Insights into the Structure of Rubisco from Dinoflagellates-In Silico Studies
Abstract
1. Introduction
2. Results and Discussion
2.1. Homologues of Form II Rubisco from Rhodospirillum Rubrum among Dinoflagellates
2.2. Analysis of the Amino Acid Sequence of Dinoflagellate Rubiscos
2.3. Instability of the Enzyme
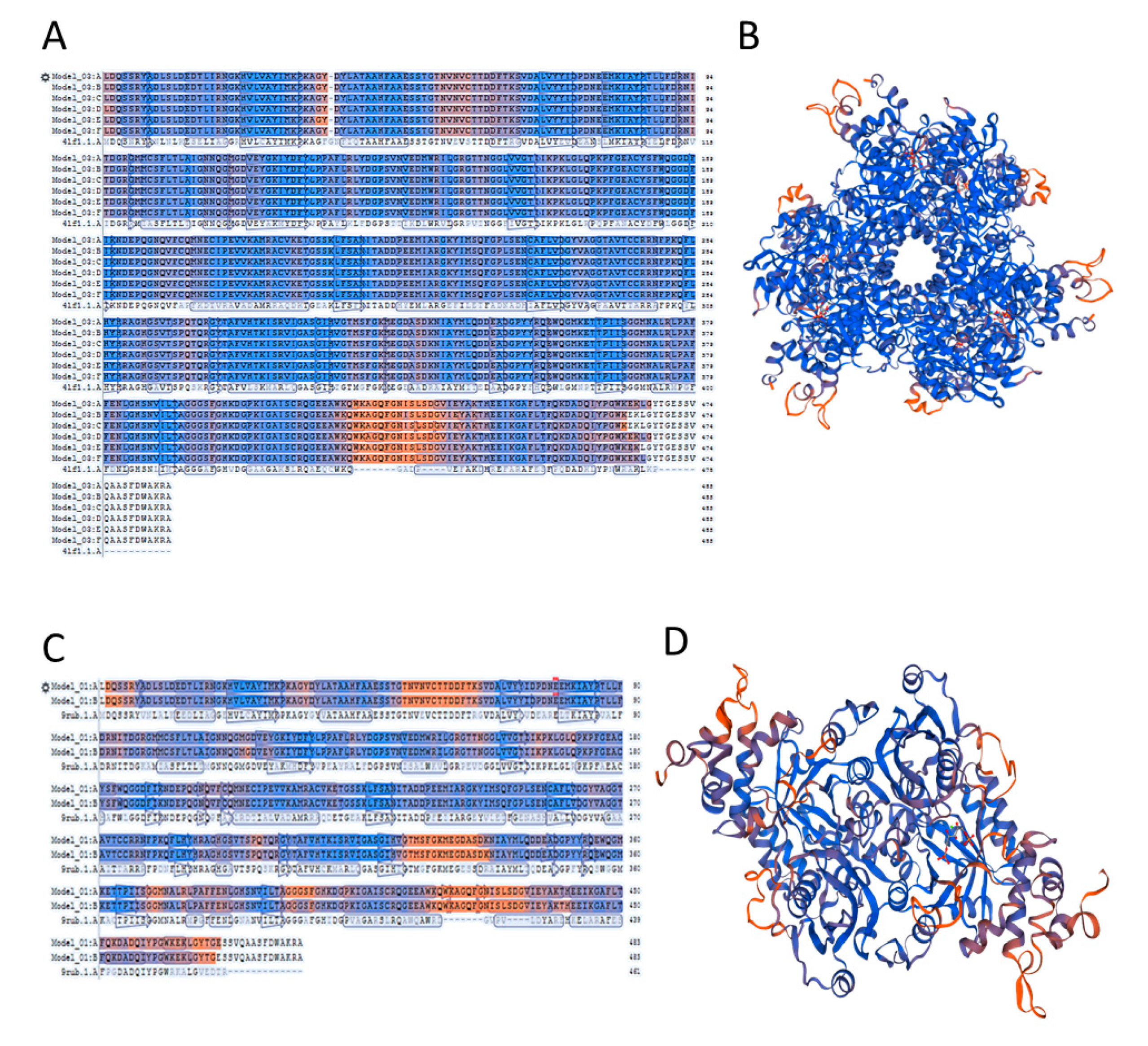
2.4. A Template Selection for Structure Modelling of Symbiodinium sp. Rubisco Using the SWISS-MODEL Software
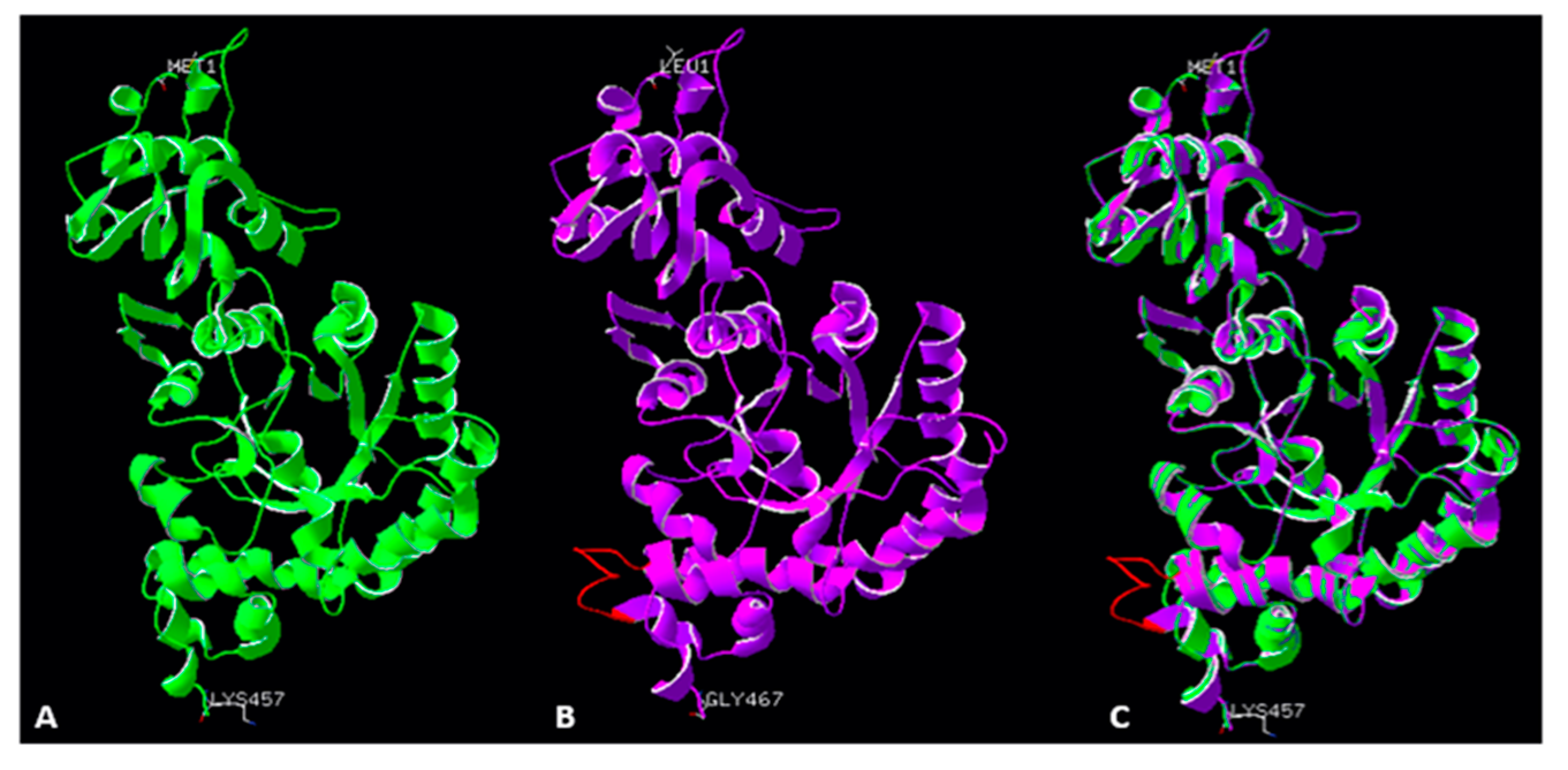
2.5. Accuracy of the Models of Symbodinium sp. Rubisco Structure Made by Homology Modelling in SWISS-MODEL
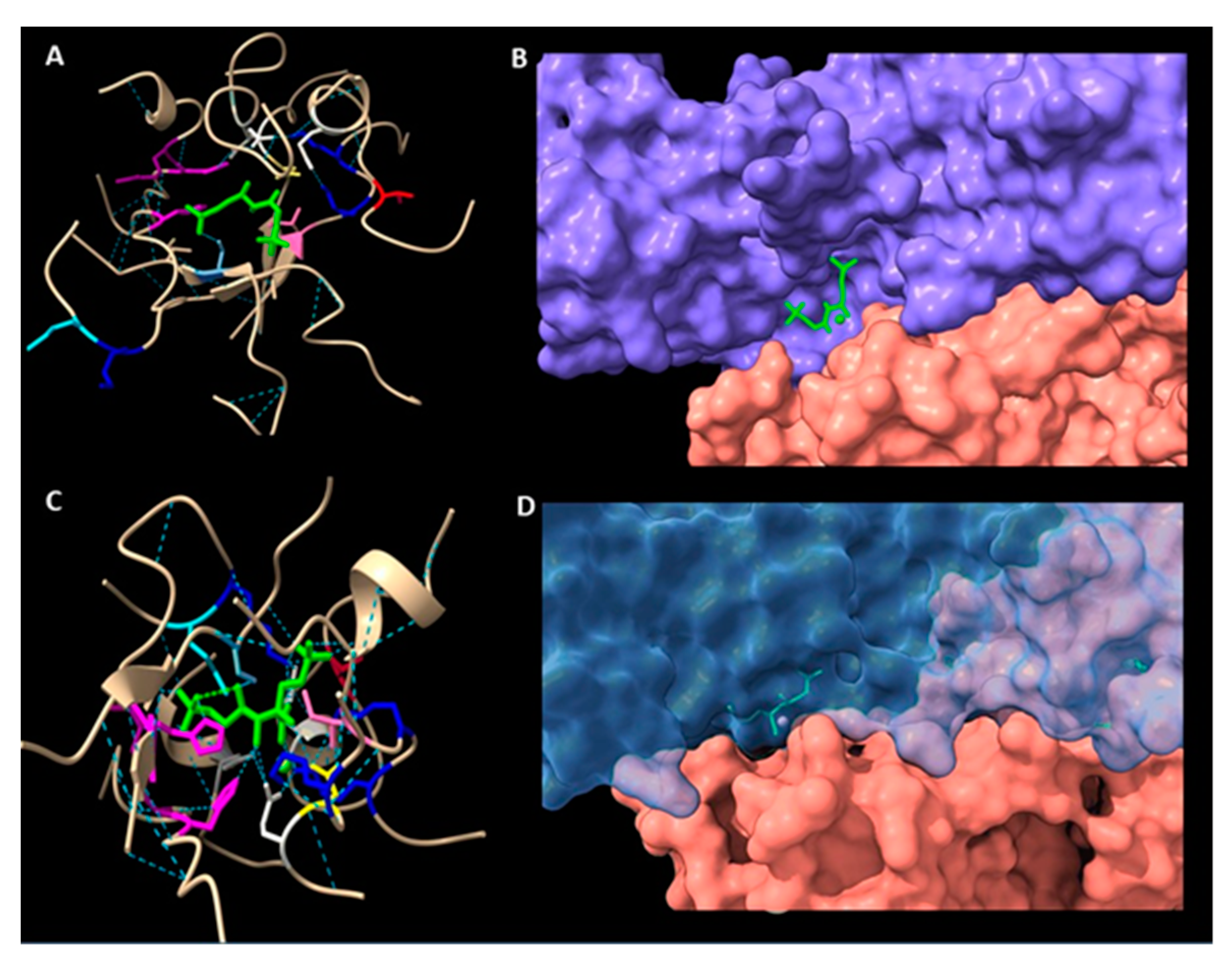
2.6. Structure of the Active Site in a Modelled Rubisco from Symbiodinium sp.
2.7. Analysis of a Possible Role of Insertions in the 413 and 425 Positions
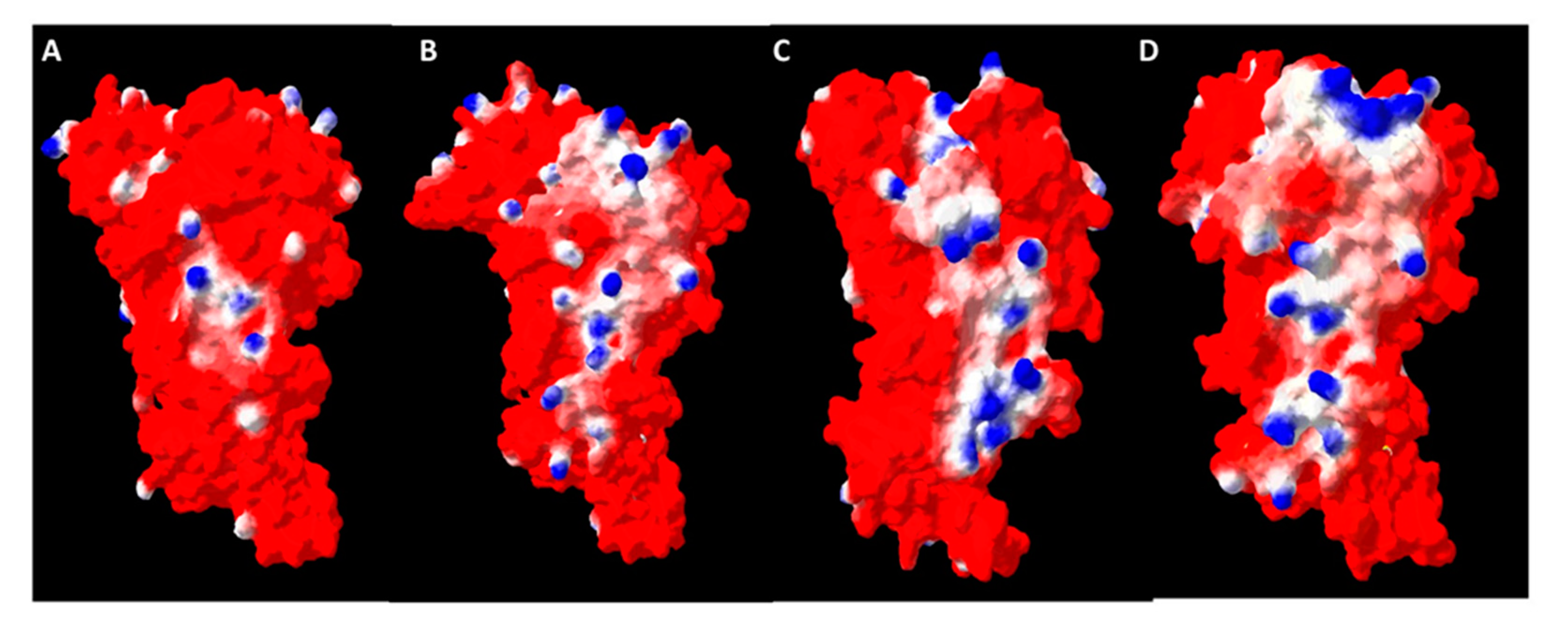
2.8. Oligomerization Interface Analysis
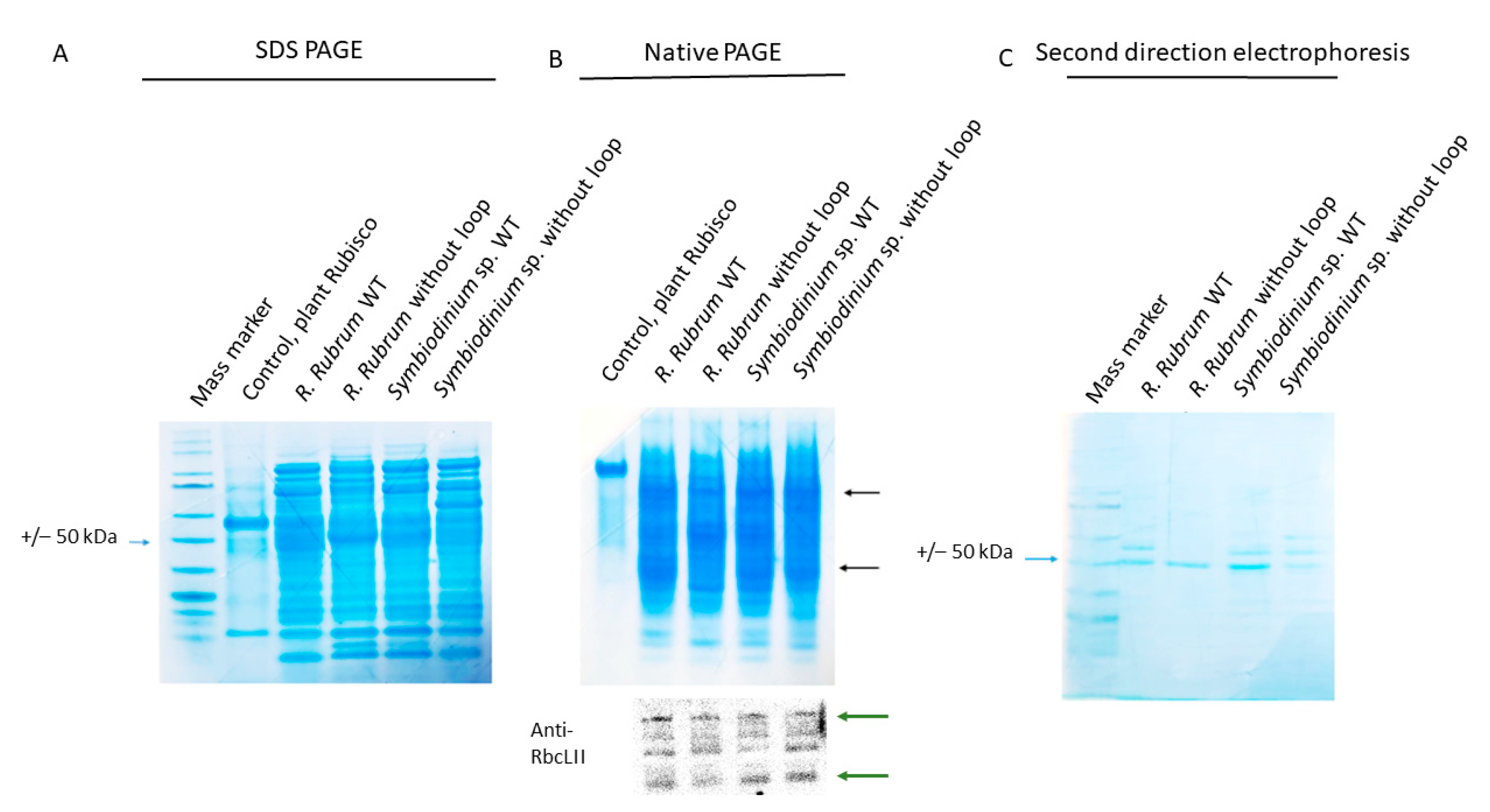
2.9. The Loop of the RbcL from Dinoflagellate Has Measurable Impact on the Enzyme’s Solubility
3. Materials and Methods
3.1. Sequence Analysis
3.2. Crystallization Prediction
3.3. Model of the Structure of Rubisco from Symbiodinium sp.
3.4. Computation of Chemical and Physical Parameters
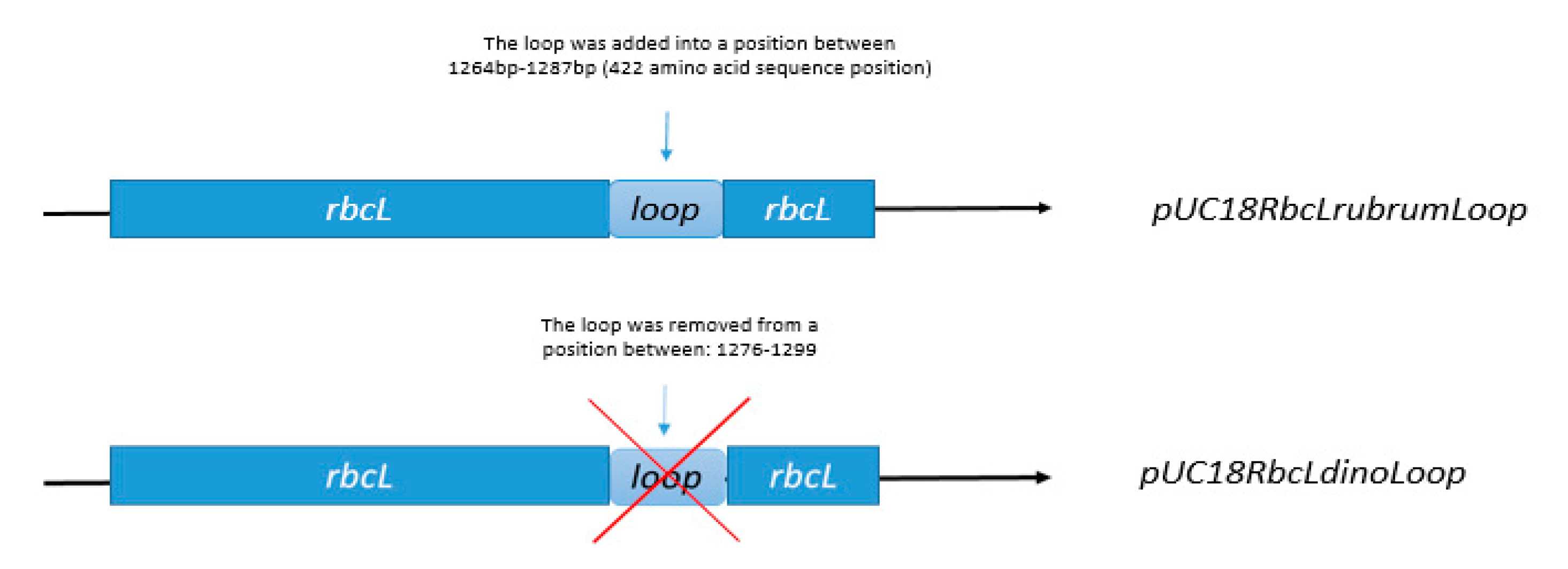
3.5. Construction of Expression Vectors pUC18RbcLrubrumLoop, pUC18RbcLdinoLoop
3.6. Expression of WT and Mutant Rubisco from R. Rubrum and Symbiodinium in E. coli
3.7. SDS-PAGE, Native-PAGE, Immunoblot Analysis, Protein Quantitation
3.8. Chemicals
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersson, I. Catalysis and regulation in Rubisco. J. Exp. Bot. 2008, 59, 1555–1568. [Google Scholar] [CrossRef] [PubMed]
- Duff, A.P.; Andrews, T.J.; Curmi, P.M. The transition between the open and closed states of rubisco is triggered by the inter-phosphate distance of the bound bisphosphate. J. Mol. Biol. 2000, 298, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.; García-Murria, M.J.; Marín-Navarro, J. Redox modulation of Rubisco conformation and activity through its cysteine residues. J. Exp. Bot. 2008, 59, 1605–1614. [Google Scholar] [CrossRef] [PubMed]
- Tabita, F.R.; Satagopan, S.; Hanson, T.E.; Kreel, N.E.; Scott, S.S. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 2008, 59, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.; Salois, P.; Markovic, P.; Hastings, J.W. A nuclear-encoded form II RuBisCO in dinoflagellates. Science 1995, 268, 1622–1624. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, S. Complex gene structure of the form II Rubisco in the dinoflagellate Prorocentrum minimum (dinophyceae). J. Phycol. 2003, 39, 1160–1171. [Google Scholar] [CrossRef]
- Palmer, J.D. Rubisco surprises in dinoflagellates. Plant Cell 1996, 8, 343. [Google Scholar]
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef]
- Rowan, R.; Whitney, S.M.; Fowler, A.; Yellowlees, D. Rubisco in marine symbiotic dinoflagellates: Form II enzymes in eukaryotic oxygenic phototrophs encoded by a nuclear multigene family. Plant Cell 1996, 8, 539–553. [Google Scholar]
- Whitney, S.M.; Yellowlees, D. Preliminary investigations into the structure and activity of ribulose bisphosphate carboxylase from two photosynthetic dinoflagellates. J. Phycol. 1995, 31, 138–146. [Google Scholar] [CrossRef]
- Bathellier, C.; Tcherkez, G.; Lorimer, G.H.; Farquhar, G.D. Rubisco is not really so bad. Plant Cell Environ. 2018, 41, 705–716. [Google Scholar] [CrossRef]
- Tessier, L.; Paulus, F.; Keller, M.; Vial, C.; Imbault, P. Structure and expression of Euglena gracilis nuclear rbcS genes encoding the small subunits of the ribulose 1, 5-bisphoshate carboxylase/oxygenase: A novel splicing process for unusual intervening sequences? J. Mol. Biol. 1995, 245, 22–33. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Hsiao, Y.-Y.; Chen, H.-K.; Chen, C.-S. Rubisco expression in the dinoflagellate Symbiodinium sp. is influenced by both photoperiod and endosymbiotic lifestyle. Mar. Biotechnol. 2014, 16, 371–384. [Google Scholar] [CrossRef]
- Aigner, H.; Wilson, R.; Bracher, A.; Calisse, L.; Bhat, J.; Hartl, F.; Hayer-Hartl, M. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 2017, 358, 1272–1278. [Google Scholar] [CrossRef]
- Hayer-Hartl, M.; Hartl, F.U. Chaperone machineries of rubisco–the most abundant enzyme. Trends Biochem. Sci 2020, 45, 748–763. [Google Scholar] [CrossRef]
- Levin, R.A.; Beltran, V.H.; Hill, R.; Kjelleberg, S.; McDougald, D.; Steinberg, P.D.; Van Oppen, M.J. Sex, scavengers, and chaperones: Transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 2016, 33, 2201–2215. [Google Scholar] [CrossRef]
- Gabruk, M.; Grzyb, J.; Kruk, J.; Mysliwa-Kurdziel, B. Light-dependent and light-independent protochlorophyllide oxidoreductases share similar sequence motifs -in silico studies. Photosynthetica 2012, 50, 529–540. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- García-Murria, M.J.; Sudhani, H.P.; Marín-Navarro, J.; Del Pino, M.M.S.; Moreno, J. Dissecting the individual contribution of conserved cysteines to the redox regulation of RubisCO. Photosynth. Res. 2018, 137, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, J.; Takahashi, S.; Sakamoto, A.; Shimada, H. Arabidopsis BSD2 reveals a novel redox regulation of Rubisco physiology in vivo. Plant Signal. Behav. 2020, 15, 1740873. [Google Scholar] [CrossRef] [PubMed]
- Jahandideh, S.; Jaroszewski, L.; Godzik, A. Improving the chances of successful protein structure determination with a random forest classifier. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Lindqvist, Y.; Lundqvist, T. Crystallographic refinement and structure of ribulose-1, 5-bisphosphate carboxylase from Rhodospirillum rubrum at 1.7 Å resolution. J. Mol. Biol. 1990, 211, 989–1008. [Google Scholar] [CrossRef]
- Schneider, D.; Schmidt, C.L. Multiple Rieske proteins in prokaryotes: Where and why? Biochim. Biophys. Acta Bioenerg. 2005, 1710, 1–12. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo-and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Slabinski, L.; Jaroszewski, L.; Rychlewski, L.; Wilson, I.A.; Lesley, S.A.; Godzik, A. XtalPred: A web server for prediction of protein crystallizability. Bioinformatics 2007, 23, 3403–3405. [Google Scholar] [CrossRef]
- Parry, M.A.; Keys, A.J.; Madgwick, P.J.; Carmo-Silva, A.E.; Andralojc, P.J. Rubisco regulation: A role for inhibitors. J. Exp. Bot. 2008, 59, 1569–1580. [Google Scholar] [CrossRef]
- Banda, D.M.; Pereira, J.H.; Liu, A.K.; Orr, D.J.; Hammel, M.; He, C.; Parry, M.A.; Carmo-Silva, E.; Adams, P.D.; Banfield, J.F. Novel bacterial clade reveals origin of form I Rubisco. Nat. Plants 2020, 6, 1158–1166. [Google Scholar] [CrossRef]
- Wilson, R.H.; Martin-Avila, E.; Conlan, C.; Whitney, S.M. An improved Escherichia coli screen for Rubisco identifies a protein–protein interface that can enhance CO2-fixation kinetics. J. Biol. Chem. 2018, 293, 18–27. [Google Scholar] [CrossRef]
- Satagopan, S.; North, J.A.; Arbing, M.A.; Varaljay, V.A.; Haines, S.N.; Wildenthal, J.A.; Byerly, K.M.; Shin, A.; Tabita, F.R. Structural Perturbations of Rhodopseudomonas palustris Form II RuBisCO Mutant Enzymes That Affect CO2 Fixation. Biochemistry 2019, 58, 3880–3892. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.; Potter, S.C.; Finn, R.D. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Slabinski, L.; Jaroszewski, L.; Rodrigues, A.P.; Rychlewski, L.; Wilson, I.A.; Lesley, S.A.; Godzik, A. The challenge of protein structure determination—lessons from structural genomics. Protein Sci. 2007, 16, 2472–2482. [Google Scholar] [CrossRef] [PubMed]
- Jaroszewski, L.; Slabinski, L.; Wooley, J.; Deacon, A.M.; Lesley, S.A.; Wilson, I.A.; Godzik, A. Genome pool strategy for structural coverage of protein families. Structure 2008, 16, 1659–1667. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Humama Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Hnasko, T.S.; Hnasko, R.M. The western blot. In ELISA; Springer: Berlin/Heidelberg, Germany, 2015; pp. 87–96. [Google Scholar]
- Schägger, H.; Von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

| Accession Number | Organism | Query Cover [%] | E Value | Percent Identity [%] |
|---|---|---|---|---|
| Q5ENN5.1 | Heterocapsa triquetra | 97 | 0.0 | 67.67 |
| OLP97682.1 | Symbiodinium microadriaticum | 97 | 0.0 | 65.95 |
| AAO13031.1 | Prorocentrum minimum | 78 | 0.0 | 64.38 |
| Q42813.2 | Lingulodinium polyedra | 97 | 0.0 | 64.24 |
| Organism | EP Class | RF Class | Length [a.a] | Gravy Index | Instability Index | Isoelectric Point | Coils | Longest Disorder Regions |
|---|---|---|---|---|---|---|---|---|
| Symbiodinium sp. | 2 | 3 | 485 | −0.33 | 36.97 | 5.56 | 0 | 4 |
| R. palustris | 1 | 3 | 461 | −0.25 | 38.26 | 6.28 | 0 | 4 |
| Organism | ID | Sequence Identity | Sequence Similarity | Coverage | Resolution | Method | GMQE | QSQE |
|---|---|---|---|---|---|---|---|---|
| Riftia pachyptila | 6ius.1.A | 68.928 | 0.515 | 0.942 | 2.120 | X-ray | 0.828 | 0.79 |
| Rhodospirillum rubrum | 9rub.1.A | 66.377 | 0.508 | 0.951 | 2.600 | X-ray | 0.820 | 0.75 |
| Rhodospirillum rubrum | 2rus.1.A | 66.522 | 0.508 | 0.948 | 2.300 | X-ray | 0.831 | 0.71 |
| Rhodopseudomonas palustris | 5hqm.1.A | 65.076 | 0.505 | 0.951 | 1.950 | X-ray | 0.872 | 0.79 |
| Rhodopseudomonas palustris | 4lf1.1.A | 65.427 | 0.506 | 0.942 | 2.380 | X-ray | 0.868 | 0.77 |
| Gallionellasp. | 5c2c.1.A | 63.377 | 0.495 | 0.940 | 2.090 | X-ray | 0.843 | 0.75 |
| Protein | Organism | Query Cover [%] | E Value | Percent Identity [%] | Accession Number |
|---|---|---|---|---|---|
| Cubilin homolog | Drosophila willistoni | 100 | 167 | 100 | XP_023033857.1 |
| Hypothetical protein | Proteobacteria bacterium | 100 | 167 | 100 | NDC23151.1 |
| MFS transporter | Gemmatimonadetes bacterium | 100 | 167 | 100 | RMH69784.1 |
| Heat shock protein 71 kDa protein | Fasciola hepatica | 100 | 168 | 100 | THD27816.1 |
| Response regulator | Bacteroidetes bacterium | 100 | 170 | 100 | NQT59094.1 |
| Name | Sequence |
|---|---|
| DinoCLoop-F | GGCGTTATTGAATATGCAAAAACCC |
| DinoCLoop-R | CTGGCCTGCTTTCCACTGTTTCCATGC |
| RubrumCLoopF | aggcatggcgcgatggcgtgtttggcaatattagcctgagtgatCCTGTTCTGGATTATGCCCGTGAAC |
| RubrumCLoopR | CACGCCATCGCGCCATGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rydzy, M.; Tracz, M.; Szczepaniak, A.; Grzyb, J. Insights into the Structure of Rubisco from Dinoflagellates-In Silico Studies. Int. J. Mol. Sci. 2021, 22, 8524. https://doi.org/10.3390/ijms22168524
Rydzy M, Tracz M, Szczepaniak A, Grzyb J. Insights into the Structure of Rubisco from Dinoflagellates-In Silico Studies. International Journal of Molecular Sciences. 2021; 22(16):8524. https://doi.org/10.3390/ijms22168524
Chicago/Turabian StyleRydzy, Małgorzata, Michał Tracz, Andrzej Szczepaniak, and Joanna Grzyb. 2021. "Insights into the Structure of Rubisco from Dinoflagellates-In Silico Studies" International Journal of Molecular Sciences 22, no. 16: 8524. https://doi.org/10.3390/ijms22168524
APA StyleRydzy, M., Tracz, M., Szczepaniak, A., & Grzyb, J. (2021). Insights into the Structure of Rubisco from Dinoflagellates-In Silico Studies. International Journal of Molecular Sciences, 22(16), 8524. https://doi.org/10.3390/ijms22168524







