Expression of Alternative Splice Variants of 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase-4 in Normoxic and Hypoxic Melanoma Cells
Abstract
1. Introduction
2. Results
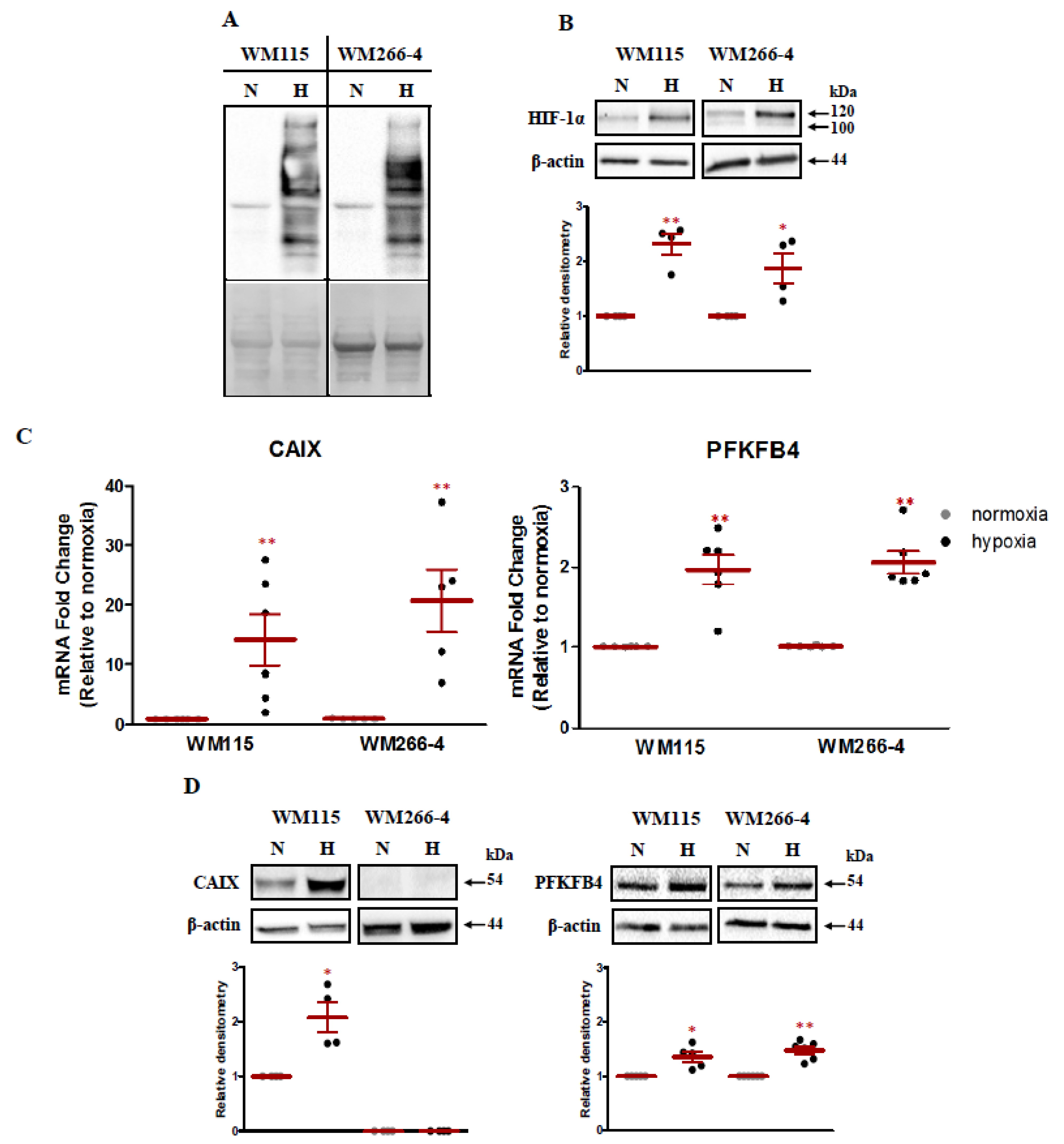
2.1. The Response of Malignant Melanoma Cells to Low Oxygen Concentration
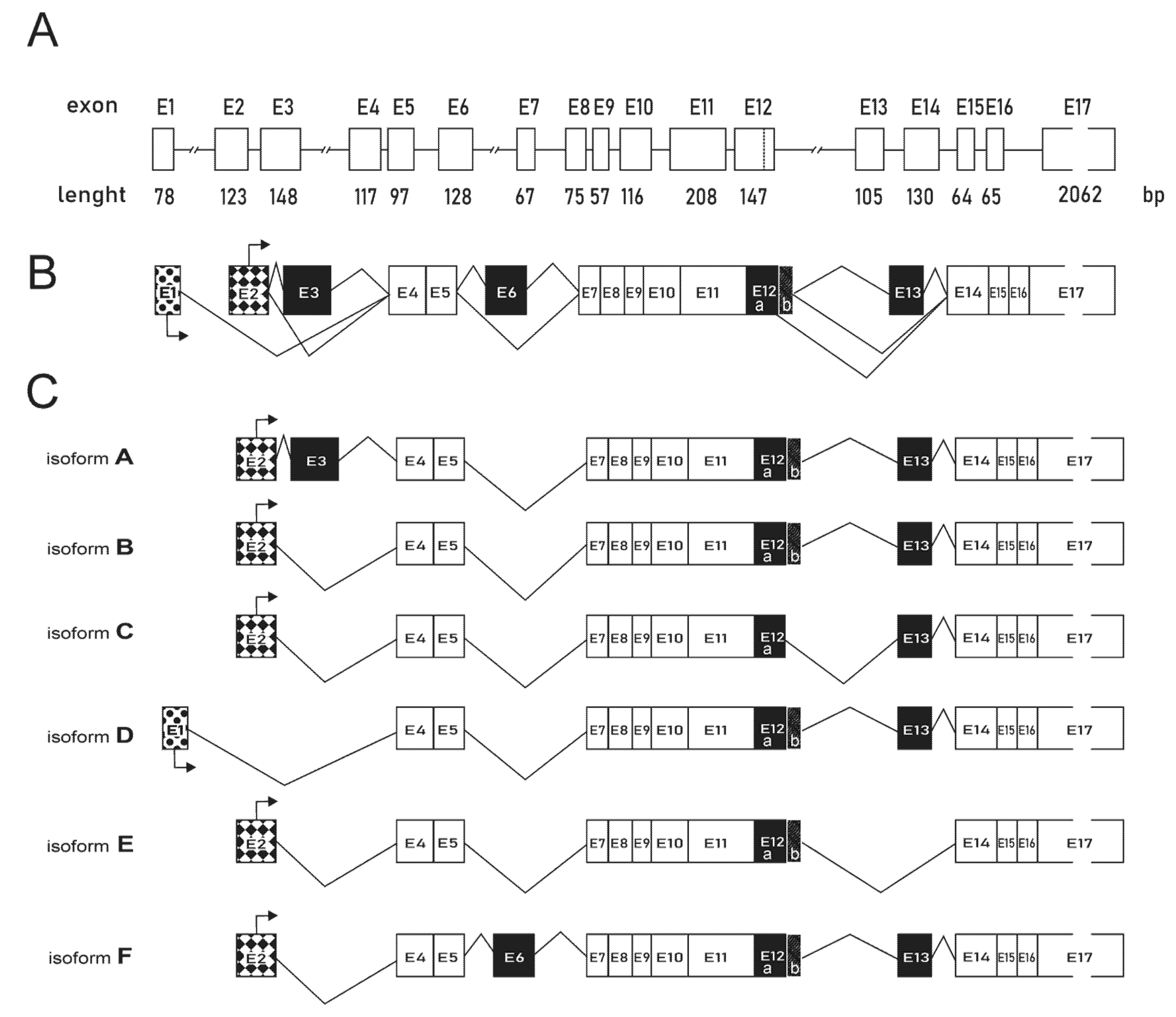
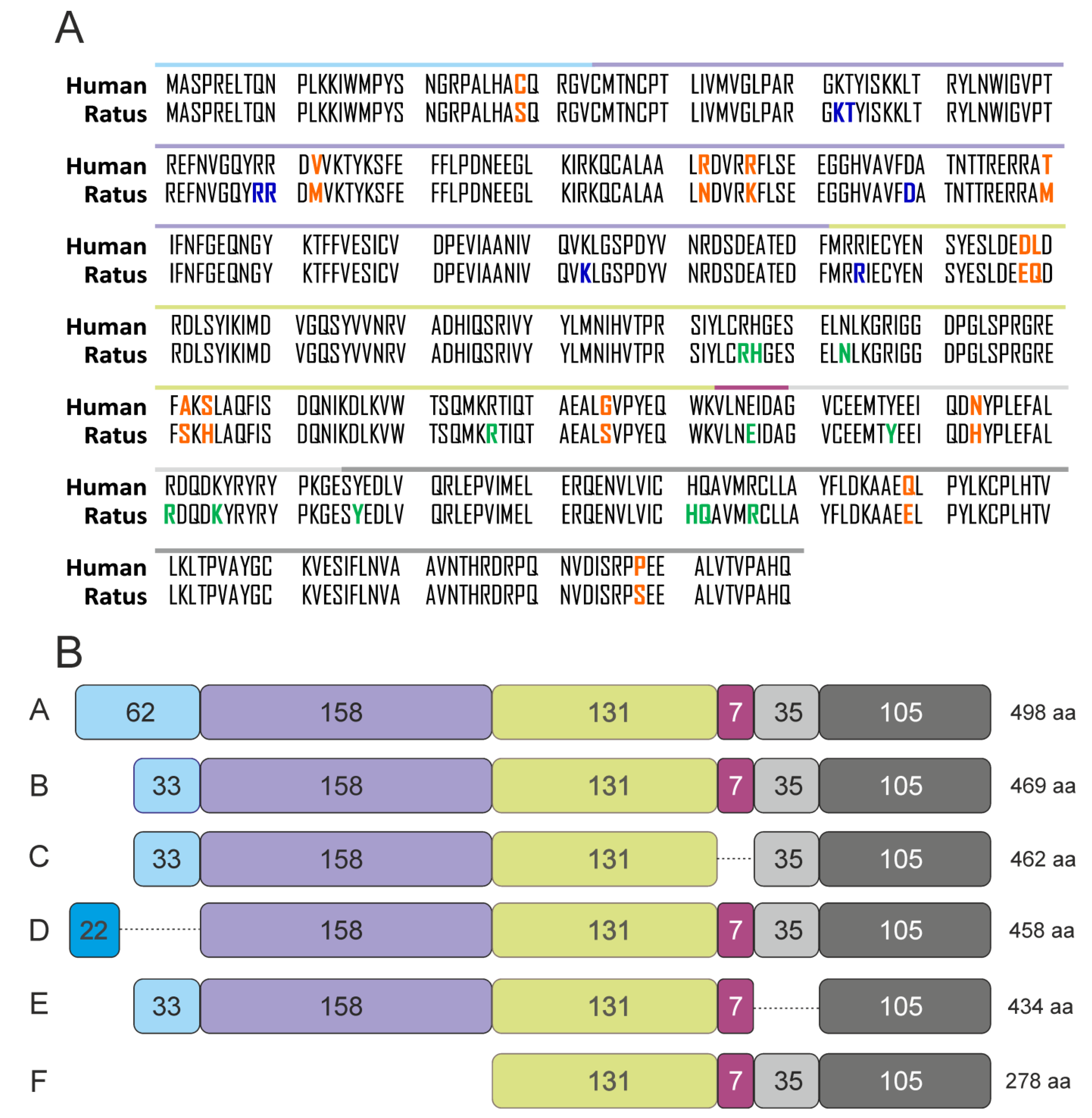
2.2. The Differences in Structure and Expression of Six Isoforms of PFKFB4 Isoenzyme
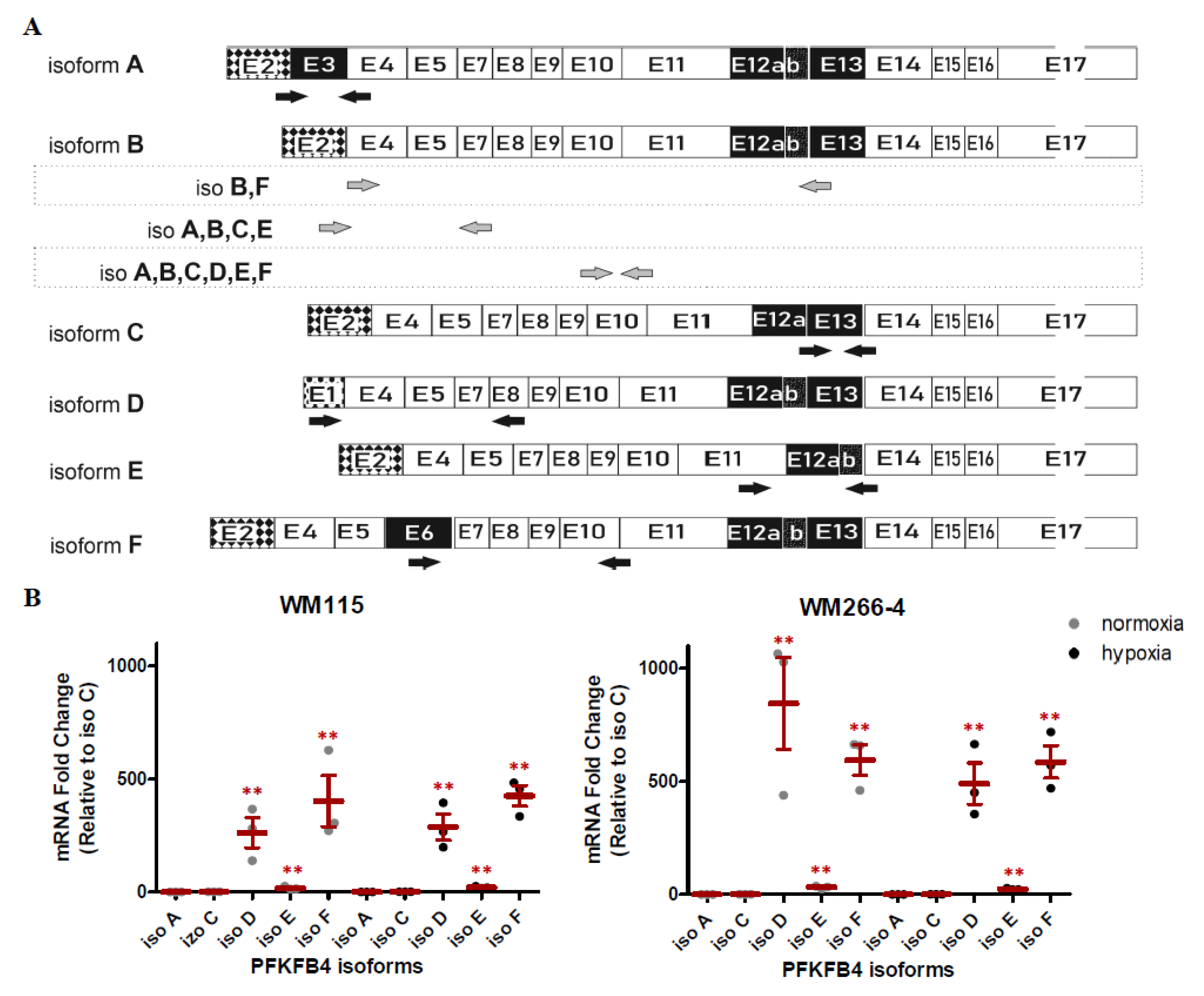
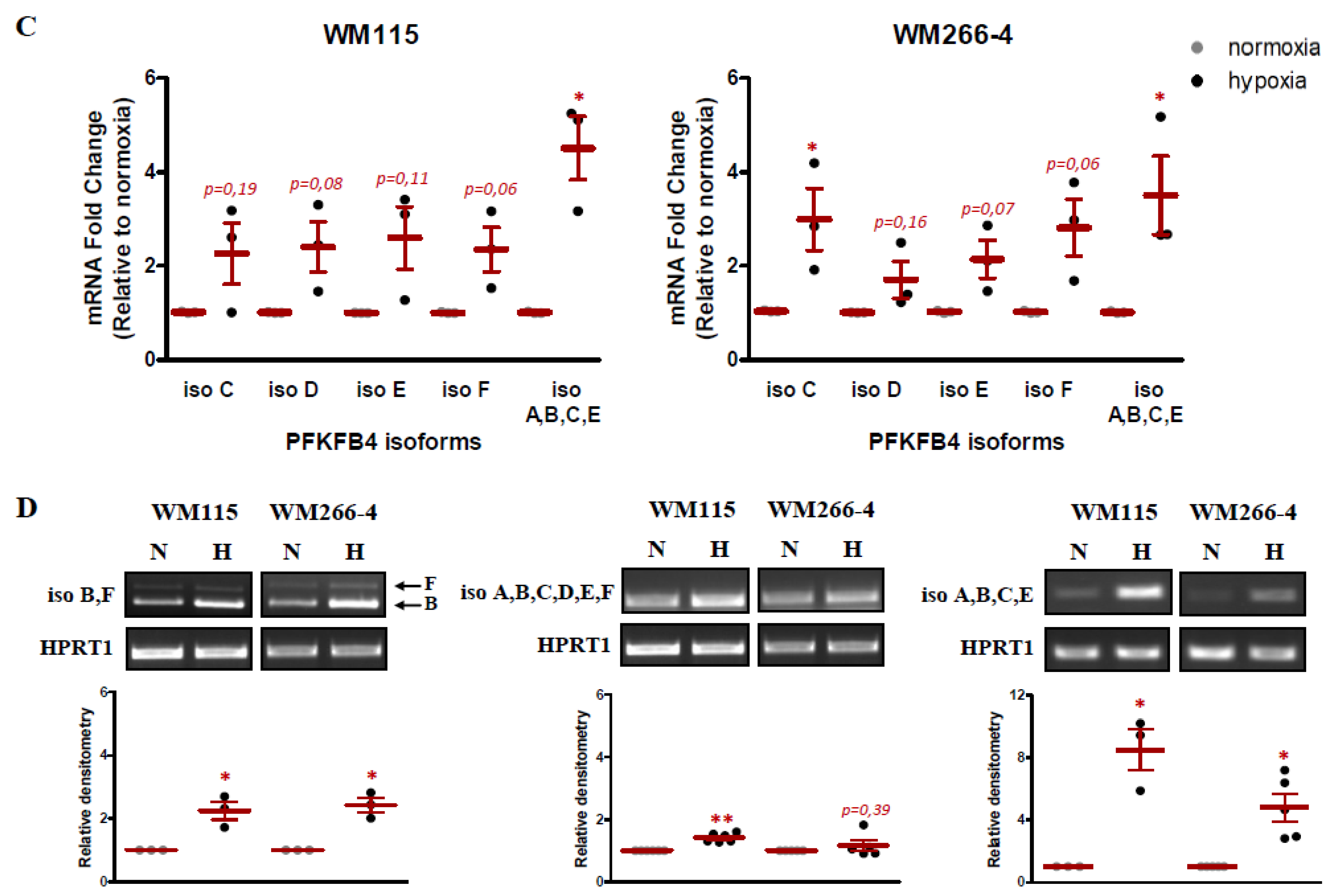
2.3. The Effect of Hypoxia on PFKFB4 Isoforms Expression in Melanoma Cell Lines
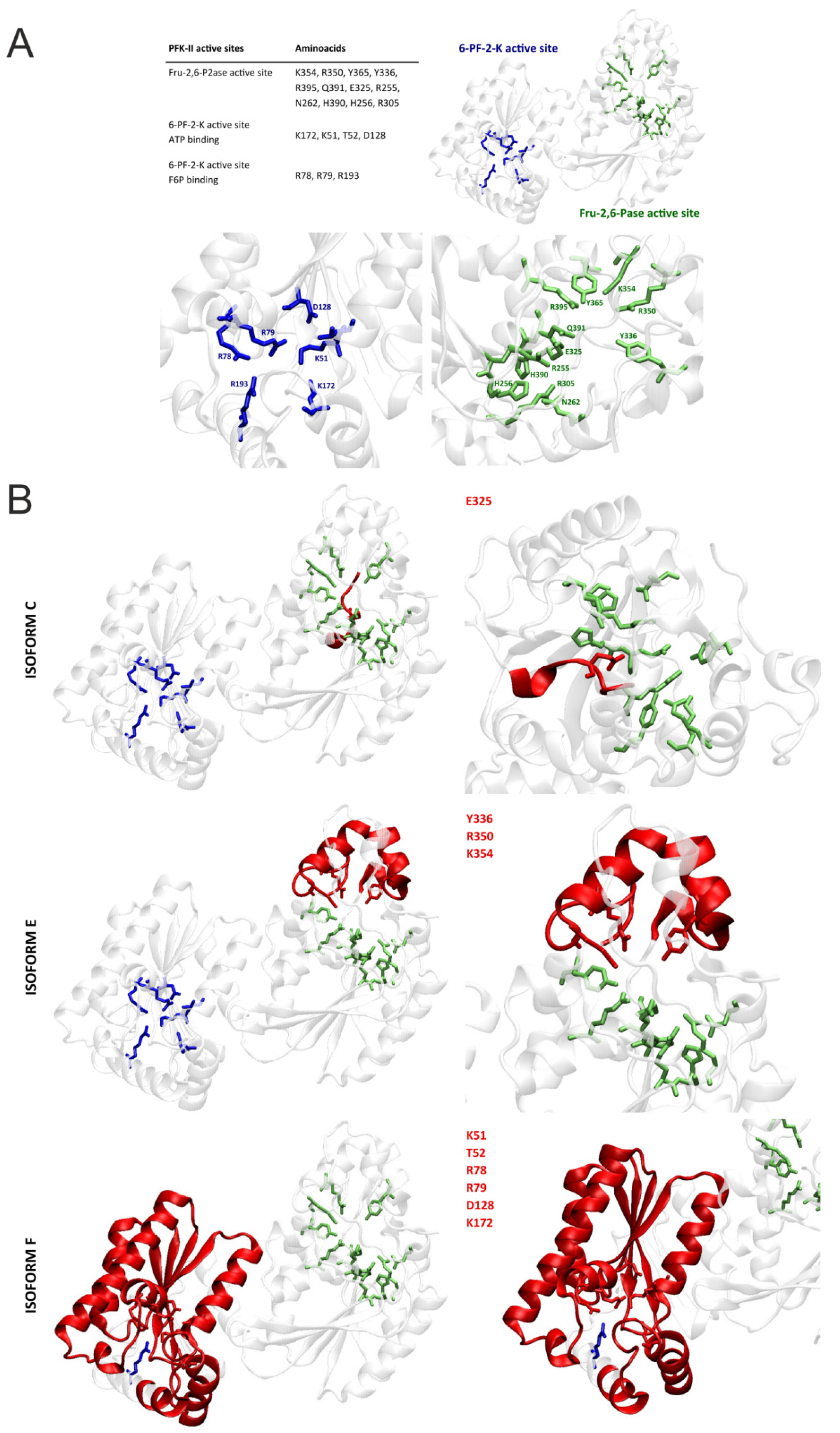
2.4. The Effect of Alternative Splicing on PFKFB4 Protein Sequence
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Cultures
4.2. RNA Isolation and cDNA Synthesis
4.3. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.4. Real-Time qPCR
4.5. Western Blot
4.6. VMD: Visual Molecular Dynamics
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rastrelli, M.; Tropea, S.; Rossi, C.R.; Alaibac, M. Melanoma: Epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo 2014, 28, 1005–1011. [Google Scholar]
- MacKie, R.M.; Hauschild, A.; Eggermont, A.M.M. Epidemiology of invasive cutaneous melanoma. Ann. Oncol. 2009, 20, vi1–vi7. [Google Scholar] [CrossRef] [PubMed]
- Heistein, J.B.; Acharya, U. Malignant Melanoma. In StatPearls; StatPearls Publishing; Posted on January 2021. Available online: https://pubmed.ncbi.nlm.nih.gov/29262210/ (accessed on 15 July 2021).
- Lenggenhager, D.; Curioni-Fontecedro, A.; Storz, M.; Shakhova, O.; Sommer, L.; Widmer, D.S.; Seifert, B.; Moch, H.; Dummer, R.; Mihic-Probst, D. An Aggressive Hypoxia Related Subpopulation of Melanoma Cells is TRP-2 Negative. Transl. Oncol. 2014, 7, 206–212. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ward, W.H.; Lambreton, F.; Goel, N.; Yu, J.Q.; Farma, J.M. Clinical Presentation and Staging of Melanoma. In Cutaneous Melanoma: Etiology and Therapy; Codon Publications: Brisbane, Australia, 2017; pp. 79–89. [Google Scholar]
- Perera, E.; Gnaneswaran, N.; Jennens, R.; Sinclair, R. Malignant Melanoma. Healthcare 2013, 2, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Bastian, B. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Soura, E.; Eliades, P.J.; Shannon, K.; Stratigos, A.J.; Tsao, H. Hereditary melanoma: Update on syndromes and management: Genetics of familial atypical multiple mole melanoma syndrome. J. Am. Acad. Dermatol. 2016, 74, 395–407, quiz 408–310. [Google Scholar] [CrossRef]
- Vaupel, P.; Kallinowski, F.; Okunieff, P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: A review. Cancer Res. 1989, 49, 6449–6465. [Google Scholar]
- Lartigau, E.; Randrianarivelo, H.; Avril, M.F.; Margulis, A.; Spatz, A.; Eschwège, F.; Guichard, M. Intratumoral oxygen tension in metastatic melanoma. Melanoma Res. 1997, 7, 400–406. [Google Scholar] [CrossRef]
- Evans, S.M.; Schrlau, A.E.; Chalian, A.A.; Zhang, P.; Koch, C.J. Oxygen Levels in Normal and Previously Irradiated Human Skin as Assessed by EF5 Binding. J. Investig. Dermatol. 2006, 126, 2596–2606. [Google Scholar] [CrossRef] [PubMed]
- Bedogni, B.; Powell, M.B. Hypoxia, melanocytes and melanoma—Survival and tumor development in the permissive microenvironment of the skin. Pigment. Cell Melanoma Res. 2009, 22, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Carreau, A.; El Hafny-Rahbi, B.; Matejuk, A.; Grillon, C.; Kieda, C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Winlove, C.P.; Michel, C.C. Oxygen Partial Pressure in Outer Layers of Skin of Human Finger Nail Folds. J. Physiol. 2003, 549, 855–863. [Google Scholar] [CrossRef]
- Trojan, S.; Piwowar, M.; Ostrowska, B.; Laidler, P.; Kocemba-Pilarczyk, K.A. Analysis of Malignant Melanoma Cell Lines Exposed to Hypoxia Reveals the Importance of PFKFB4 Overexpression for Disease Progression. Anticancer Res. 2018, 38, 6745–6752. [Google Scholar] [CrossRef]
- Minchenko, O.H.; Ogura, T.; Opentanova, I.L.; Minchenko, D.O.; Esumi, H. Splice isoform of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-4: Expression and hypoxic regulation. Mol. Cell. Biochem. 2005, 280, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.; Harris, A.L. Hypoxia Inducible Carbonic Anhydrase IX, Marker of Tumour: Hypoxia, Survival Pathway and Therapy Target. Cell Cycle 2004, 3, 159–162. [Google Scholar] [CrossRef]
- Kaluz, S.; Kaluzová, M.; Liao, S.-Y.; Lerman, M.; Stanbridge, E.J. Transcriptional control of the tumor- and hypoxia-marker carbonic anhydrase 9: A one transcription factor (HIF-1) show? Biochim. Biophys. Acta BBA Bioenergy 2009, 1795, 162–172. [Google Scholar] [CrossRef]
- Benej, M.; Pastorekova, S.; Pastorek, J. Carbonic anhydrase IX: Regulation and role in cancer. Subcell. Biochem. 2014, 75, 199–219. [Google Scholar] [PubMed]
- Shin, H.-J.; Rho, S.B.; Jung, D.C.; Han, I.-O.; Oh, E.-S.; Kim, J.-Y. Carbonic anhydrase IX (CA9) modulates tumor-associated cell migration and invasion. J. Cell Sci. 2011, 124, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Pastorekova, S.; Gillies, R.J. The role of carbonic anhydrase IX in cancer development: Links to hypoxia, acidosis, and beyond. Cancer Metastasis Rev. 2019, 38, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Chafe, S.C.; McDonald, P.C.; Saberi, S.; Nemirovsky, O.; Venkateswaran, G.; Burugu, S.; Gao, D.; Delaidelli, A.; Kyle, A.H.; Baker, J.H.E.; et al. Targeting Hypoxia-Induced Carbonic Anhydrase IX Enhances Immune-Checkpoint Blockade Locally and Systemically. Cancer Immunol. Res. 2019, 7, 1064–1078. [Google Scholar] [CrossRef]
- Andreucci, E.; Peppicelli, S.; Carta, F.; Brisotto, G.; Biscontin, E.; Ruzzolini, J.; Bianchini, F.; Biagioni, A.; Supuran, C.T.; Calorini, L. Carbonic anhydrase IX inhibition affects viability of cancer cells adapted to extracellular acidosis. J. Mol. Med. 2017, 95, 1341–1353. [Google Scholar] [CrossRef]
- Federici, C.; Lugini, L.; Marino, M.L.; Carta, F.; Iessi, E.; Azzarito, T.; Supuran, C.T.; Fais, S. Lansoprazole and carbonic anhydrase IX inhibitors sinergize against human melanoma cells. J. Enzym. Inhib. Med. Chem. 2016, 31, 119–125. [Google Scholar] [CrossRef]
- Hsin, M.-C.; Hsieh, Y.-H.; Hsiao, Y.-H.; Chen, P.-N.; Wang, P.-H.; Yang, S.-F. Carbonic Anhydrase IX Promotes Human Cervical Cancer Cell Motility by Regulating PFKFB4 Expression. Cancers 2021, 13, 1174. [Google Scholar] [CrossRef]
- Manzano, A.; Pérez, J.; Nadal, M.; Estivill, X.; Lange, A.; Bartrons, R. Cloning, expression and chromosomal localization of a human testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene. Gene 1999, 229, 83–89. [Google Scholar] [CrossRef]
- Chesney, J.; Clark, J.; Lanceta, L.; Trent, J.O.; Telang, S. Targeting the sugar metabolism of tumors with a first-in-class 6-phosphofructo-2-kinase (PFKFB4) inhibitor. Oncotarget 2015, 6, 18001–18011. [Google Scholar] [CrossRef] [PubMed]
- Kotowski, K.; Rosik, J.; Machaj, F.; Supplitt, S.; Wiczew, D.; Jabłońska, K.; Wiechec, E.; Ghavami, S.; Dzięgiel, P. Role of PFKFB3 and PFKFB4 in Cancer: Genetic Basis, Impact on Disease Development/Progression, and Potential as Therapeutic Targets. Cancers 2021, 13, 909. [Google Scholar] [CrossRef] [PubMed]
- Hasemann, C.A.; Istvan, E.S.; Uyeda, K.; Deisenhofer, J. The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies. Structure 1996, 4, 1017–1029. [Google Scholar] [CrossRef]
- Bartrons, R.; Simon, H.; Rodríguez-García, A.; Castaño, E.; Navarro-Sabate, A.; Manzano, A.; Martinez-Outschoorn, U. Fructose 2,6-Bisphosphate in Cancer Cell Metabolism. Front. Oncol. 2018, 8, 331. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Z.; Hoshino, A.; Zheng, H.; Morley, M.; Arany, Z.; Rabinowitz, J.D. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat. Metab. 2019, 1, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Pan, H.; Liu, Z.; Xie, J.; Han, W. Roles of PFKFB3 in cancer. Signal Transduct. Target. Ther. 2017, 2, 17044. [Google Scholar] [CrossRef]
- Ros, S.; Schulze, A. Balancing glycolytic flux: The role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 2013, 1, 8. [Google Scholar] [CrossRef]
- Chesney, J.; Clark, J.; Klarer, A.C.; Imbert-Fernandez, Y.; Lane, A.N.; Telang, S. Fructose-2,6-Bisphosphate synthesis by 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase 4 (PFKFB4) is required for the glycolytic response to hypoxia and tumor growth. Oncotarget 2014, 5, 6670–6686. [Google Scholar] [CrossRef]
- Ros, S.; Santos, C.R.; Moco, S.; Baenke, F.; Kelly, G.; Howell, M.; Zamboni, N.; Schulze, A. Functional Metabolic Screen Identifies 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 4 as an Important Regulator of Prostate Cancer Cell Survival. Cancer Discov. 2012, 2, 328–343. [Google Scholar] [CrossRef]
- Yi, M.; Ban, Y.; Tan, Y.; Xiong, W.; Li, G.; Xiang, B. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: A pair of valves for fine-tuning of glucose metabolism in human cancer. Mol. Metab. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Telang, S.; Yalcin, A.; Clem, A.L.; Bucala, R.; Lane, A.N.; Eaton, J.W.; Chesney, J. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene 2006, 25, 7225–7234. [Google Scholar] [CrossRef] [PubMed]
- Kessler, R.; Fleischer, M.; Springsguth, C.; Bigl, M.; Warnke, J.-P.; Eschrich, K. Prognostic Value of PFKFB3 to PFKFB4 mRNA Ratio in Patients With Primary Glioblastoma (IDH-Wildtype). J. Neuropathol. Exp. Neurol. 2019, 78, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Sharoyko, V.V.; Spegel, P.; Gullberg, U.; Mulder, H.; Olsson, I.; Ajore, R. The transcriptional co-repressor myeloid translocation gene 16 inhibits glycolysis and stimulates mitochondrial respiration. PLoS ONE 2013, 8, e68502. [Google Scholar] [CrossRef]
- Minchenko, O.; Opentanova, I.; Minchenko, D.; Ogura, T.; Esumi, H. Hypoxia induces transcription of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase-4 gene via hypoxia-inducible factor-1α activation. FEBS Lett. 2004, 576, 14–20. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, C.; Fang, M.; Yan, W.; Chen, M.; Ji, Y.; He, S.; Liu, T.; Chen, T.; Xiao, J. HIF-1α activates hypoxia-induced PFKFB4 expression in human bladder cancer cells. Biochem. Biophys. Res. Commun. 2016, 476, 146–152. [Google Scholar] [CrossRef]
- Dasgupta, S.; Rajapakshe, K.; Zhu, B.; Nikolai, B.; Yi, P.; Putluri, N.; Choi, J.M.; Jung, S.Y.; Coarfa, C.; Westbrook, T.F.; et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature 2018, 556, 249–254. [Google Scholar] [CrossRef]
- Wang, Q.; Zeng, F.; Sun, Y.; Qiu, Q.; Zhang, J.; Huang, W.; Huang, J.; Huang, X.; Guo, L. Etk Interaction with PFKFB4 Modulates Chemoresistance of Small-cell Lung Cancer by Regulating Autophagy. Clin. Cancer Res. 2017, 24, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Strohecker, A.M.; Joshi, S.P.; Possemato, R.; Abraham, R.T.; Sabatini, D.M.; White, E. Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel autophagy regulator by high content shRNA screening. Oncogene 2015, 34, 5662–5676. [Google Scholar] [CrossRef] [PubMed]
- Sittewelle, M.; Kappès, V.; Lécuyer, D.; Monsoro-Burq, A.H. The glycolysis regulator PFKFB4 interacts with ICMT and activates RAS/AKT signaling-dependent cell migration in melanoma. Preprint 2021. [Google Scholar] [CrossRef]
- Kocemba-Pilarczyk, K.A.; Ostrowska, B.; Trojan, S.; Aslan, E.; Kusior, D.; Lasota, M.; Lenouvel, C.; Dulińska-Litewka, J. Targeting the hypoxia pathway in malignant plasma cells by using 17-allylamino-17-demethoxygeldanamycin. Acta Biochim. Pol. 2018, 65, 101–109. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Kocemba-Pilarczyk, K.A.; Trojan, S.; Ostrowska, B.; Lasota, M.; Dudzik, P.; Kusior, D.; Kot, M. Influence of metformin on HIF-1 pathway in multiple myeloma. Pharmacol. Rep. 2020, 72, 1407–1417. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
| PFKFB4 Isoform | NCBI Gene ID | Forward and Reverse Primers | Product Length |
|---|---|---|---|
| Isoform A | NM_001317134.2 | F: CAGCGCGGTGAACTTTCAAA | 168 bp |
| R: GTCATGCACAATGTCCCGGG | |||
| Isoform C | NM_001317135.2 | F: CAGTGGAAGGGCGTCTGTG | 126 bp |
| R: GTCCTCGTAGGACTCCCCTT | |||
| Isoform D | NM_001317136.2 | F: TGGACAGAGGCTCGTTAGGA | 383 bp |
| R: CGCTCTCCGTTCTCGGGTG | |||
| Isoform E | NM_001317137.2 | F: TCAAGATCATGGATGTGGGC | 351 bp |
| R: CCTCGTAGGACGCATCGATC | |||
| Isoform F | NM_001317138.2 | F: AATCTAAGCCCATCCACCGC | 386 bp |
| R: GGACAGGTCCCTATCCAGGT | |||
| Isoforms A, B, C, D, E, F | F: TCAAGATCATGGATGTGGGC | 214 bp | |
| R: TCTTGGCAAACTCCCTGC | |||
| Isoforms A, B, C, E | F: CAGCGCGGTGTGTGCATGAC | 233 bp | |
| R: GCACACTGCTTCCTGATTTT | |||
| Isoforms B and F | NM_004567.4 | F: CAGCGCGGTGTGTGCATGAC | Iso B 911 bp |
| NM_001317138.2 | R: TCACAGACGCCCGCATCGATC | Iso F 1039 bp |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trojan, S.E.; Dudzik, P.; Totoń-Żurańska, J.; Laidler, P.; Kocemba-Pilarczyk, K.A. Expression of Alternative Splice Variants of 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase-4 in Normoxic and Hypoxic Melanoma Cells. Int. J. Mol. Sci. 2021, 22, 8848. https://doi.org/10.3390/ijms22168848
Trojan SE, Dudzik P, Totoń-Żurańska J, Laidler P, Kocemba-Pilarczyk KA. Expression of Alternative Splice Variants of 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase-4 in Normoxic and Hypoxic Melanoma Cells. International Journal of Molecular Sciences. 2021; 22(16):8848. https://doi.org/10.3390/ijms22168848
Chicago/Turabian StyleTrojan, Sonia E., Paulina Dudzik, Justyna Totoń-Żurańska, Piotr Laidler, and Kinga A. Kocemba-Pilarczyk. 2021. "Expression of Alternative Splice Variants of 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase-4 in Normoxic and Hypoxic Melanoma Cells" International Journal of Molecular Sciences 22, no. 16: 8848. https://doi.org/10.3390/ijms22168848
APA StyleTrojan, S. E., Dudzik, P., Totoń-Żurańska, J., Laidler, P., & Kocemba-Pilarczyk, K. A. (2021). Expression of Alternative Splice Variants of 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase-4 in Normoxic and Hypoxic Melanoma Cells. International Journal of Molecular Sciences, 22(16), 8848. https://doi.org/10.3390/ijms22168848






