Integrated Metabolomic and Transcriptomic Analysis Reveals the Flavonoid Regulatory Network by Eutrema EsMYB90
Abstract
:1. Introduction
2. Results
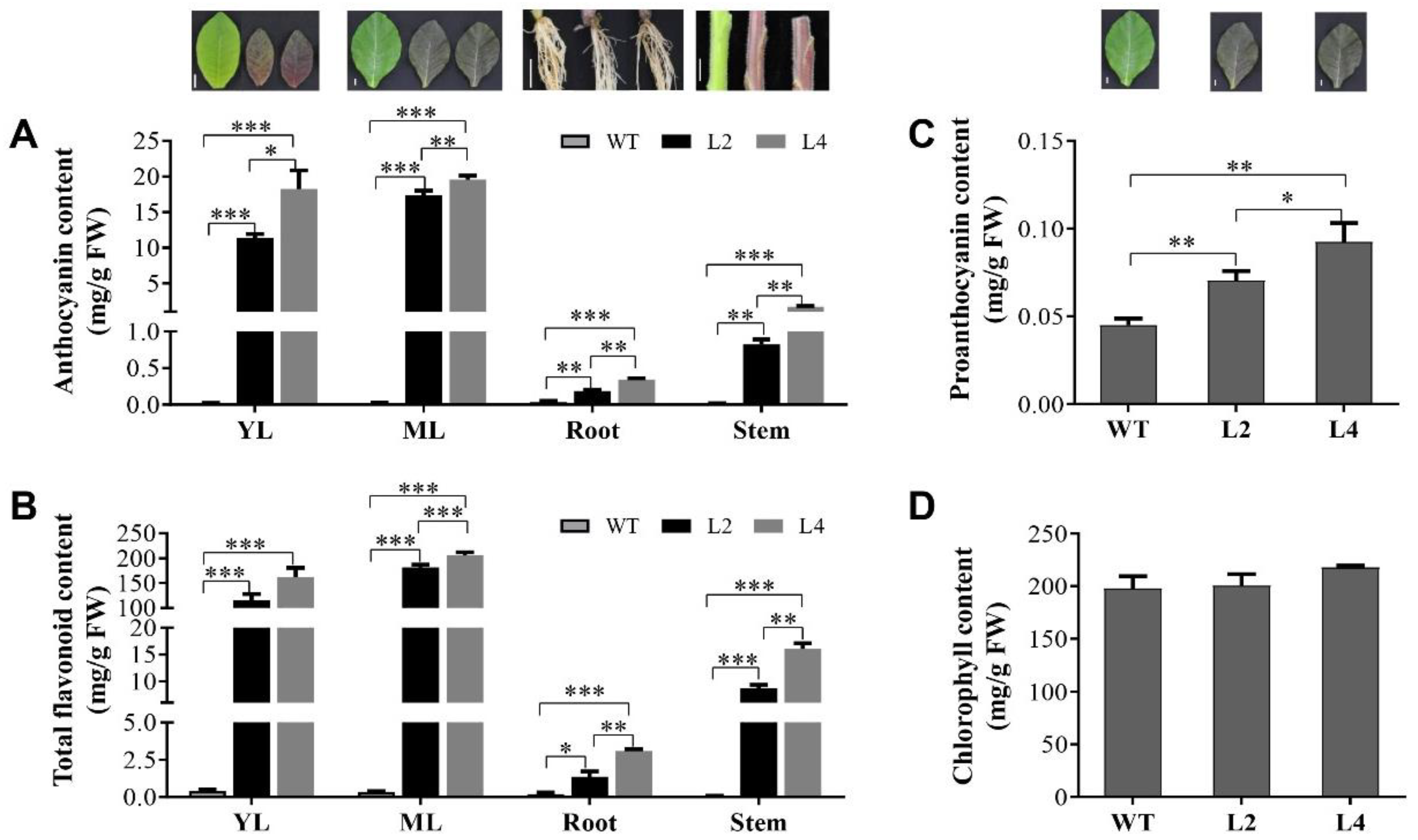
2.1. Increased Flavonoid Compounds in EsMYB90 Transgenic Tobacco Leaves
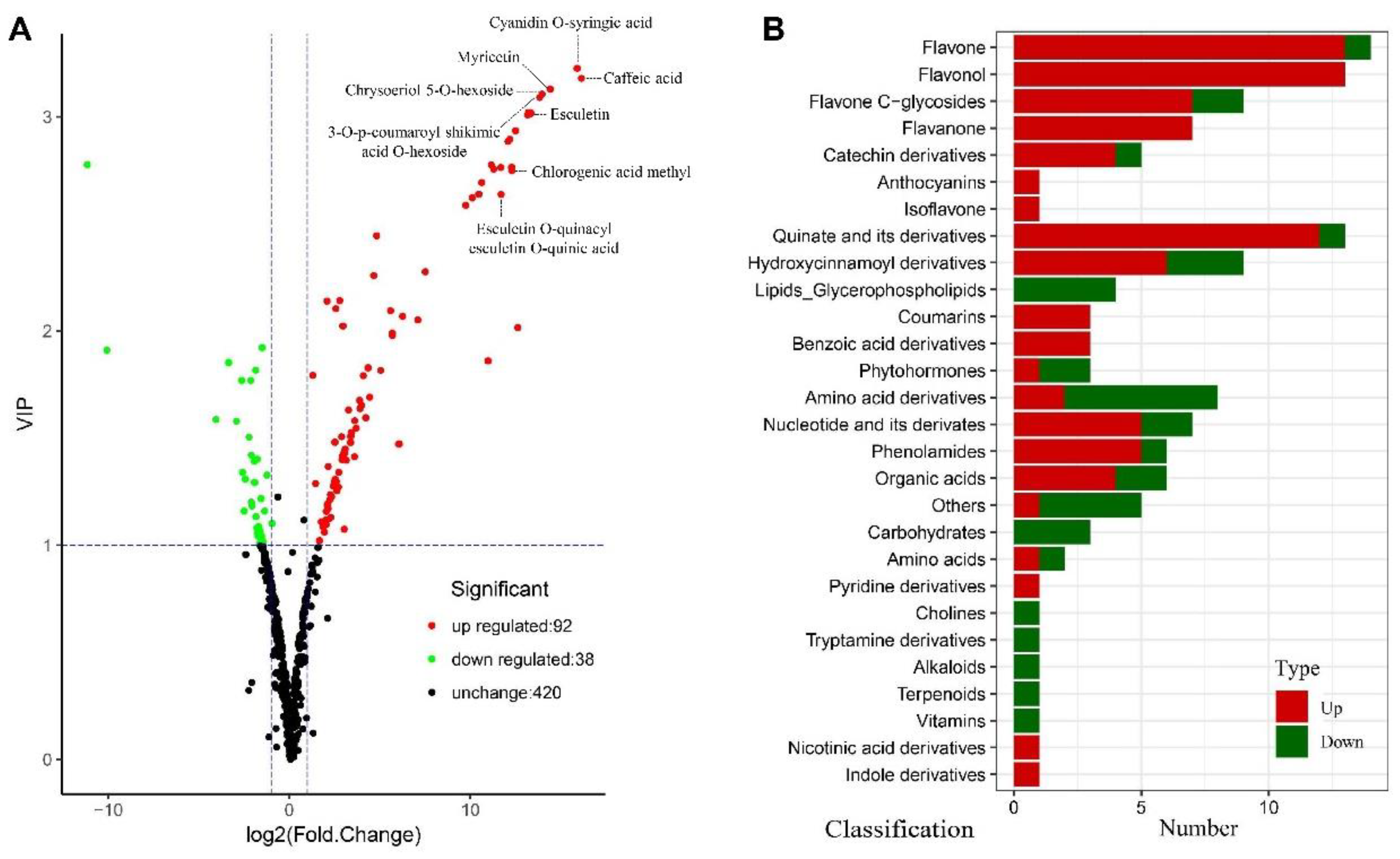
2.2. Metabolite Analysis Based on OPLS-DA Model
2.3. Metabolic Profiling and Significantly Differential Metabolite Analysis
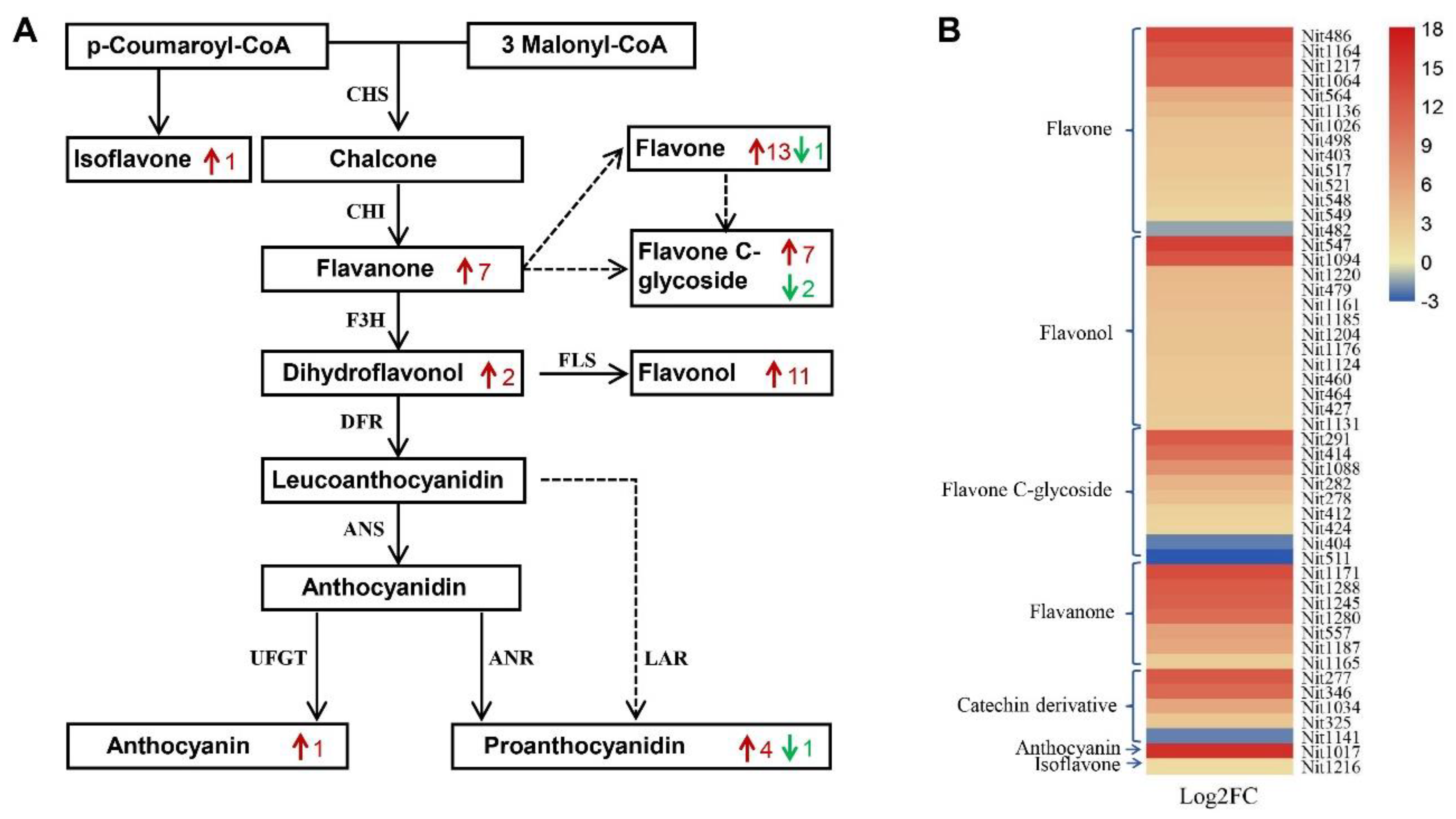
2.4. Integrated Analysis of Metabolite Profiling and RNA-seq in Phenylpropanoid/Flavonoid Biosynthesis Pathways
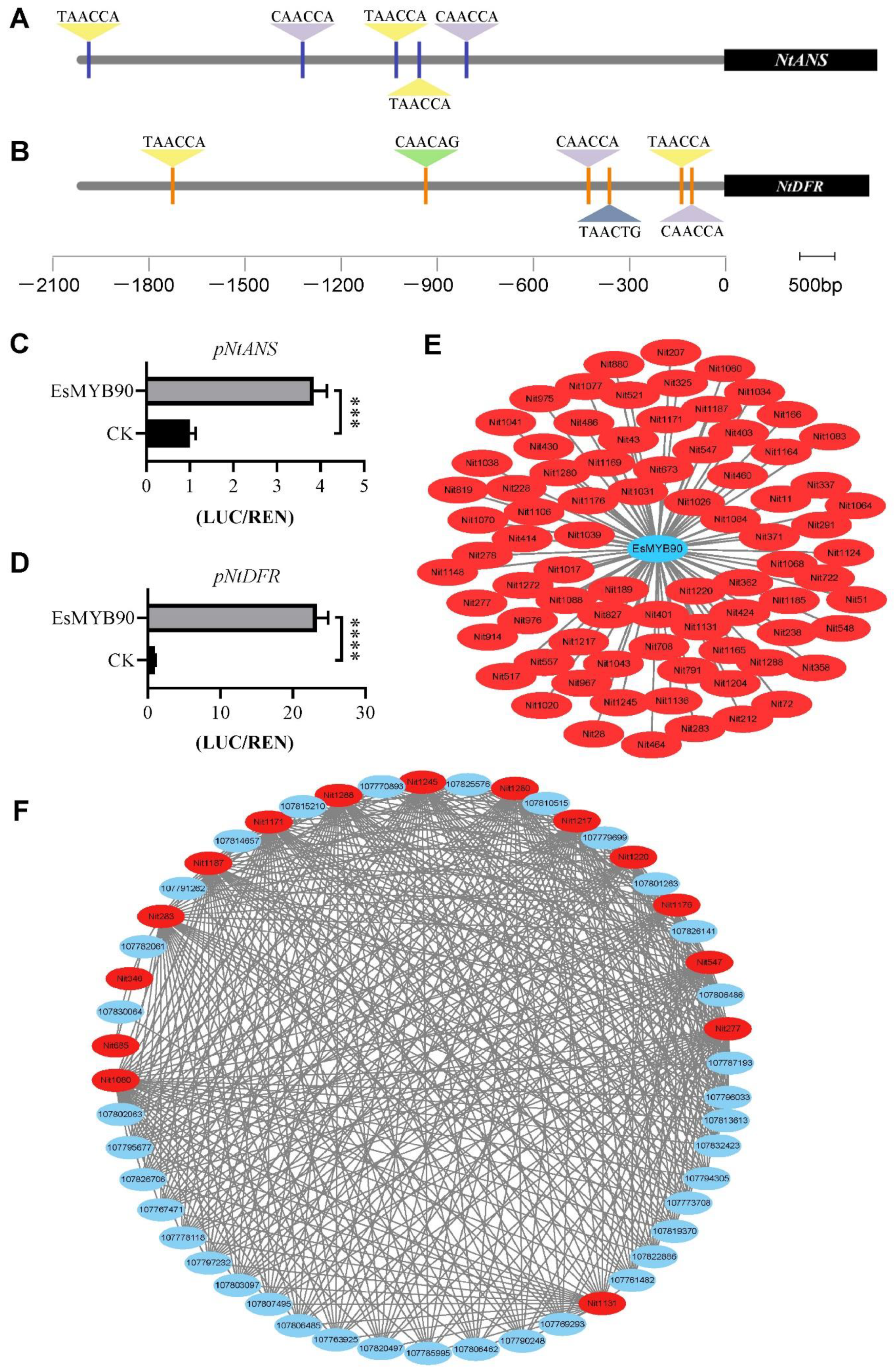
2.5. EsMYB90 Enhanced Flavonoid Metabolite Level via Activating Transcription of Flavonoid Biosynthesis Genes
3. Discussion
3.1. Key Role of EsMYB90 in Enhancing Antioxidative Metabolite Level
3.2. Regulating Mechanism of EsMYB90 in Phenylpropanoid/Flavonoid Pathways and Its Novelty
3.3. EsMYB90—A Potential Important Gene for Genetic Breeding to Improve the Flavonoid Level of Crops
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Measurement of Anthocyanin, Total Flavonoid, PA and Chlorophyll
4.3. RNA-Seq and Bioinformatic Analysis
4.4. Analysis of Metabolite Profiling
4.5. Differential Metabolite Analysis and Metabolic Pathways Construction
4.6. Integrated Analysis of Metabolome and Transcriptome
4.7. Dual Luciferase Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 4CL | 4-coumarate-CoA ligase |
| ANR | Anthocyanidin reductase |
| C4H | Trans-cinnamate 4-monooxygenase |
| CCoAOMT | Caffeoyl-CoA O-methyltransferase |
| CHI | Chalcone isomerase |
| CHS | Chalcone synthase |
| DEG | Differential expressed gene |
| DFR | Dihydroflavonol reductase |
| DHK | Dihydrokaempferol |
| DHM | Dihydromyricetin |
| DHQ | Dihydroquercetin |
| EBGs | Early biosynthesis genes |
| EGC | Epigallocatechin |
| F3′5′H | Flavonoid 3′5′- hydroxylase |
| F3′H | Flavonoid 3′-hydroxylase |
| F3H | Flavanone 3-hydroxylase |
| FLS | Flavonol synthase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LAR | Leucoanthocyanidin reductase |
| LBGs | Late biosynthetic genes |
| LDOX/ANS | Leucoanthocyanidin dioxygenase/anthocyanidin synthase |
| MBW | MYB-bHLH-WD40 |
| ML | Mature leaves |
| MS/MS | Tandem mass spectrometry |
| MWDB | MetWare database |
| MYB | v-myb avian myeloblastosis viral oncogene homolog |
| OPLS-DA | Orthogonal partial least squares-discriminant analysis |
| PA | Proanthocyanidin |
| PAL | Phenylalanine ammonia lyase |
| PCA | Principal component analysis |
| PCC | Pearson correlation coefficient |
| PCCP | p-value of Pearson correlation coefficient |
| UFGT | UDP-glucose:flavonoid 3-glucosyltransferase |
| UPLC | Ultra-performance liquid chromatography |
| VIP | Variable importance in projection |
| YL | Young leaves |
References
- Ni, J.; Zhao, Y.; Tao, R.; Yin, L.; Gao, L.; Strid, A.; Qian, M.; Li, J.; Li, Y.; Shen, J. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2019, 18, 1223–1240. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Dubos, C.; Lepiniec, L.C. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buer, C.S.; Imin, N.; Djordjevic, M.A. Flavonoids: New roles for old molecules. J. Integr. Plant Biol. 2010, 52, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Bac-Molenaar, J.A.; Fradin, E.F.; Rienstra, J.A.; Vreugdenhil, D.; Keurentjes, J.J. GWA mapping of anthocyanin accumulation reveals balancing selection of MYB90 in Arabidopsis thaliana. PLoS ONE 2015, 10, e0143212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Taylor, L.P.; Grotewold, E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005, 8, 317–323. [Google Scholar] [CrossRef]
- Peer, W.A.; Murphy, A.S. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef]
- Duthie, G.; Crozier, A. Plant-derived phenolic antioxidants. Curr. Opin. Lipidol. 2000, 11, 43–47. [Google Scholar] [CrossRef]
- Lee, K.W.; Lee, H.J. The roles of polyphenols in cancer chemoprevention. Biofactors 2006, 26, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, D.L.; Gonzali, S.; Loreti, E.; Pucciariello, C.; Degl’Innocenti, E.; Guidi, L.; Alpi, A.; Perata, P. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct. Plant Biol. 2008, 35, 606–618. [Google Scholar] [CrossRef]
- Peng, T.; Moriguchi, T. The molecular network regulating the coloration in apple. Sci. Hortic. 2013, 163, 1–9. [Google Scholar] [CrossRef]
- Dela, G.; Or, E.; Ovadia, R.; Nissim-Levi, A.; Weiss, D.; Oren-Shamir, M. Changes in anthocyanin concentration and composition in ‘Jaguar’rose flowers due to transient high-temperature conditions. Plant Sci. 2003, 164, 333–340. [Google Scholar] [CrossRef]
- Steyn, W.J.; Wand, S.; Holcroft, D.; Jacobs, G. In Red colour development and loss in pears. Acta Hortic. 2004, 671, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Mori, K.; Goto-Yamamoto, N.; Kitayama, M.; Hashizume, K. Loss of anthocyanins in red-wine grape under high temperature. J. Exp. Bot. 2007, 58, 1935–1945. [Google Scholar] [CrossRef]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.I.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Terrier, N.; Torregrosa, L.; Ageorges, A.; Vialet, S.; Verries, C.; Cheynier, V.; Romieu, C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009, 149, 1028–1041. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Pang, Y.; Dixon, R.A. The mysteries of proanthocyanidin transport and polymerization. Plant Physiol. 2010, 153, 437–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, S.; Matsuda, F.; Tohge, T.; Yonekura-Sakakibara, K.; Yamazaki, M.; Saito, K.; Narumi, I. Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J. 2010, 62, 549–559. [Google Scholar] [CrossRef]
- Debeaujon, I.; Nesi, N.; Perez, P.; Devic, M.; Grandjean, O.; Caboche, M.; Lepiniec, L. Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 2003, 15, 2514–2531. [Google Scholar] [CrossRef] [Green Version]
- Kidd, P.S.; Llugany, M.; Poschenrieder, C.; Gunsé, B.; Barceló, J. The role of root exudates in aluminium resistance and silicon-induced amelioration of aluminium toxicity in three varieties of maize (Zea mays L.). J. Exp. Bot. 2001, 359, 1339–1352. [Google Scholar]
- Pollastri, S.; Tattini, M. Flavonols: Old compounds for old roles. Ann. Bot. 2011, 108, 1225–1233. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, M.K.; Murrell, J.R.; Shirley, B.W. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis (Further evidence for differential regulation of “early” and “late” genes). Plant Physiol. 1997, 113, 1437–1445. [Google Scholar] [CrossRef] [Green Version]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Park, J.-S.; Kim, J.-B.; Cho, K.-J.; Cheon, C.-I.; Sung, M.-K.; Choung, M.-G.; Roh, K.-H. Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep. 2008, 27, 985–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracke, R.; Jahns, O.; Keck, M.; Tohge, T.; Niehaus, K.; Fernie, A.R.; Weisshaar, B. Analysis of Production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010, 188, 985–1000. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Xu, W. Deciphering the regulatory network controlling flavonoid biosynthesis by MYB-bHLH-WDR complexes in Arabidopsis seed. Rev. Sci. Instrum. 2014, 85, 02A929. [Google Scholar]
- Gatica-Arias, A.; Farag, M.; Stanke, M.; Matoušek, J.; Wessjohann, L.; Weber, G. Flavonoid production in transgenic hop (Humulus lupulus L.) altered by PAP1/MYB75 from Arabidopsis thaliana L. Plant Cell Rep. 2012, 31, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-S.; Zhang, J.; Li, M.-X.; Shi, L.-X. Metabolomics analysis reveals the salt-tolerant mechanism in Glycine soja. J. Plant Growth Regul. 2017, 36, 460–471. [Google Scholar] [CrossRef]
- Lu, Y.; Lam, H.; Pi, E.; Zhan, Q.; Tsai, S.; Wang, C.; Kwan, Y.; Ngai, S. Comparative metabolomics in Glycine max and Glycine soja under salt stress to reveal the phenotypes of their offspring. J. Agric. Food Chem. 2013, 61, 8711–8721. [Google Scholar] [CrossRef]
- Qi, Y.; Gu, C.; Wang, X.; Gao, S.; Li, C.; Zhao, C.; Li, C.; Ma, C.; Zhang, Q. Identification of the Eutrema salsugineum EsMYB90 gene important for anthocyanin biosynthesis. BMC Plant Biol. 2020, 20, 186. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Wang, R.; Xian, B.; Ren, C.; Liu, Q.; Wu, Q.; Pei, J. Integrated metabolomics and transcriptome analysis on flavonoid biosynthesis in safflower (Carthamus tinctorius L.) under MeJA treatment. BMC Plant Biol. 2020, 20, 353. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [Green Version]
- Steyn, W. Prevalence and functions of anthocyanins in fruits. Anthocyanins 2008, 86–105. [Google Scholar] [CrossRef]
- Hoch, W.A.; Zeldin, E.L.; McCown, B.H. Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiol. 2001, 21, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Xie, D.Y.; Sharma, S.B. Proanthocyanidins—A final frontier in flavonoid research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef] [Green Version]
- Hale, K.L.; McGrath, S.P.; Lombi, E.; Stack, S.M.; Terry, N.; Pickering, I.J.; George, G.N.; Pilon-Smits, E.A. Molybdenum sequestration in Brassica species. A role for anthocyanins? Plant Physiol. 2001, 126, 1391–1402. [Google Scholar] [CrossRef] [Green Version]
- Feild, T.S.; Lee, D.W.; Holbrook, N.M. Why leaves turn red in autumn. The role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol. 2001, 127, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-M.; Yu, H.-J.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Xia, X.-J. Photoprotective roles of anthocyanins in Begonia semperflorens. Plant Sci. 2010, 179, 202–208. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Environmental significance of anthocyanins in plant stress responses. Photoch. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Do anthocyanins function as osmoregulators in leaf tissues? Adv. Bot. Res. 2002, 37, 103–127. [Google Scholar]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef] [Green Version]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef]
- Rowan, D.D.; Cao, M.; Lin-Wang, K.; Cooney, J.M.; Jensen, D.J.; Austin, P.T.; Hunt, M.B.; Norling, C.; Hellens, R.P.; Schaffer, R.J. Environmental regulation of leaf colour in red 35S: PAP1 Arabidopsis thaliana. New Phytol. 2009, 182, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.B.; Mcnear, D.H. Induced transcriptional profiling of phenylpropanoid pathway genes increased flavonoid and lignin content in Arabidopsis leaves in response to microbial products. BMC Plant Biol. 2014, 14, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Fukushima, A.; Saito, K. Transcriptome data modeling for targeted plant metabolic engineering. Curr. Opin. Biotechnol. 2013, 24, 285–290. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F.; Chandler, S. Recent progress of flower colour modification by biotechnology. Int. J. Mol. Sci. 2009, 10, 5350–5369. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F.; Kalc, G.; Senior, M.; Dyson, B.; Nakamura, N.; Katsumoto, Y.; Chandler, S. Flower color modification by engineering of the flavonoid biosynthetic pathway: Practical perspectives. Biosci. Biotechnol. Biochem. 2010, 1007282070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 2011, 33, 433–441. [Google Scholar] [CrossRef]
- Liu, X.; Xiang, L.; Yin, X.; Grierson, D.; Li, F.; Chen, K. The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation in Chrysanthemum flowers. Sci. Hortic. 2015, 194, 278–285. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Saito, K. Review: Genetically modified plants for the promotion of human health. Biotechnol. Lett. 2006, 28, 1983–1991. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Evers, D.L.; Chao, C.F.; Wang, X.; Zhang, Z.; Huong, S.M.; Huang, E.S. Human cytomegalovirus inhibitory flavonoids: Studies on antiviral activity and mechanism of action. Antivir. Res. 2005, 68, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Keddy, P.G.; Dunlop, K.; Warford, J.; Samson, M.L.; Jones, Q.R.; Rupasinghe, H.P.; Robertson, G.S. Neuroprotective and anti-infl ammatory effects of the fl avonoid-enriched fraction AF4 in a mouse model of hypoxic-ischemic brain injury. PLoS ONE 2012, 7, e51324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neff, M.M. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998, 118, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Dere, S.; Gunes, T.; Sivaci, R. Spectrophotometric determination of chlorophyll-A, B and total carotenoid contents of some algae species using different solvents. Botany 1998, 22, 13–17. [Google Scholar]
- Sun, B.; Ricardo-da-Silva, J.M.; Spranger, I. Critical factors of vanillin assay for catechins and proanthocyanidins. J. Agric. Food Chem. 1998, 46, 4267–4274. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, Y.; Li, C.; Duan, C.; Gu, C.; Zhang, Q. Integrated Metabolomic and Transcriptomic Analysis Reveals the Flavonoid Regulatory Network by Eutrema EsMYB90. Int. J. Mol. Sci. 2021, 22, 8751. https://doi.org/10.3390/ijms22168751
Qi Y, Li C, Duan C, Gu C, Zhang Q. Integrated Metabolomic and Transcriptomic Analysis Reveals the Flavonoid Regulatory Network by Eutrema EsMYB90. International Journal of Molecular Sciences. 2021; 22(16):8751. https://doi.org/10.3390/ijms22168751
Chicago/Turabian StyleQi, Yuting, Chuanshun Li, Chonghao Duan, Caihong Gu, and Quan Zhang. 2021. "Integrated Metabolomic and Transcriptomic Analysis Reveals the Flavonoid Regulatory Network by Eutrema EsMYB90" International Journal of Molecular Sciences 22, no. 16: 8751. https://doi.org/10.3390/ijms22168751
APA StyleQi, Y., Li, C., Duan, C., Gu, C., & Zhang, Q. (2021). Integrated Metabolomic and Transcriptomic Analysis Reveals the Flavonoid Regulatory Network by Eutrema EsMYB90. International Journal of Molecular Sciences, 22(16), 8751. https://doi.org/10.3390/ijms22168751





