Paradoxical Pro-angiogenic Effect of Low-Dose Ellipticine Identified by In Silico Drug Repurposing
Abstract
1. Introduction
2. Results
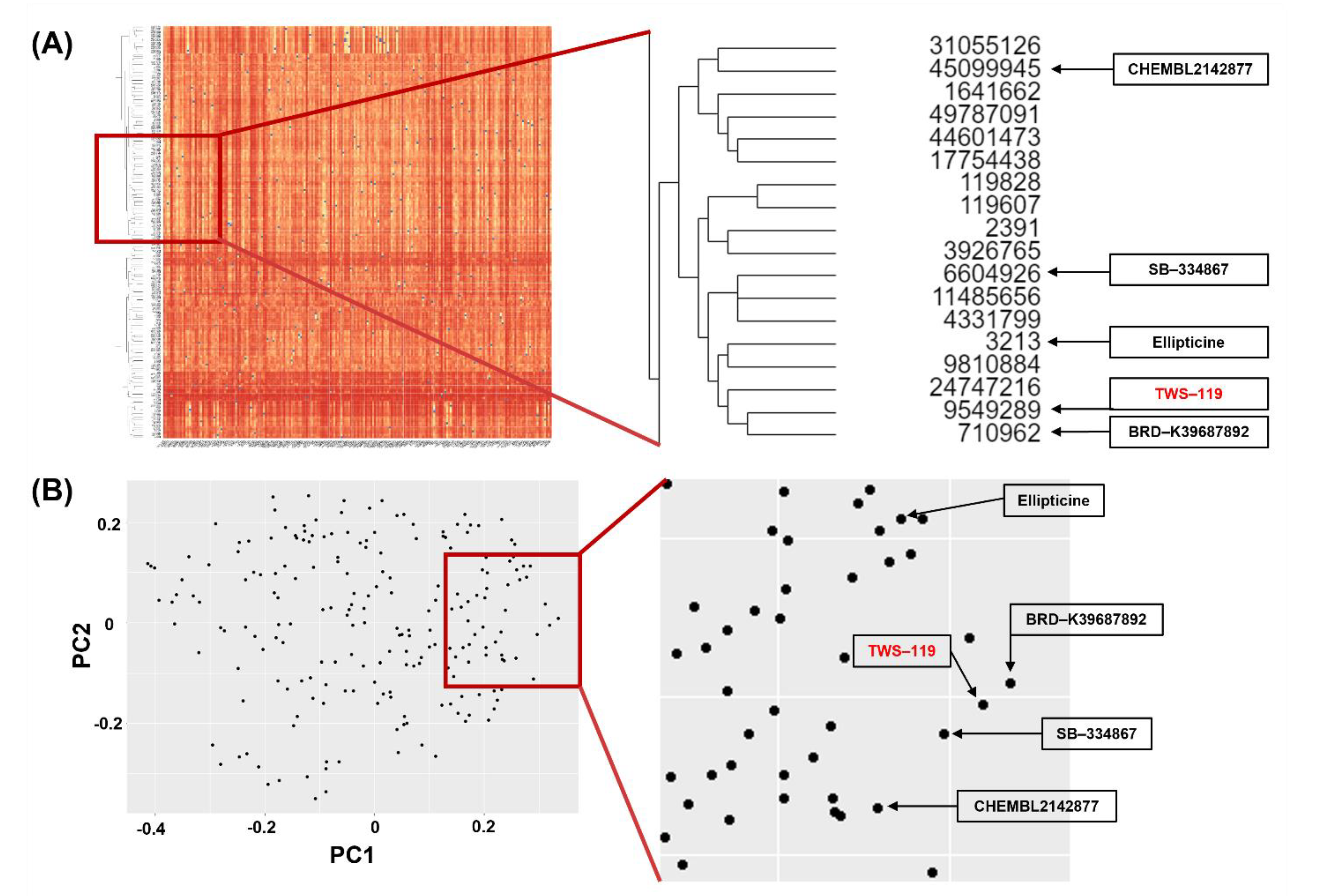
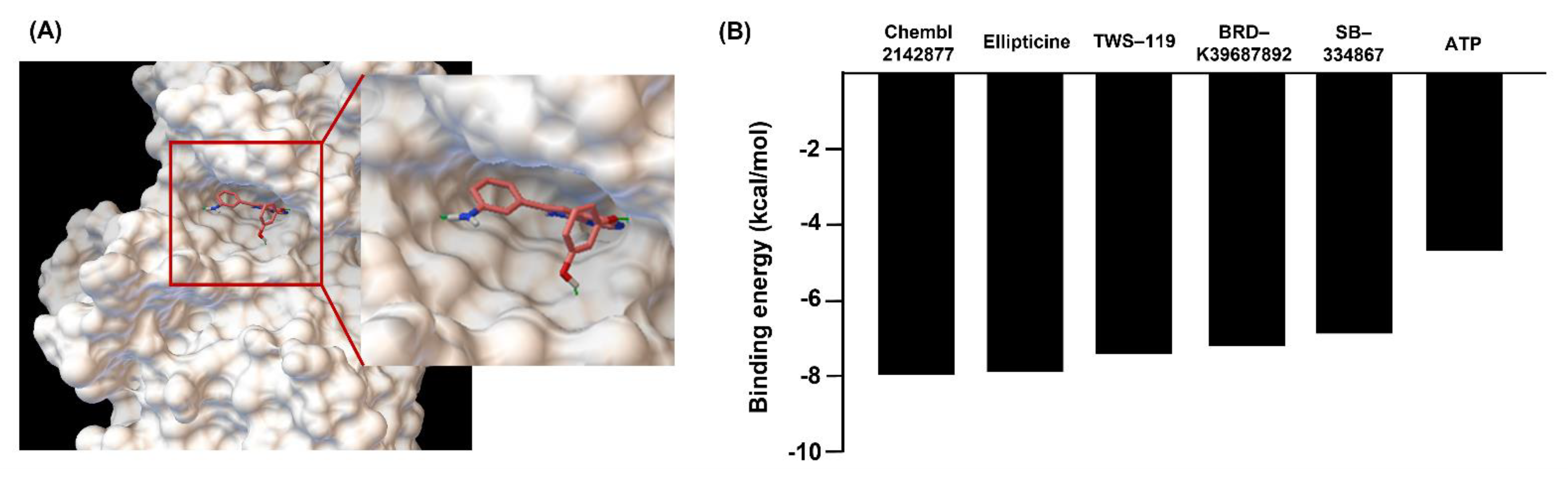
2.1. Initial Identification of Candidate Drugs to Enhance Angiogenesis by In Silico Virtual Screening
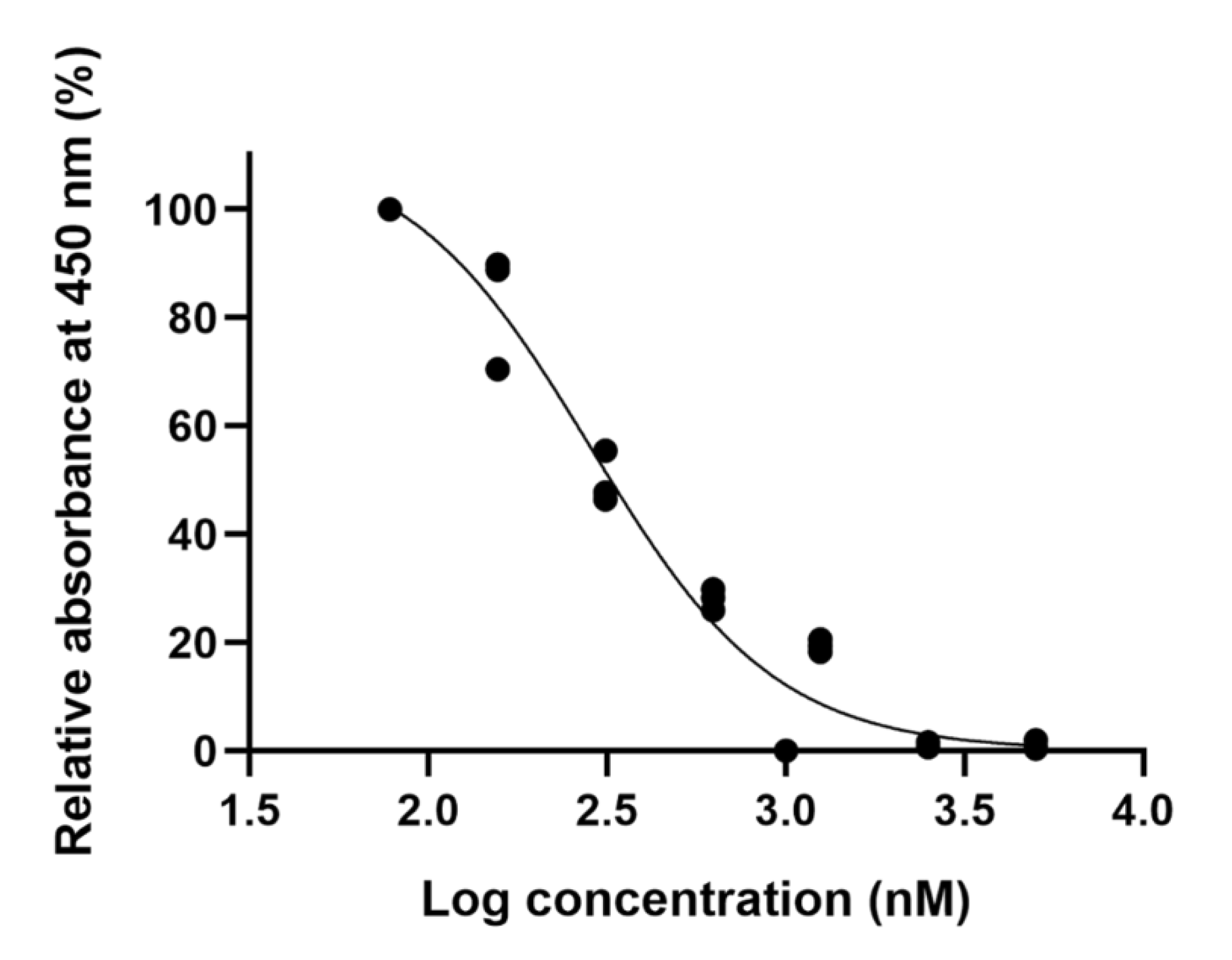
2.2. The Noncytotoxic Concentration of Ellipticine in a Dose–Response Curve Is 156.25 nM
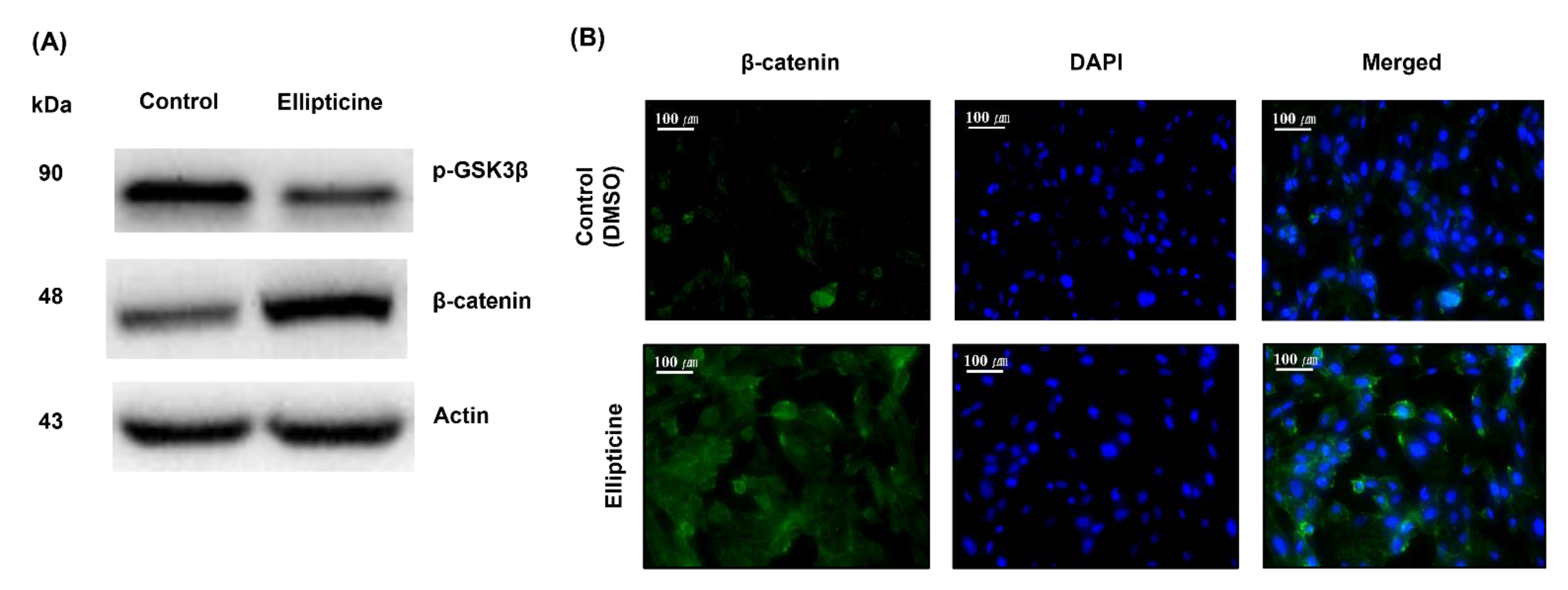
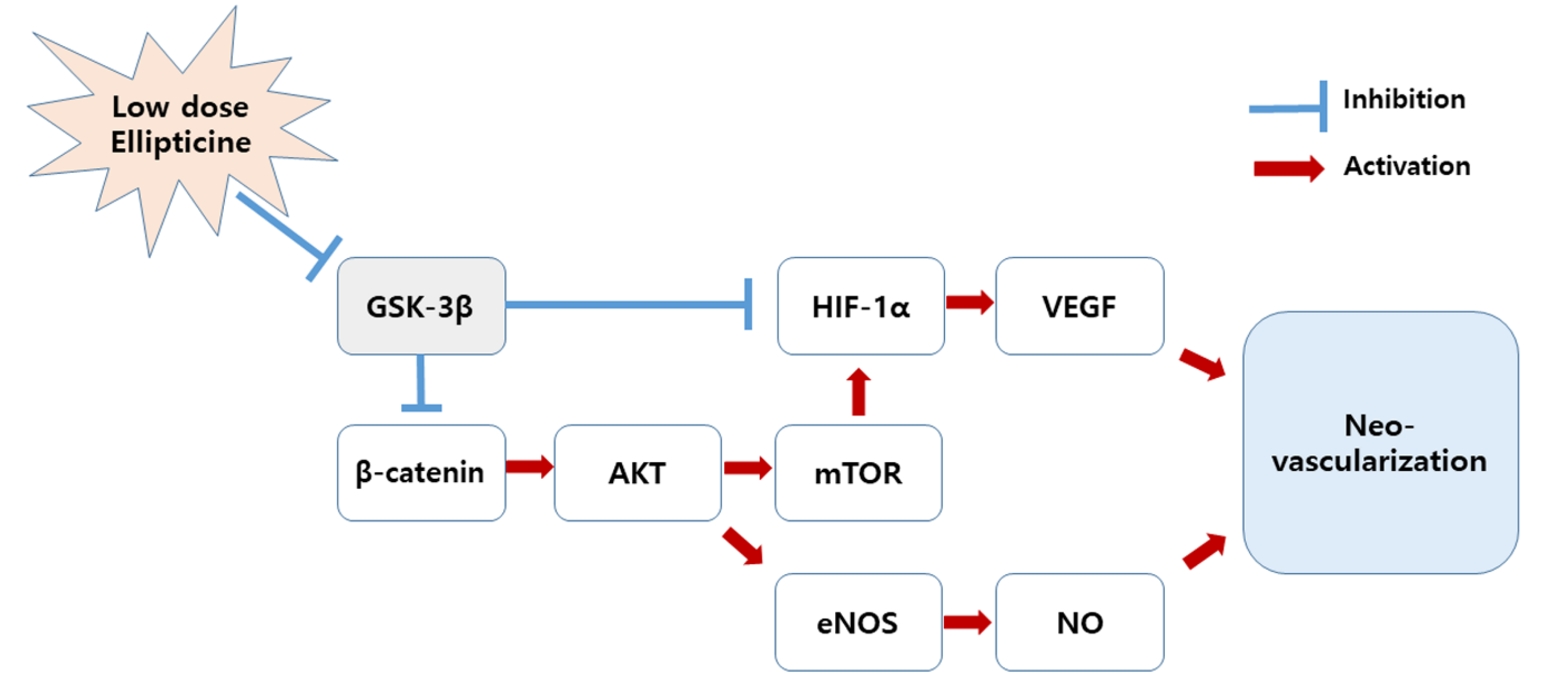
2.3. Low-Dose Ellipticine Reduces the Phosphorylation of GSK-3β and Increases β-Catenin Expression
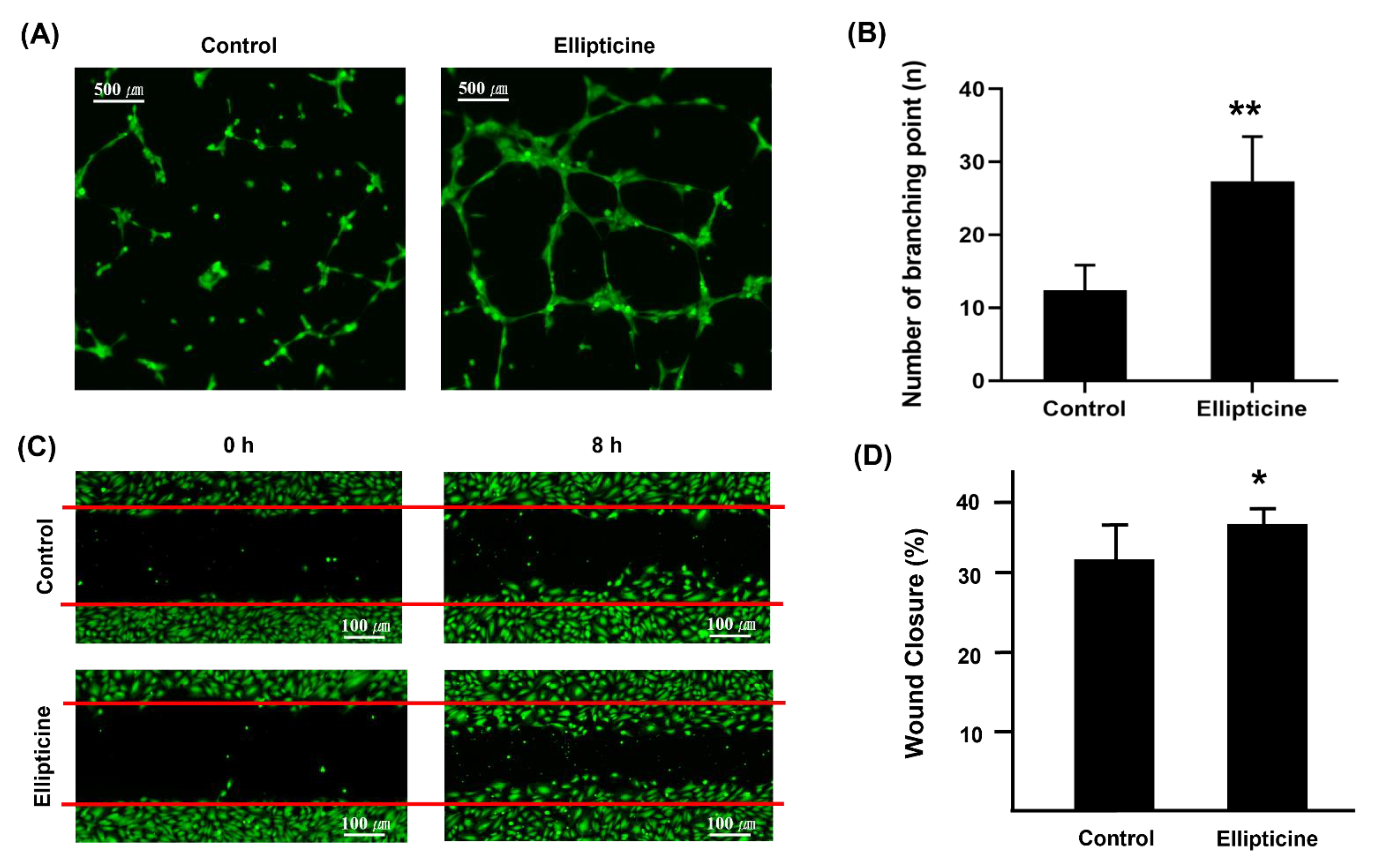
2.4. Low-Dose Ellipticine Enhances Endothelial Cell Migration
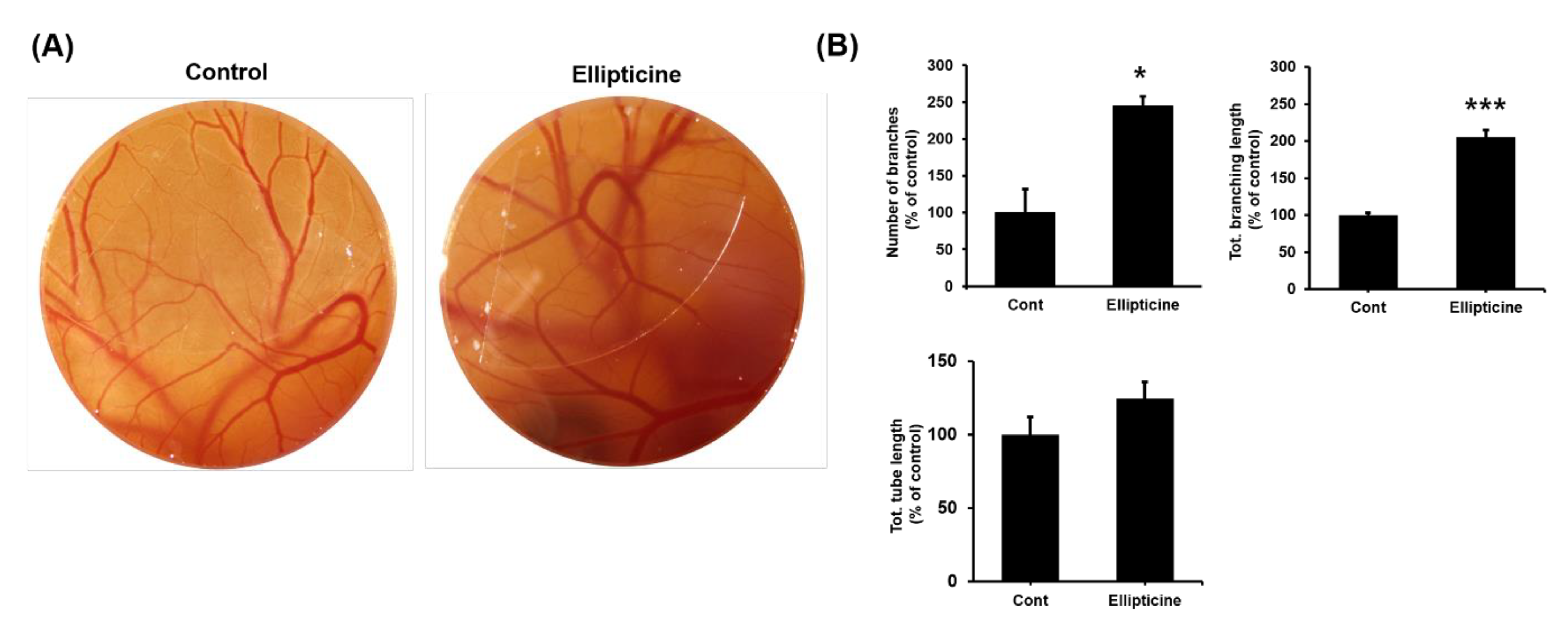
2.5. Low-Dose Ellipticine Has a Pro-angiogenic Property
3. Discussion
4. Materials and Methods
4.1. Screening Procedure
4.2. Chemicals
4.3. Cell Culture and Chemical Treatment
4.4. Cell Viability Assay
4.5. Western Blot
4.6. Immunofluorescence
4.7. Tube Formation Assay
4.8. Migration Assay
4.9. Chorioallantoic Membrane (CAM) Assay
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Folkman, J. Anggiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1995, 1, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y. Thromboangiitis obliterans (Buerger’s disease). Int. J. Cardiol. 1996, 54 (Suppl. S1), S185–S187. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Breckenridge, A.; Jacob, R. Overcoming the legal and regulatory barriers to drug repurposing. Nat. Rev. Drug Discov. 2019, 18, 1–2. [Google Scholar] [CrossRef] [PubMed]
- McInnes, C. Virtual screening strategies in drug discovery. Curr. Opin. Chem. Biol. 2007, 11, 494–502. [Google Scholar] [CrossRef]
- Hodos, R.A.; Kidd, B.A.; Shameer, K.; Readhead, B.P.; Dudley, J.T. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip Rev. Syst. Biol. Med. 2016, 8, 186–210. [Google Scholar] [CrossRef]
- Fritsche, M.; Haessler, C.; Brandner, G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene 1993, 8, 307–318. [Google Scholar]
- Paoletti, C.; Le Pecq, J.B.; Dat-Xuong, N.; Juret, P.; Garnier, H.; Amiel, J.L.; Rouesse, J. Antitumor activity, pharmacology, and toxicity of ellipticines, ellipticinium, and 9-hydroxy derivatives: Preliminary clinical rials of 2-methyl-9-hydroxy ellipticinium (NSC 264-137). Recent Results Cancer Res. 1980, 74, 107–123. [Google Scholar]
- Duan, Q.; Reid, S.P.; Clark, N.R.; Wang, Z.; Fernandez, N.F.; Rouillard, A.D.; Readhead, B.; Tritsch, S.R.; Hodos, R.; Hafner, M.; et al. L1000CDS(2): LINCS L1000 characteristic direction signatures search engine. NPJ Syst. Biol. Appl. 2016, 2, 1–12. [Google Scholar] [CrossRef]
- Song, D.; Zhang, X.; Chen, J.; Liu, X.; Xue, J.; Zhang, L.; Lan, X. Wnt canonical pathway activator TWS119 drives microglial anti-inflammatory activation and facilitates neurological recovery following experimental stroke. J. Neuroinflam. 2019, 16, 256. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, M.; Wang, Y.; Li, Q.; Deng, G.; Wan, J.; Yang, Q.; Chen, Q.; Wang, J. GSK-3beta inhibitor TWS119 attenuates rtPA-induced hemorrhagic transformation and activates the Wnt/beta-catenin signaling pathway after acute ischemic stroke in rats. Mol. Neurobiol. 2016, 53, 7028–7036. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, M.; Wang, Y.; Wang, Z.; Zhang, W.; Guan, F.; Chen, Q.; Wang, J. GSK-3beta as a target for protection against transient cerebral ischemia. Int. J. Med. Sci. 2017, 14, 333–339. [Google Scholar] [CrossRef]
- Backman, T.W.; Cao, Y.; Girke, T. ChemMine tools: An online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011, 39, W486–W491. [Google Scholar] [CrossRef] [PubMed]
- Garcia, B.L.; Skaff, D.A.; Chatterjee, A.; Hanning, A.; Walker, J.K.; Wyckoff, G.J.; Geisbrecht, B.V. Identification of C3b-Binding Small-Molecule Complement Inhibitors Using Cheminformatics. J. Immunol. 2017, 198, 3705–3718. [Google Scholar] [CrossRef] [PubMed]
- Hockel, M.; Schlenger, K.; Doctrow, S.; Kissel, T.; Vaupel, P. Therapeutic angiogenesis. Arch Surg. 1993, 128, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, S.; Zheng, L.P.; Brogi, E.; Kearney, M.; Pu, L.Q.; Bunting, S.; Ferrara, N.; Symes, J.F.; Isner, J.M. Therapeutic angiogenesis. A single intraarterial bolus of vascular endothelial growth factor augments revascularization in a rabbit ischemic hind limb model. J. Clin. Investig. 1994, 93, 662–670. [Google Scholar] [CrossRef]
- Semenza, G.L. Therapeutic angiogenesis: Another passing phase? Circ. Res. 2006, 98, 1115–1116. [Google Scholar] [CrossRef]
- Katsila, T.; Spyroulias, G.A.; Patrinos, G.P.; Matsoukas, M.T. Computational approaches in target identification and drug discovery. Comput. Struct. Biotechnol. J. 2016, 14, 177–184. [Google Scholar] [CrossRef]
- Lavecchia, A.; di Giovanni, C. Virtual screening strategies in drug discovery: A critical review. Curr. Med. Chem. 2013, 20, 2839–2860. [Google Scholar] [CrossRef]
- Dudley, J.T.; Sirota, M.; Shenoy, M.; Pai, R.K.; Roedder, S.; Chiang, A.P.; Morgan, A.A.; Sarwal, M.M.; Pasricha, P.J.; Butte, A.J. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 2011, 3, 96ra76. [Google Scholar] [CrossRef]
- Wagner, A.; Cohen, N.; Kelder, T.; Amit, U.; Liebman, E.; Steinberg, D.M.; Radonjic, M.; Ruppin, E. Drugs that reverse disease transcriptomic signatures are more effective in a mouse model of dyslipidemia. Mol. Syst. Biol. 2015, 11, 791. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Smith, S.; Thorpe, A.; Barratt, M.J.; Karim, F. Evaluation of phenoxybenzamine in the CFA model of pain following gene expression studies and connectivity mapping. Mol. Pain 2010, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Josset, L.; Textoris, J.; Loriod, B.; Ferraris, O.; Moules, V.; Lina, B.; N’Guyen, C.; Diaz, J.J.; Rosa-Calatrava, M. Gene expression signature-based screening identifies new broadly effective influenza a antivirals. PLoS ONE 2010, 5, e13169. [Google Scholar] [CrossRef] [PubMed]
- Jahchan, N.S.; Dudley, J.T.; Mazur, P.K.; Flores, N.; Yang, D.; Palmerton, A.; Zmoos, A.F.; Vaka, D.; Tran, K.Q.; Zhou, M.; et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013, 3, 1364–1377. [Google Scholar] [CrossRef]
- Zerbini, L.F.; Bhasin, M.K.; de Vasconcellos, J.F.; Paccez, J.D.; Gu, X.; Kung, A.L.; Libermann, T.A. Computational repositioning and preclinical validation of pentamidine for renal cell cancer. Mol. Cancer 2014, 13, 1929–1941. [Google Scholar] [CrossRef]
- McClendon, A.K.; Osheroff, N. DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res. 2007, 623, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Stiborova, M.; Bieler, C.A.; Wiessler, M.; Frei, E. The anticancer agent ellipticine on activation by cytochrome P450 forms covalent DNA adducts. Biochem. Pharm. 2001, 62, 1675–1684. [Google Scholar]
- Stiborova, M.; Cerna, V.; Moserova, M.; Mrizova, I.; Arlt, V.M.; Frei, E. The anticancer drug ellipticine activated with cytochrome P450 mediates DNA damage determining its pharmacological efficiencies: Studies with rats, Hepatic Cytochrome P450 Reductase Null (HRN) mice and pure enzymes. Int. J. Mol. Sci. 2014, 16, 284–306. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.L.; Hsu, Y.L.; Chang, C.H.; Lin, C.C. The mechanism of ellipticine-induced apoptosis and cell cycle arrest in human breast MCF-7 cancer cells. Cancer Lett. 2005, 223, 293–301. [Google Scholar] [CrossRef]
- Kuo, Y.C.; Kuo, P.L.; Hsu, Y.L.; Cho, C.Y.; Lin, C.C. Ellipticine induces apoptosis through p53-dependent pathway in human hepatocellular carcinoma HepG2 cells. Life Sci. 2006, 78, 2550–2557. [Google Scholar] [CrossRef]
- Kizek, R.; Adam, V.; Hrabeta, J.; Eckschlager, T.; Smutny, S.; Burda, J.V.; Frei, E.; Stiborova, M. Anthracyclines and ellipticines as DNA-damaging anticancer drugs: Recent advances. Pharm. Ther. 2012, 133, 26–39. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, S.G.; Chung, J.Y.; Kim, Y.J.; Park, J.E.; Koh, H.; Han, M.S.; Park, Y.C.; Yoo, Y.H.; Kim, J.M. Ellipticine induces apoptosis in human endometrial cancer cells: The potential involvement of reactive oxygen species and mitogen-activated protein kinases. Toxicology 2011, 289, 91–102. [Google Scholar] [CrossRef]
- Poljakova, J.; Eckschlager, T.; Hrabeta, J.; Hrebackova, J.; Smutny, S.; Frei, E.; Martinek, V.; Kizek, R.; Stiborova, M. The mechanism of cytotoxicity and DNA adduct formation by the anticancer drug ellipticine in human neuroblastoma cells. Biochem. Pharm. 2009, 77, 1466–1479. [Google Scholar] [CrossRef]
- Wen, H.L.; Yang, G.; Dong, Q.R. Ellipticine inhibits the proliferation and induces apoptosis in rheumatoid arthritis fibroblast-like synoviocytes via the STAT3 pathway. Immunopharmacol. Immunotoxicol. 2017, 39, 219–224. [Google Scholar] [CrossRef]
- Stiborova, M.; Poljakova, J.; Martinkova, E.; Borek-Dohalska, L.; Eckschlager, T.; Kizek, R.; Frei, E. Ellipticine cytotoxicity to cancer cell lines—A comparative study. Interdiscip. Toxicol. 2011, 4, 98–105. [Google Scholar] [CrossRef]
- Prajongtat, P.; Suramitr, S.; Nokbin, S.; Nakajima, K.; Mitsuke, K.; Hannongbua, S. Density functional theory study of adsorption geometries and electronic structures of azo-dye-based molecules on anatase TiO2 surface for dye-sensitized solar cell applications. J. Mol. Graph. Model. 2017, 76, 551–561. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/beta-Catenin Signaling, Disease, and Emerging. Ther. Modalities Cell 2017, 169, 985–999. [Google Scholar]
- Billadeau, D.D. Primers on molecular pathways. The glycogen synthase kinase-3beta. Pancreatology 2007, 7, 398–402. [Google Scholar] [CrossRef][Green Version]
- Ougolkov, A.V.; Fernandez-Zapico, M.E.; Savoy, D.N.; Urrutia, R.A.; Billadeau, D.D. Glycogen synthase kinase-3beta participates in nuclear factor kappaB-mediated gene transcription and cell survival in pancreatic cancer cells. Cancer Res. 2005, 65, 2076–2081. [Google Scholar] [CrossRef]
- Doble, B.W.; Woodgett, J.R. GSK-3: Tricks of the trade for a multi-tasking kinase. J. Cell Sci. 2003, 116, 1175–1186. [Google Scholar] [CrossRef]
- Farago, M.; Dominguez, I.; Landesman-Bollag, E.; Xu, X.; Rosner, A.; Cardiff, R.D.; Seldin, D.C. Kinase-inactive glycogen synthase kinase 3beta promotes Wnt signaling and mammary tumorigenesis. Cancer Res. 2005, 65, 5792–5801. [Google Scholar] [CrossRef]
- Voskas, D.; Ling, L.S.; Woodgett, J.R. Does GSK-3 provide a shortcut for PI3K activation of Wnt signalling? F1000 Biol. Rep. 2010, 2, 82. [Google Scholar] [CrossRef] [PubMed]
- Denham, M.; Bye, C.; Leung, J.; Conley, B.J.; Thompson, L.H.; Dottori, M. Glycogen synthase kinase 3beta and activin/nodal inhibition in human embryonic stem cells induces a pre-neuroepithelial state that is required for specification to a floor plate cell lineage. Stem. Cells 2012, 30, 2400–2411. [Google Scholar] [CrossRef]
- Kwon, C.; Cheng, P.; King, I.N.; Andersen, P.; Shenje, L.; Nigam, V.; Srivastava, D. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat. Cell Biol. 2011, 13, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Doble, B.W.; MacAulay, K.; Sinclair, E.M.; Drucker, D.J.; Woodgett, J.R. Tissue-specific role of glycogen synthase kinase 3beta in glucose homeostasis and insulin action. Mol. Cell Biol. 2008, 28, 6314–6328. [Google Scholar] [CrossRef] [PubMed]
- Tajadura, V.; Hansen, M.H.; Smith, J.; Charles, H.; Rickman, M.; Farrell-Dillon, K.; Claro, V.; Warboys, C.; Ferro, A. Beta-catenin promotes endothelial survival by regulating eNOS activity and flow-dependent anti-apoptotic gene expression. Cell Death Dis. 2020, 11, 493. [Google Scholar] [CrossRef]
- Arsham, A.M.; Plas, D.R.; Thompson, C.B.; Simon, M.C. Akt and hypoxia-inducible factor-1 independently enhance tumor growth and angiogenesis. Cancer Res. 2004, 64, 3500–3507. [Google Scholar] [CrossRef] [PubMed]
- Carhart, R.E.; Smith, D.H.; Venkataraghavan, R. Atom Pairs as Molecular-Features in Structure Activity Studies—Definition and Applications. J. Chem. Inf. Comp. Sci. 1985, 25, 64–73. [Google Scholar] [CrossRef]
- Chen, X.; Reynolds, C.H. Performance of similarity measures in 2D fragment-based similarity searching: Comparison of structural descriptors and similarity coefficients. J. Chem. Inf. Comp. Sci. 2002, 42, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Mardia, K.V. Some properties of clasical multi-dimesional scaling. Commun. Stat.—Theory Methods 2009, 7, 1233–1241. [Google Scholar] [CrossRef]
- Weibel, N.; Grunder, S.; Mayor, M. Functional molecules in electronic circuits. Org. Biomol. Chem. 2007, 5, 2343–2353. [Google Scholar]

| GEO Accession Number | Experimental Group | Sample Number | Species | Study Summary |
|---|---|---|---|---|
| GSE837 | High serum PlGF 24 h High serum VEGF-A 24 h Low serum PlGF 24 h Low serum VEGF-A 4 h Low serum VEGF-A 24 h | GSM12894–GSM12926 | Homo sapience | HUVECs are treated with the angiogenic factors VEGF-A and PlGF in low or high serum media |
| GSE9677 | Angiopoietin-1 (Sparse)Angiopoietin-1 (Confluence) | GSM244647–GSM244654 | Homo sapience | Angiopoietin-1 stimulation on HUVECs |
| GSE19098 | VEGF (spread) VEGF (unspread) | GSM473026–GSM473037 | Homo sapience | 50 ng/mL VEGF or no growth factor was treated on HUVEC for 18 h |
| GSE22695 | VEGF 30 min VEGF 60 min VEGF 150 min | GSM385333–GSM385353 | Homo sapience | HUVEC and CEP were treated with VEGF-A for 30, 60, 150 min |
| GSE71216 | VEGF | GSM1830134–GSM1830137 | Homo sapience | HUVECs were treated with 16 ng/mL VEGF165 for 4 days |
| Compounds | IUPAC Name | Known MOA | CID | MW |
|---|---|---|---|---|
| TWS-119 | 3-[[6-(3-aminophenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]oxy]phenol | GSK-3β inhibitor | 9549289 | 318.3 |
| Ellipticine | 5,11-dimethyl-6H-pyrido[4,3-b]carbazole | Topoisomerase II inhibitor | 3213 | 246.31 |
| BRD-K39687892 | N-(3-fluorophenyl)-2-pyridin-4-ylquinazolin-4-amine | GSK-3β inhibitor | 710962 | 316.3 |
| SB-334867 | 1-(2-methyl-1,3-benzoxazol-6-yl)-3-(1,5-naphthyridin-4-yl)urea | Orexin antagonist | 6604926 | 319.32 |
| CHEMBL2142877 | 4-phenyl-N-(6-phenyl-3-pyridin-2-yl-1,2,4-triazin-5-yl)-1,3-thiazol-2-amine | Unknown | 45099945 | 408.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, J.; Lee, H.H.; Jeong, Y.; Yoon, S.; An, H.-J.; Baek, M.; Kim, D.K.; Lee, S. Paradoxical Pro-angiogenic Effect of Low-Dose Ellipticine Identified by In Silico Drug Repurposing. Int. J. Mol. Sci. 2021, 22, 9067. https://doi.org/10.3390/ijms22169067
Oh J, Lee HH, Jeong Y, Yoon S, An H-J, Baek M, Kim DK, Lee S. Paradoxical Pro-angiogenic Effect of Low-Dose Ellipticine Identified by In Silico Drug Repurposing. International Journal of Molecular Sciences. 2021; 22(16):9067. https://doi.org/10.3390/ijms22169067
Chicago/Turabian StyleOh, Jisu, Hyeon Hae Lee, Yunhui Jeong, Siyeong Yoon, Hyun-Ju An, Minjung Baek, Do Kyung Kim, and Soonchul Lee. 2021. "Paradoxical Pro-angiogenic Effect of Low-Dose Ellipticine Identified by In Silico Drug Repurposing" International Journal of Molecular Sciences 22, no. 16: 9067. https://doi.org/10.3390/ijms22169067
APA StyleOh, J., Lee, H. H., Jeong, Y., Yoon, S., An, H.-J., Baek, M., Kim, D. K., & Lee, S. (2021). Paradoxical Pro-angiogenic Effect of Low-Dose Ellipticine Identified by In Silico Drug Repurposing. International Journal of Molecular Sciences, 22(16), 9067. https://doi.org/10.3390/ijms22169067







