Complementary Functions of Plant AP Endonucleases and AP Lyases during DNA Repair of Abasic Sites Arising from C:G Base Pairs
Abstract
:1. Introduction
2. Results
2.1. Design of DNA Substrates
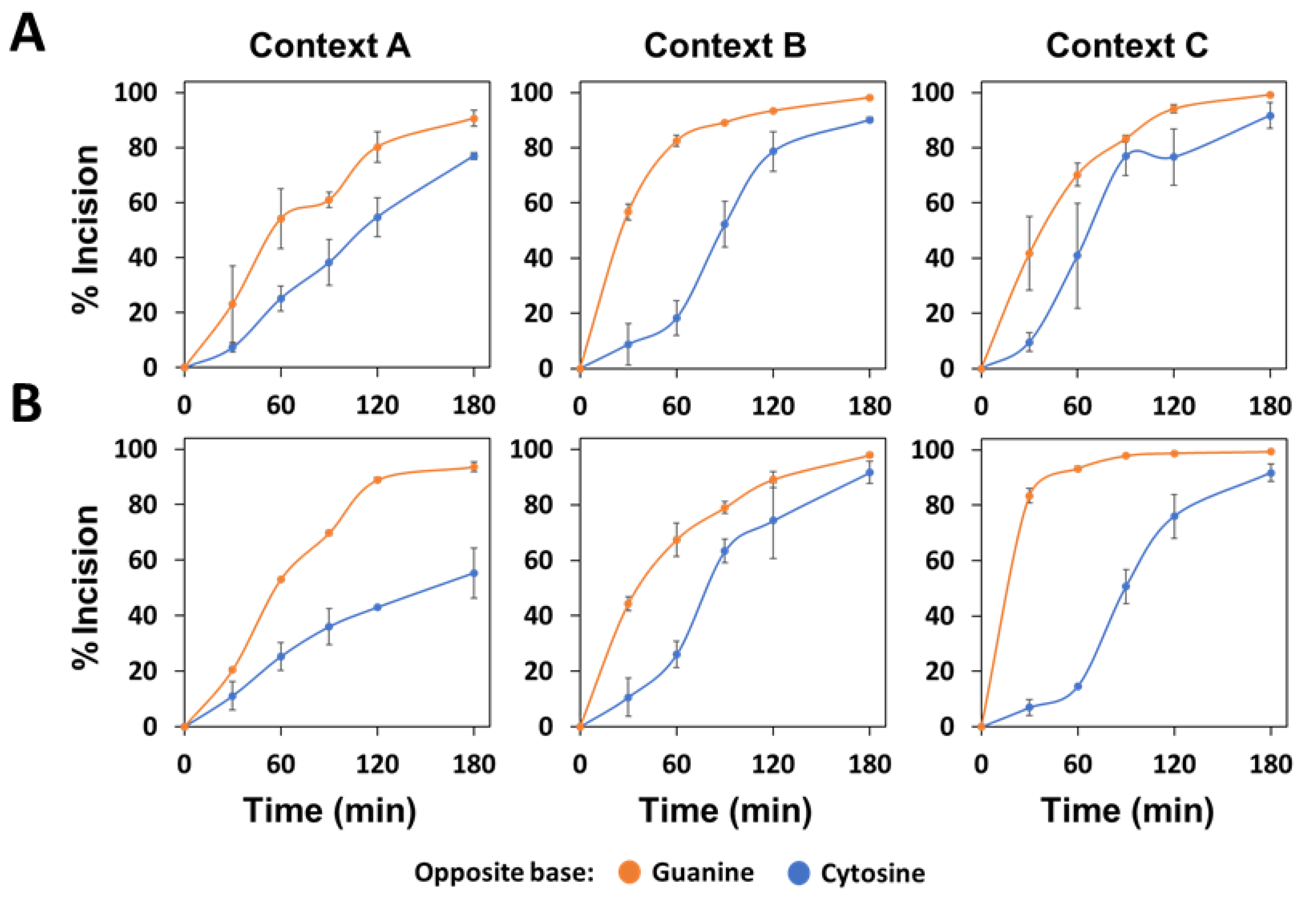
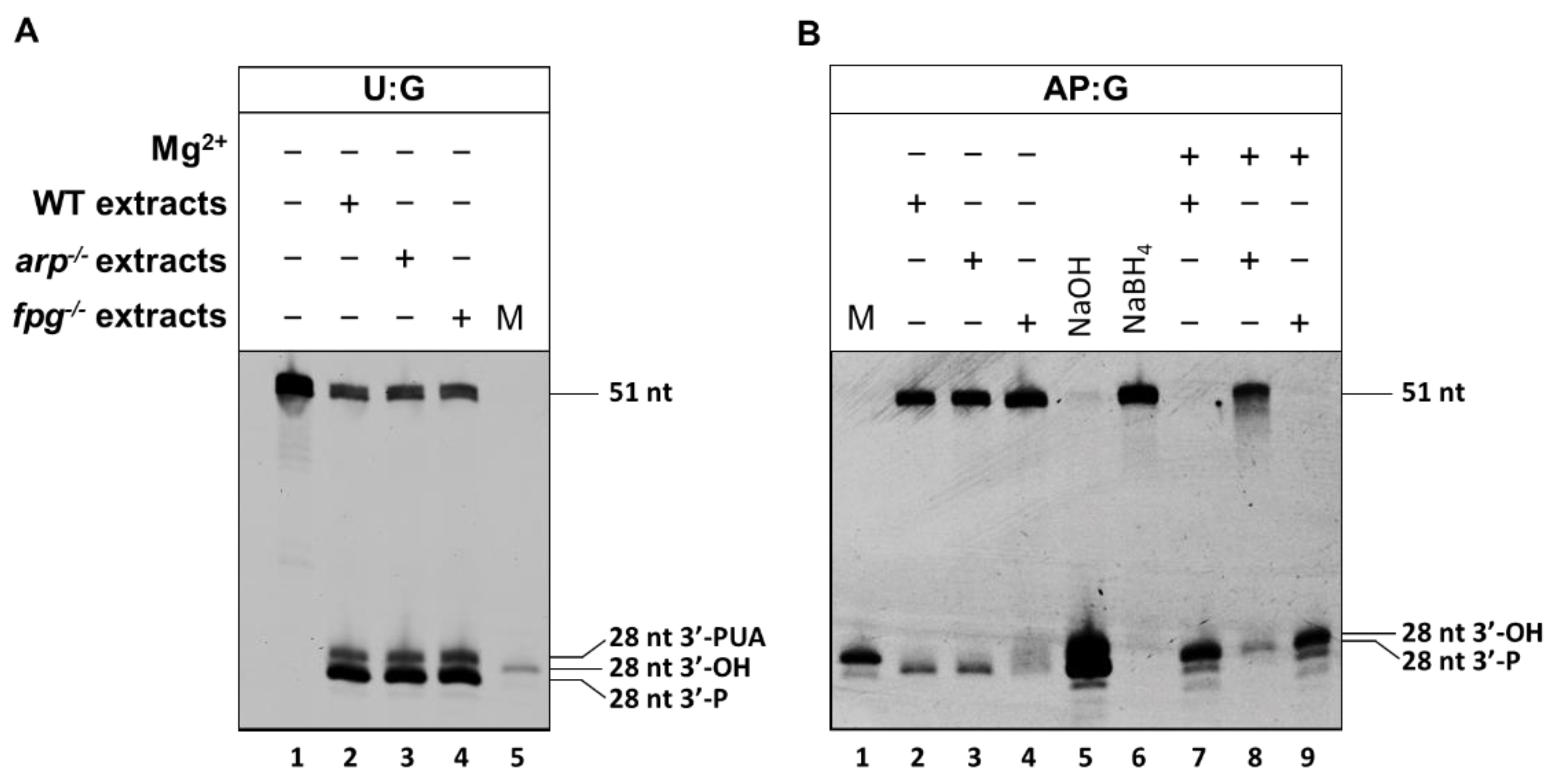
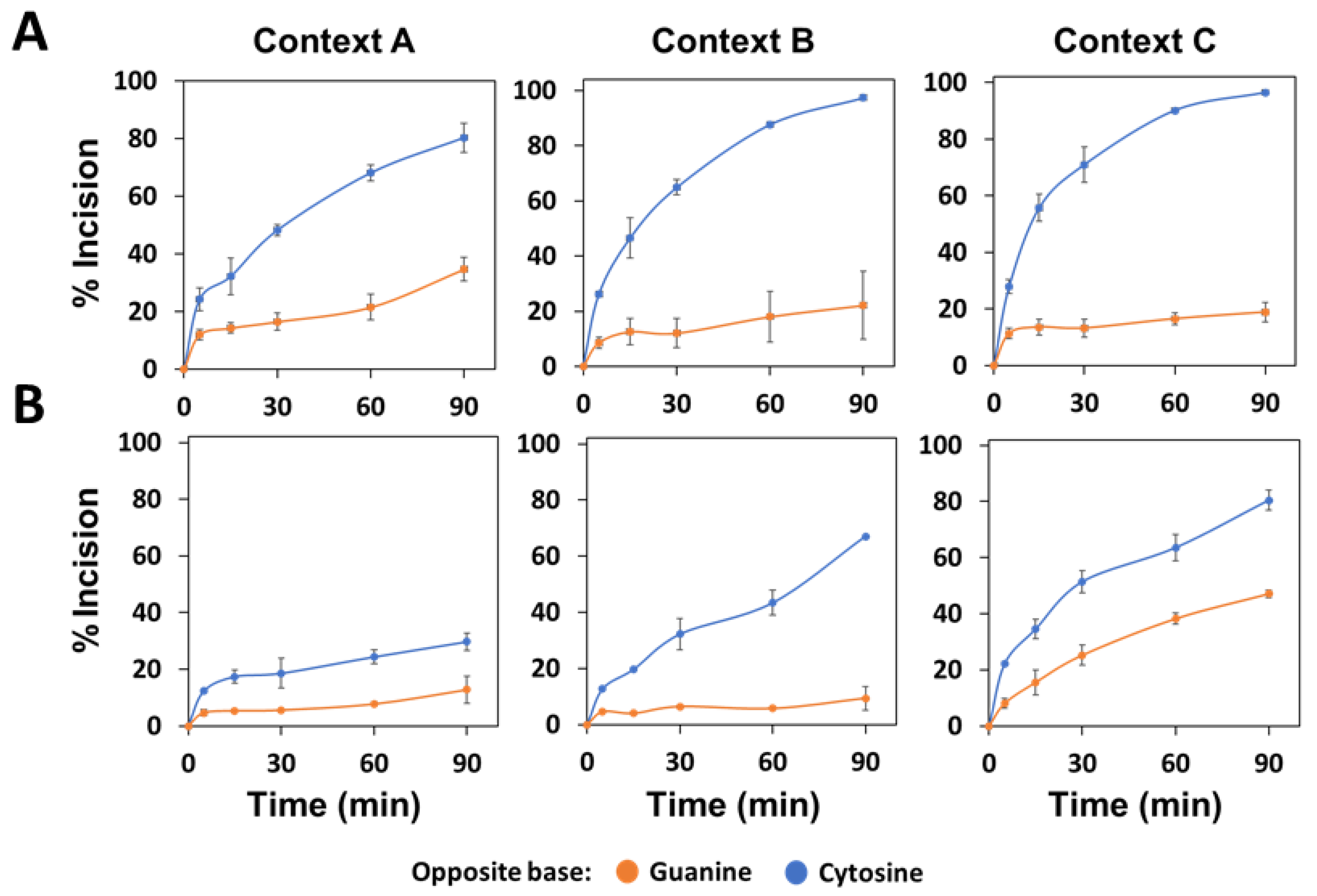
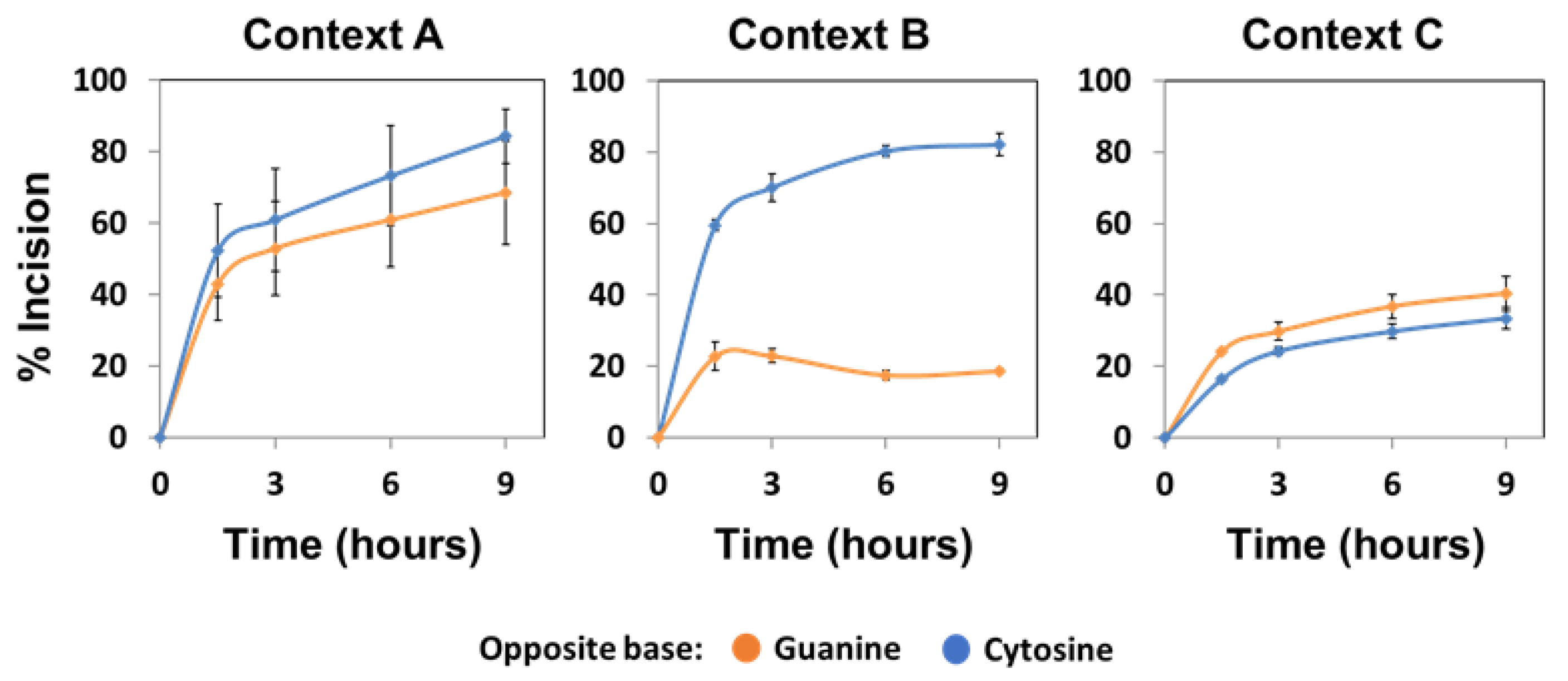
2.2. The Main Arabidopsis AP Endonuclease, ARP, Prefers G to C as the Base Opposite the Abasic Site
2.3. The Main Arabidopsis AP Lyase, FPG, Prefers G to C as the Base Opposite the Abasic Site
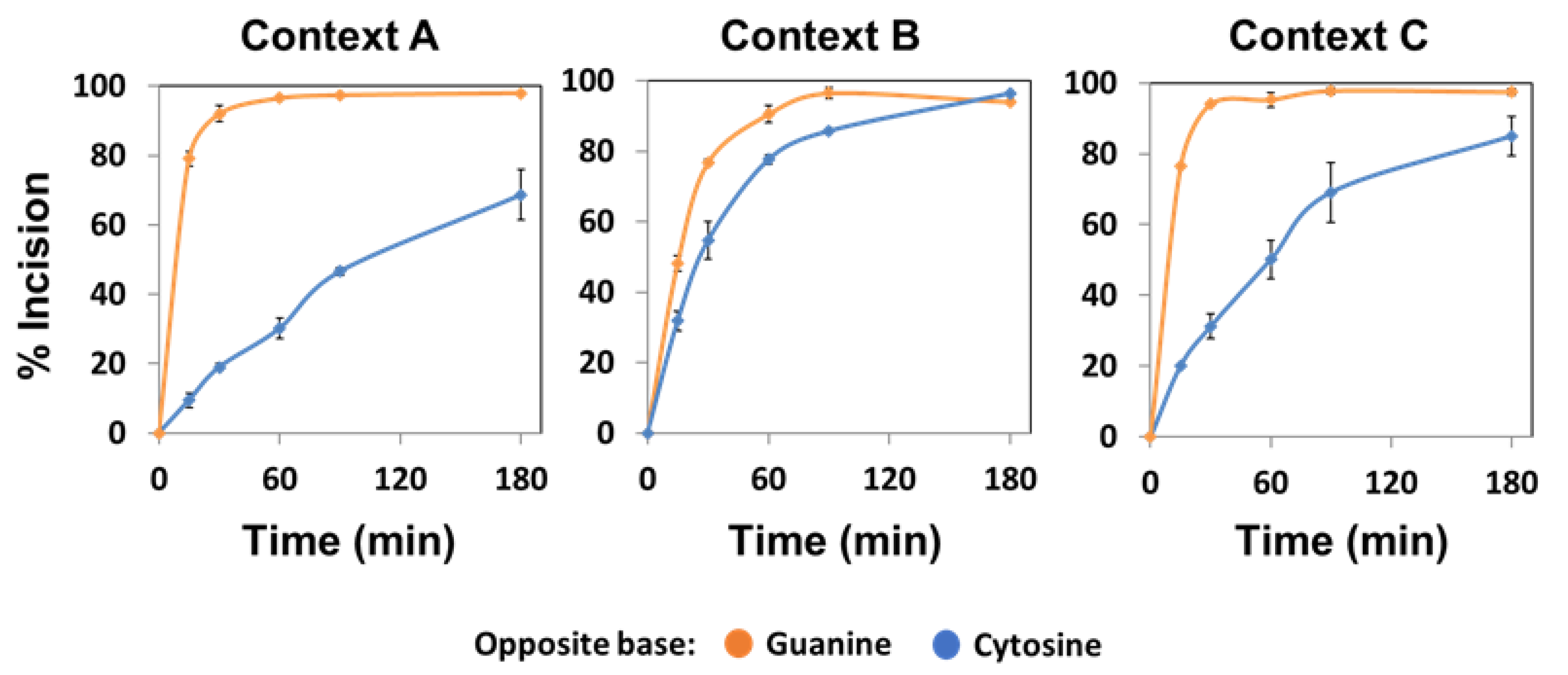
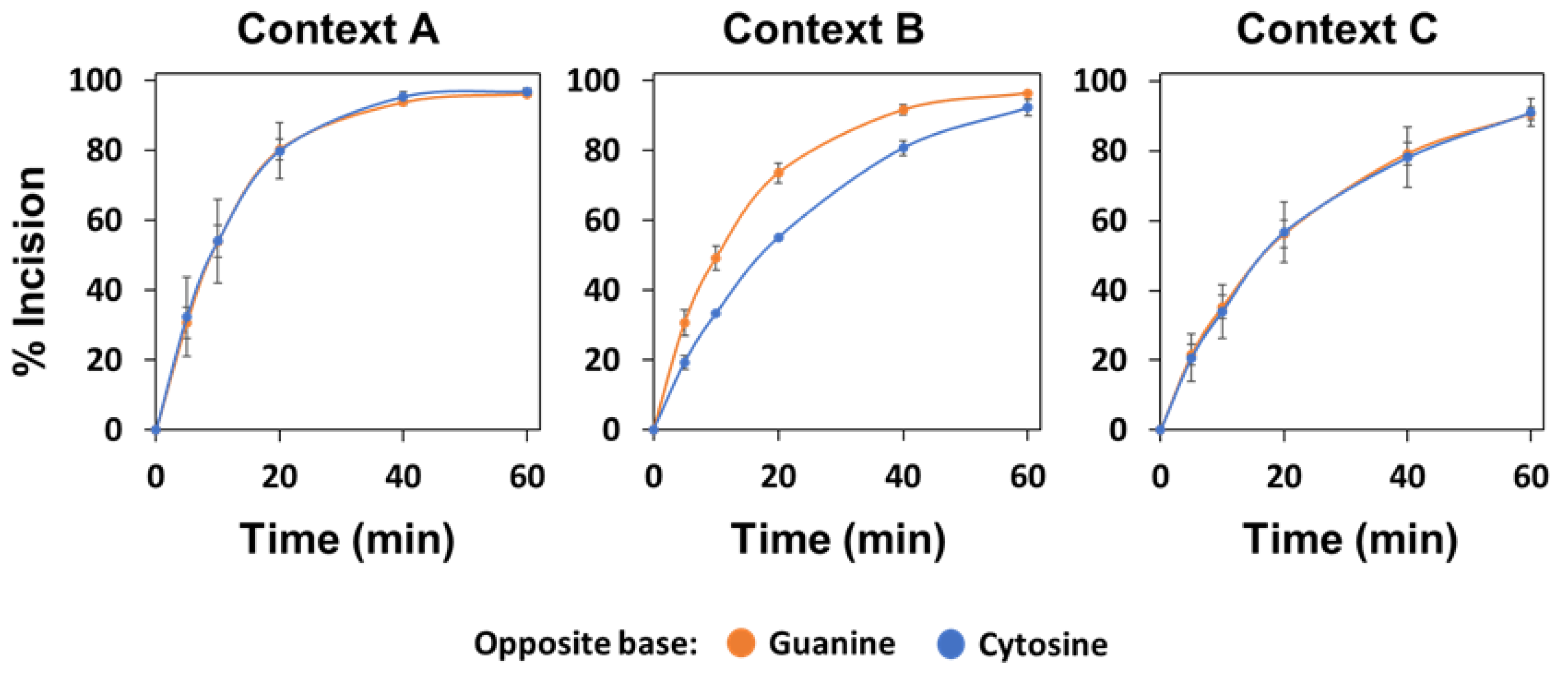
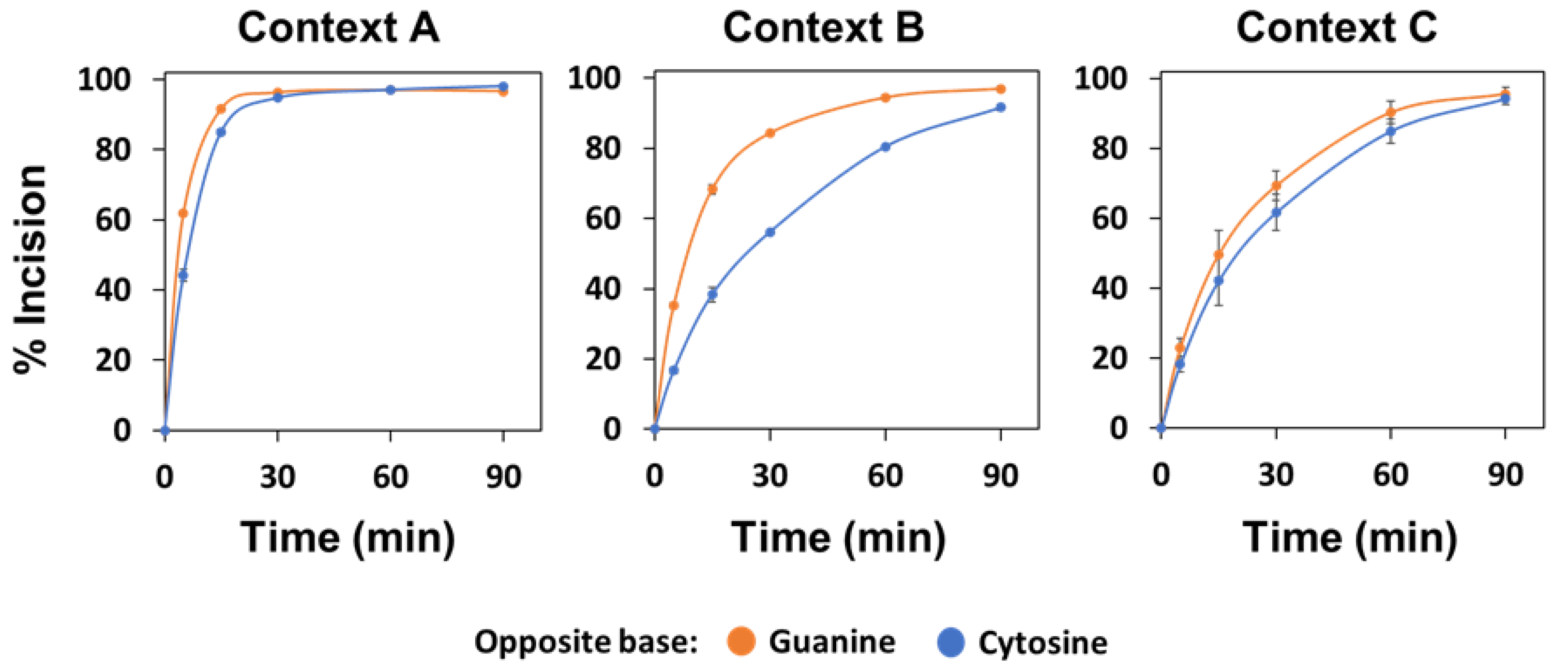
2.4. The Major Human AP Endonuclease, APE1, Exhibits a Preference for G as the Orphan Base, but Not in All Sequence Contexts
3. Discussion
4. Materials and Methods
4.1. Plant Material and Cell Extract Preparation
4.2. Cell Culture and Cell Extract Preparation
4.3. Protein Expression and Purification
4.4. Reagents and Enzymes
4.5. DNA Substrates
4.6. DNA Incision Assay
4.7. Kinetic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Strauss, B.; Hill, T. The intermediate in the degradation of DNA alkylated with a monofunctional alkylating agent. Biochim. Biophys. Acta 1970, 213, 14–25. [Google Scholar] [CrossRef]
- Talpaert-Borle, M. Formation, detection and repair of AP sites. Mutat. Res. 1987, 181, 45–56. [Google Scholar] [CrossRef]
- Gates, K.S.; Nooner, T.; Dutta, S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem. Res. Toxicol. 2004, 17, 839–856. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog. Nucleic Acid Res. Mol. Biol. 1979, 22, 135–192. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjoras, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Roldán-Arjona, T.; Ariza, R.R.; Cordoba-Canero, D. DNA Base Excision Repair in Plants: An Unfolding Story with Familiar and Novel Characters. Front. Plant Sci. 2019, 10, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Córdoba-Cañero, D.; Ariza, R.R.; Roldán-Arjona, T. Base Excision Repair in Plants: Variations on a Theme. In DNA Damage, DNA Repair and Disease, 1st ed.; Dizdaroglu, M., Lloyd, R.S., Eds.; Royal Society of Chemistry: London, UK, 2020; Volume 2, pp. 48–74. [Google Scholar] [CrossRef]
- Nakamura, J.; Swenberg, J.A. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999, 59, 2522–2526. [Google Scholar]
- Beger, R.D.; Bolton, P.H. Structures of apurinic and apyrimidinic sites in duplex DNAs. J. Biol. Chem. 1998, 273, 15565–15573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lhomme, J.; Constant, J.F.; Demeunynck, M. Abasic DNA structure, reactivity, and recognition. Biopolymers 1999, 52, 65–83. [Google Scholar] [CrossRef]
- Loeb, L.A.; Preston, B.D. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986, 20, 201–230. [Google Scholar] [CrossRef] [PubMed]
- Boiteux, S.; Guillet, M. Abasic sites in DNA: Repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 2004, 3, 1–12. [Google Scholar] [CrossRef]
- Demple, B.; Harrison, L. Repair of oxidative damage to DNA: Enzymology and biology. Annu. Rev. Biochem. 1994, 63, 915–948. [Google Scholar] [CrossRef] [PubMed]
- Seeberg, E.; Luna, L.; Morland, I.; Eide, L.; Johnsen, B.; Larsen, E.; Alseth, I.; Dantzer, F.; Baynton, K.; Aamodt, R.; et al. Base removers and strand scissors: Different strategies employed in base excision and strand incision at modified base residues in DNA. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 135–142. [Google Scholar] [CrossRef]
- Kitsera, N.; Rodriguez-Alvarez, M.; Emmert, S.; Carell, T.; Khobta, A. Nucleotide excision repair of abasic DNA lesions. Nucleic Acids Res. 2019, 47, 8537–8547. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.D.; Demple, B. Analysis of class II (hydrolytic) and class I (beta-lyase) apurinic/apyrimidinic endonucleases with a synthetic DNA substrate. Nucleic Acids Res. 1990, 18, 5069–5075. [Google Scholar] [CrossRef] [Green Version]
- Krokan, H.E.; Standal, R.; Slupphaug, G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997, 325, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Swanson, R.L.; Morey, N.J.; Doetsch, P.W.; Jinks-Robertson, S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell Biol. 1999, 19, 2929–2935. [Google Scholar] [CrossRef] [Green Version]
- Dianov, G.L.; Sleeth, K.M.; Dianova, I.I.; Allinson, S.L. Repair of abasic sites in DNA. Mutat. Res. 2003, 531, 157–163. [Google Scholar] [CrossRef]
- Alseth, I.; Korvald, H.; Osman, F.; Seeberg, E.; Bjoras, M. A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2004, 32, 5119–5125. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, T.; Igawa, E.; Tanihigashi, H.; Matsubara, M.; Ide, H.; Ikeda, S. Roles of base excision repair enzymes Nth1p and Apn2p from Schizosaccharomyces pombe in processing alkylation and oxidative DNA damage. DNA Repair 2005, 4, 1270–1280. [Google Scholar] [CrossRef]
- Hanna, M.; Chow, B.L.; Morey, N.J.; Jinks-Robertson, S.; Doetsch, P.W.; Xiao, W. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair 2004, 3, 51–59. [Google Scholar] [CrossRef]
- Maher, R.L.; Wallace, S.S.; Pederson, D.S. The lyase activity of bifunctional DNA glycosylases and the 3′-diesterase activity of APE1 contribute to the repair of oxidized bases in nucleosomes. Nucleic Acids Res. 2019, 47, 2922–2931. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Chapman, J.H.; Custance, M.F.; Tricola, G.M.; Jones, C.E.; Furano, A.V. Perturbation of base excision repair sensitizes breast cancer cells to APOBEC3 deaminase-mediated mutations. Elife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Barbado, C.; Córdoba-Cañero, D.; Ariza, R.R.; Roldán-Arjona, T. Nonenzymatic release of N7-methylguanine channels repair of abasic sites into an AP endonuclease-independent pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E916–E924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Córdoba-Cañero, D.; Dubois, E.; Ariza, R.R.; Doutriaux, M.P.; Roldán-Arjona, T. Arabidopsis uracil DNA glycosylase (UNG) is required for base excision repair of uracil and increases plant sensitivity to 5-fluorouracil. J. Biol. Chem. 2010, 285, 7475–7483. [Google Scholar] [CrossRef] [Green Version]
- Ramiro-Merina, A.; Ariza, R.R.; Roldán-Arjona, T. Molecular characterization of a putative plant homolog of MBD4 DNA glycosylase. DNA Repair 2013, 12, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Morales-Ruiz, T.; Ortega-Galisteo, A.P.; Ponferrada-Marin, M.I.; Martinez-Macias, M.I.; Ariza, R.R.; Roldán-Arjona, T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. USA 2006, 103, 6853–6858. [Google Scholar] [CrossRef] [Green Version]
- Ponferrada-Marin, M.I.; Roldán-Arjona, T.; Ariza, R.R. ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 2009, 37, 4264–4274. [Google Scholar] [CrossRef]
- Lister, R.; O′Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef] [Green Version]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [Green Version]
- Wilson, D.M.; Barsky, D. The major human abasic endonuclease: Formation, consequences and repair of abasic lesions in DNA. Mutat. Res. DNA Repair 2001, 485, 283–307. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schar, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donigan, K.A.; Sweasy, J.B. Sequence context-specific mutagenesis and base excision repair. Mol. Carcinog. 2009, 48, 362–368. [Google Scholar] [CrossRef] [Green Version]
- Murphy, T.M.; Gao, M.J. Multiple forms of formamidopyrimidine-DNA glycosylase produced by alternative splicing in Arabidopsis thaliana. J. Photochem. Photobiol. B 2001, 61, 87–93. [Google Scholar] [CrossRef]
- Boiteux, S.; Coste, F.; Castaing, B. Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic. Biol. Med. 2017, 107, 179–201. [Google Scholar] [CrossRef] [PubMed]
- Córdoba-Cañero, D.; Roldán-Arjona, T.; Ariza, R.R. Arabidopsis ZDP DNA 3′-phosphatase and ARP endonuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant. J. 2014, 79, 824–834. [Google Scholar] [CrossRef] [Green Version]
- Castaing, B.; Geiger, A.; Seliger, H.; Nehls, P.; Laval, J.; Zelwer, C.; Boiteux, S. Cleavage and binding of a DNA fragment containing a single 8-oxoguanine by wild type and mutant FPG proteins. Nucleic Acids Res. 1993, 21, 2899–2905. [Google Scholar] [CrossRef] [Green Version]
- Tchou, J.; Bodepudi, V.; Shibutani, S.; Antoshechkin, I.; Miller, J.; Grollman, A.P.; Johnson, F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J. Biol. Chem. 1994, 269, 15318–15324. [Google Scholar] [CrossRef]
- Alexeeva, M.; Guragain, P.; Tesfahun, A.N.; Tomkuviene, M.; Arshad, A.; Gerasimaite, R.; Ruksenaite, A.; Urbanaviciute, G.; Bjoras, M.; Laerdahl, J.K.; et al. Excision of the doubly methylated base N(4),5-dimethylcytosine from DNA by Escherichia coli Nei and Fpg proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [Green Version]
- Eide, L.; Luna, L.; Gustad, E.C.; Henderson, P.T.; Essigmann, J.M.; Demple, B.; Seeberg, E. Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry 2001, 40, 6653–6659. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.M.; Takeshita, M.; Grollman, A.P.; Demple, B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 1995, 270, 16002–16007. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Svilar, D.; McClellan, S.; Kim, J.H.; Ahn, E.E.; Vens, C.; Wilson, D.M., 3rd; Sobol, R.W. DNA Repair Molecular Beacon assay: A platform for real-time functional analysis of cellular DNA repair capacity. Oncotarget 2018, 9, 31719–31743. [Google Scholar] [CrossRef]
- Visnes, T.; Akbari, M.; Hagen, L.; Slupphaug, G.; Krokan, H.E. The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair 2008, 7, 1869–1881. [Google Scholar] [CrossRef] [Green Version]
- Serre, L.; Pereira de Jesus, K.; Boiteux, S.; Zelwer, C.; Castaing, B. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002, 21, 2854–2865. [Google Scholar] [CrossRef] [Green Version]
- Manoharan, M.; Ransom, S.C.; Mazumder, A.; Gerlt, J.A.; Wilde, J.A.; Withka, J.A.; Bolton, P.H. The characterization of abasic sites in DNA heteroduplexes by site specific labeling with carbon-13. J. Am. Chem. Soc. 1988, 110, 1620–1622. [Google Scholar] [CrossRef]
- Córdoba-Cañero, D.; Roldán-Arjona, T.; Ariza, R.R. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011, 68, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Córdoba-Cañero, D.; Morales-Ruiz, T.; Roldán-Arjona, T.; Ariza, R.R. Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 2009, 60, 716–728. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Li, Y.; Córdoba-Cañero, D.; Qian, W.; Zhu, X.; Tang, K.; Zhang, H.; Ariza, R.R.; Roldán-Arjona, T.; Zhu, J.K. An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis [corrected]. PLoS Genet. 2015, 11, e1004905. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, U.; Bentele, M.; Jiricny, J.; Schär, P. The versatile thymine DNA-glycosylase: A comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003, 31, 2261–2271. [Google Scholar] [CrossRef] [PubMed]
- Ponferrada-Marin, M.I.; Roldán-Arjona, T.; Ariza, R.R. Demethylation initiated by ROS1 glycosylase involves random sliding along DNA. Nucleic Acids Res. 2012, 40, 11554–11562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
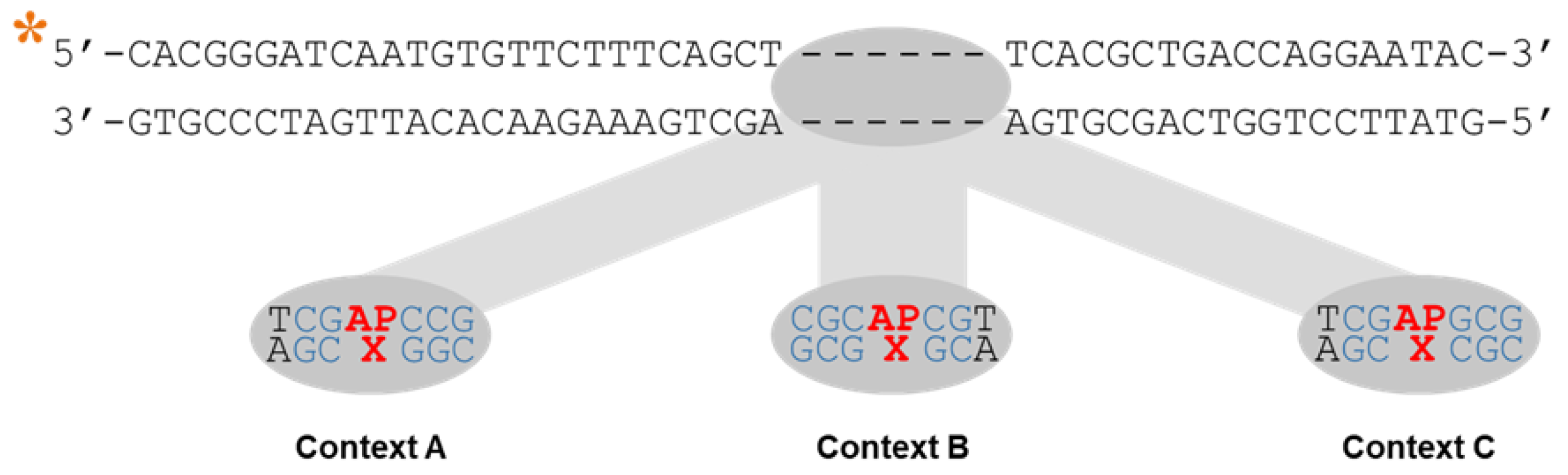

| Name a | DNA Sequence b | Strand c |
|---|---|---|
| Fl_UGCG-F | CACGGGATCAATGTGTTCTTTCAGCTCGUGCGTCACGCTGACCAGGAATAC | U |
| UGCG-RG | GTATTCCTGGTCAGCGTGACGCGCGAGCTGAAAGAACACATTGATCCCGTG | L |
| UGCG-RC | GTATTCCTGGTCAGCGTGACGCCCGAGCTGAAAGAACACATTGATCCCGTG | L |
| Fl_CUCG-F | CACGGGATCAATGTGTTCTTTCAGCTCGCUCGTCACGCTGACCAGGAATAC | U |
| CUCG-RG | GTATTCCTGGTCAGCGTGACGGGCGAGCTGAAAGAACACATTGATCCCGTG | L |
| Fl_UCCG-F | CACGGGATCAATGTGTTCTTTCAGCTCGUCCGTCACGCTGACCAGGAATAC | U |
| UCCG-RG | GTATTCCTGGTCAGCGTGACGGGCGAGCTGAAAGAACACATTGATCCCGTG | L |
| UCCG-RC | GTATTCCTGGTCAGCGTGACGGCCGAGCTGAAAGAACACATTGATCCCGTG | L |
| Fl-UGF | TCACGGGATCAATGTGTTCTTTCAGCTCUGGTCACGCTGACCAGGAATACC | U |
| CGR_G | GGTATTCCTGGTCAGCGTGACCGGAGCTGAAAGAACACATTGATCCCGTGA | L |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jordano-Raya, M.; Beltrán-Melero, C.; Moreno-Recio, M.D.; Martínez-Macías, M.I.; Ariza, R.R.; Roldán-Arjona, T.; Córdoba-Cañero, D. Complementary Functions of Plant AP Endonucleases and AP Lyases during DNA Repair of Abasic Sites Arising from C:G Base Pairs. Int. J. Mol. Sci. 2021, 22, 8763. https://doi.org/10.3390/ijms22168763
Jordano-Raya M, Beltrán-Melero C, Moreno-Recio MD, Martínez-Macías MI, Ariza RR, Roldán-Arjona T, Córdoba-Cañero D. Complementary Functions of Plant AP Endonucleases and AP Lyases during DNA Repair of Abasic Sites Arising from C:G Base Pairs. International Journal of Molecular Sciences. 2021; 22(16):8763. https://doi.org/10.3390/ijms22168763
Chicago/Turabian StyleJordano-Raya, Marina, Cristina Beltrán-Melero, M. Dolores Moreno-Recio, M. Isabel Martínez-Macías, Rafael R. Ariza, Teresa Roldán-Arjona, and Dolores Córdoba-Cañero. 2021. "Complementary Functions of Plant AP Endonucleases and AP Lyases during DNA Repair of Abasic Sites Arising from C:G Base Pairs" International Journal of Molecular Sciences 22, no. 16: 8763. https://doi.org/10.3390/ijms22168763
APA StyleJordano-Raya, M., Beltrán-Melero, C., Moreno-Recio, M. D., Martínez-Macías, M. I., Ariza, R. R., Roldán-Arjona, T., & Córdoba-Cañero, D. (2021). Complementary Functions of Plant AP Endonucleases and AP Lyases during DNA Repair of Abasic Sites Arising from C:G Base Pairs. International Journal of Molecular Sciences, 22(16), 8763. https://doi.org/10.3390/ijms22168763






