 and 6-BA treatments were systematically analyzed in the present study. These 10 AsKNOX genes showed different expression patterns among garlic tissues and most of them had responsiveness under the treatments of exogenous GA3 and 6-BA, which might indicate that the involvement of AsKNOX genes in the gibberellin and cytokinin signaling pathways. Overall, our work laid a foundation for the further functional study of AsKNOX genes in garlic, which might also enlighten the potential roles of AsKNOX genes in regulating the GA3-promoted garlic clove differentiations and developments.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22179237/s1, Table S1: Informations of 10 AsKNOX genes. Table S2: Information of KNOX proteins used for phenlogenitc tree construction. Table S3: Primers used for qPCR analyis in this study. Table S4: Sequencesns of primers used for gene cloning and vector construction.
Author Contributions
Conceptualization, S.Z. and Y.P.; methodology, S.Z.; software, S.Z. and C.Z.; validation, S.Z., Y.Z. and X.W.; formal analysis, S.Z. and Y.P.; investigation, S.Z. and C.Z.; resources, S.Z., Y.Z., X.W., X.L. and Y.P.; data curation, S.Z. and X.L.; writing—original draft preparation, S.Z. and Y.P.; writing—review and editing, S.Z. and Y.P.; visualization, S.Z., Y.Z., X.L. and X.W.; supervision, Z.C. and Y.P.; project administration, Z.C and Y.P.; funding acquisition, Z.C. and Y.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Education Development Fund Project of Northwest A&F University (2017) to Z.C. and the Chinese Universities Scientific Fund (2452019017) to Y.P.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to acknowledge the financial support of the College of Horticulture, Northwest A&F University, Yangling, China and funding agencies. Additionally, we are grateful to Muhammad Ali for proofreading and revising this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Etoh, T.; Watanabe, H.; Iwai, S. RAPD variation of garlic clones in the center of origin and the westernmost area of distribution. Mem. Fac. Agric. 2001, 37, 21–27. [Google Scholar]
- Sun, X.; Zhu, S.; Li, N.; Cheng, Y.; Liu, T. A chromosome-level genome assembly of garlic (Allium sativum L.) provides insights into genome evolution and allicin biosynthesis. Mol. Plant 2020, 13, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Kamenetsky, R. Garlic: Botany and Horticulture; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Ni, J.; Gao, C.; Chen, M.S.; Pan, B.Z.; Ye, K.; Xu, Z.F. Gibberellin promotes shoot branching in the perennial woody plant Jatropha curcas. Plant Cell Physiol. 2015, 8, 1655–1666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexopoulos, A.A.; Akoumianakis, K.A.; Passam, H.C. The effect of the time and mode of application of gibberellic acid on the growth and yield of potato plants derived from true potato seed. Russ. J. Appl. Chem. 2010, 86, 2189–2195. [Google Scholar] [CrossRef]
- Yamazaki, H.; Shiraiwa, N.; Itai, A.; Honda, I. Involvement of gibberellins in the regulation of tillering in welsh onion (Allium fistulosum L.). Hortic. J. 2015, 84, MI-050. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Deng, R.; Huang, C.; Cheng, Z.; Meng, H. Exogenous gibberellins alter morphology and nutritional traits of garlic (Allium sativum L.) bulb. Sci. Hortic. 2019, 246, 298–306. [Google Scholar] [CrossRef]
- Liu, H.J.; Huang, C.P.; Tong, P.J.; Yang, X.; Cheng, Z.H. Response of axillary bud development in garlic (Allium sativum L.) to seed cloves soaked in gibberellic acid (GA3) solution. J. Integr. Agric. 2020, 19, 1044–1054. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Y. Axillary meristem initiation-a way to branch out. Curr. Opin. Plant Biol. 2018, 41, 61–66. [Google Scholar] [CrossRef]
- Giulio, T.; Emiliano, C.; Ignazio, V.; Chiara, N.; Emilia, C.; Teresa, D.M.; Elisa, V.; Leonardo, B.; Beatrice, B.M.; Giovanni, M. The peach (Prunus persica L. Batsch) genome harbours 10 KNOX genes, which are differentially expressed in stem development, and the class 1 KNOPE1 regulates elongation and lignification during primary growth. J. Exp. Bot. 2012, 63, 5417–5435. [Google Scholar]
- Bürglin, T. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997, 25, 4173. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef] [Green Version]
- Hay, A.; Tsiantis, M. KNOX genes: Versatile regulators of plant development and diversity. Development 2010, 137, 3153–3165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furumizu, C.; Alvarez, J.P.; Sakakibara, K.; Bowman, J.L.; Qu, L.J. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet. 2015, 11, e1004980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truernit, E.; Haseloff, J. A role for KNAT class II genes in root development. Plant Signal. Behav. 2007, 2, 10–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Lee, C.; Zhou, J.; Ye, M. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [Green Version]
- Jia, P.; Xing, L.; Zhang, C.; Zhang, D.; An, N. MdKNOX19, a class II knotted-like transcription factor of apple, plays roles in ABA signalling /sensitivity by targeting ABI5 during organ development. Plant Sci. 2021, 302, 110701. [Google Scholar] [CrossRef]
- Byrne, M.E.; Simorowski, J.; Martienssen, R.A. Asymmetric leaves1 reveals knox gene redundancy in Arabidopsis. Development 2002, 129, 1957–1965. [Google Scholar] [CrossRef]
- Venglat, S.P.; Dumonceaux, T.; Rozwadowski, K.; Parnell, L.; Datla, R. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 4730–4735. [Google Scholar] [CrossRef] [Green Version]
- Ragni, L.; Belles-Boix, E.; Gunl, M.; Pautot, V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 2008, 20, 888–900. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Patibanda, V.; Smith, H. A novel role of BELL1-like homeobox genes, PENNYWISE and POUND-FOOLISH, in floral patterning. Planta 2009, 229, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Mansfield, S.D.; Groover, A.T. The Populus homeobox gene Arborknox2 regulates cell differentiation during secondary growth. Plant J. 2010, 60, 1000–1014. [Google Scholar] [CrossRef]
- Cheng, X.; Li, M.; Abdullah, M.; Li, G.; Lin, Y. In silico genome-wide analysis of the pear (Pyrus bretschneideri) KNOX family and the functional characterization of PbKNOX1, an Arabidopsis Brevipedicellus orthologue gene, involved in cell wall and lignin biosynthesis. Front. Genet. 2019, 10, 632. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, E.; Reiser, L.; Hake, S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 2000, 127, 3161. [Google Scholar] [CrossRef]
- Jia, P.; Zhang, C.; Xing, L.; Li, Y.; Shah, K.; Zuo, X.; Zhang, D.; An, N.; Han, M.; Ren, X. Genome-wide identification of the MdKNOX gene family and characterization of its transcriptional regulation in Malus domestica. Front. Plant Sci. 2020, 11, 128. [Google Scholar] [CrossRef]
- Jia, P.; Xing, L.; Zhang, C.; Chen, H.; An, N. MdKNOX15, a class I knotted-like transcription factor of apple, controls flowering and plant height by regulating GA levels through promoting the MdGA2ox7 transcription. Environ. Exp. Bot. 2021, 185, 104411. [Google Scholar] [CrossRef]
- Hackbusch, J.; Richter, K.; Müller, J.; Salamini, F.; Uhrig, J.F. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, D.H. Plant transcription factors evolutionary, structural and functional aspects. In Introduction to Transcription Factor Structure and Function; Academic Press: Cambridge, MA, USA, 2016; pp. 3–11. ISBN 978-0-12-800854-6. [Google Scholar]
- Lucas, W.J.; Bouche-Pillon, S.; Jackson, D.; Nguyen, L.; Baker, L.; Ding, B.; Hake, S. Selective trafficking of KNOTTED1 Homeodomain protein and its mRNA through plasmodesmata. Science 1995, 270, 1980–1983. [Google Scholar] [CrossRef]
- Bueno, N.; Alvarez, J.M.; Ordás, R. Characterization of the Knotted1-Like Homeobox (KNOX) gene family in Pinus pinaster Ait. Plant Sci. 2020, 301, 110691. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.; Forneris, N.; Savidge, R.; Johnson, L.; Mansfield, S.; Ellis, B.; Douglas, C. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yamaguchi, M.; Grienenberger, E.; Martone, P.T.; Samuels, A.L.; Mansfield, S.D. The Class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 2020, 101, 293–309. [Google Scholar] [CrossRef]
- Huang, L.M. ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture. J. Exp. Bot. 2012, 63, 1423–1433. [Google Scholar]
- Tsuda, K.; Hake, S. Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 2015, 27, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef] [Green Version]
- Hay, A.; Kaur, H.; Phillips, A.; Hedden, P.; Tsiantis, M. The gibberellin pathway mediates knotted1-type homeobox function in plants with different body plans. Curr. Biol. 2002, 12, 1557–1565. [Google Scholar] [CrossRef] [Green Version]
- Ye, S.G.; Zai, W.S.; Xiong, Z.L.; Zhang, H.L.; Ma, Y.R. Genome-wide identification of gene family in tomato and their evolutionary relationship in Solanaceae. J. Nucl. Agric. Sci. 2017, 31, 1263–1271. [Google Scholar]
- Jie, G.; Yang, X.; Zhao, W.; Lang, T.; Tore, S. Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 2015, 6, 882. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, M.; Wu, Z.; Jiang, F. Selection and validation of garlic reference genes for quantitative real-time PCR normalization. Plant Cell Tissue Organ Cult. 2015, 122, 435–444. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).