The Role of lncRNA in the Development of Tumors, including Breast Cancer
Abstract
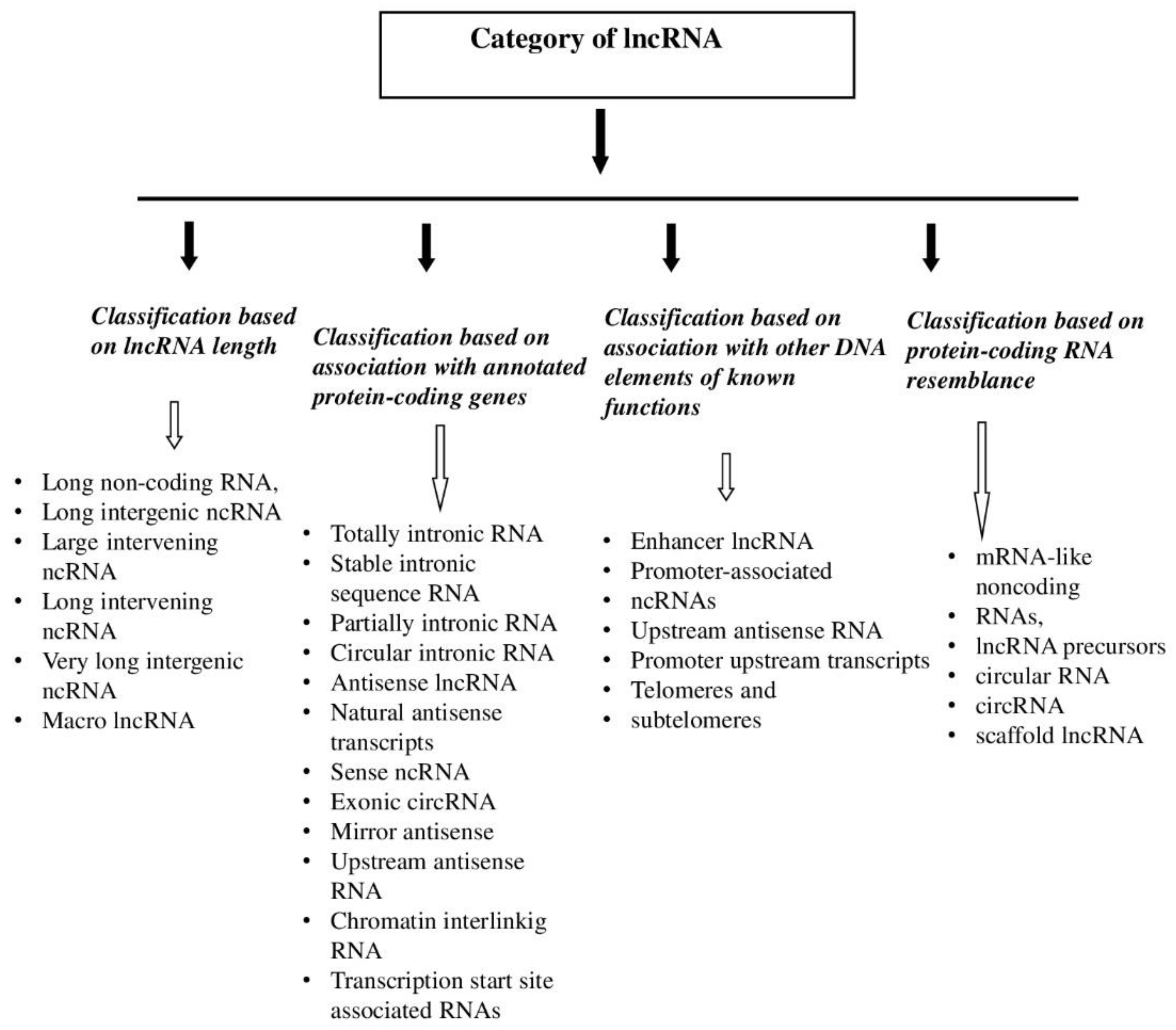
1. lncRNA—History
2. lncRNA—Characteristics
3. lncRNA—Functions
4. lncRNA and Malignant Tumors
5. lncRNA and Breast Cancer
6. lncRNA—T-UCRs
7. T-UCRs and Malignant Tumors
8. T-UCRs and Breast Cancer
9. Conclusive Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Inter-national human genome sequencing consortium. Initial sequencing and analysis of the human ge-nome. Nature 2001, 409, 860–921. [Google Scholar]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar]
- Marciniak, M. Imprinting genomowy u ssaków: Najnowsze doniesienia. Postępy Biol. Komórki 2008, 35, 243–257. [Google Scholar]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P.; Stöger, R.; Herrmann, B.G.; Saito, K.; Schweifer, N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 1991, 349, 84–87. [Google Scholar] [CrossRef]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef]
- XIST Gene—GeneCards|XIST RNA Gene. Weizmann Institute of Science. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=XIST&keywords=xist (accessed on 4 August 2021).
- Lyon, M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961, 190, 372–373. [Google Scholar] [CrossRef]
- Ayupe, A.C.; Tahira, A.C.; Camargo, L.; Beckedorff, F.C.; Verjovski-Almeida, S.; Reis, E.M. Global analysis of biogenesis, stability and sub-cellular localization of lncRNAs mapping to intragenic re-gions of the human genome. RNA Biol. 2015, 12, 877–892. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Ward, M.; McEwan, C.; Mills, J.D.; Janitz, M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J. Hum. Transcr. 2015, 1, 2–9. [Google Scholar] [CrossRef]
- Jiang, C.; Li, Y.; Zhao, Z.; Lu, J.; Chen, H.; Ding, N.; Wang, G.; Xu, J.; Li, X. Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 2016, 7, 7120–7133. [Google Scholar] [CrossRef]
- Kumar, V.; Westra, H.J.; Karjalainen, J.; Zhernakova, D.V.; Esko, T.; Hrdlickova, B.; Almeida, R.; Zhernakova, A.; Reinmaa, E.; Võsa, U.; et al. Human disease-associated genetic variation impacts large intergen-ic non-coding RNA expression. PLoS Genet. 2013, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Giannakakis, A.; Zhang, J.; Jenjaroenpun, P.; Nama, S.; Zainolabidin, N.; Aau, M.Y.; Yarmishyn, A.A.; Vaz, C.; Ivshina, A.V.; Grinchuk, O.V.; et al. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Sci. Rep. 2015, 5, 9737. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Şenergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn tran-scriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Sigova, A.A.; Abraham, B.J.; Ji, X.; Molinie, B.; Hannett, N.M.; Guo, Y.E.; Jangi, M.; Giallourakis, C.C.; Sharp, P.A.; Young, R.A.; et al. Transcription factor trapping by RNA in gene regulatory elements. Science 2015, 350, 978–981. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hämmerle, M.; Diederichs, S. MALAT1—A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.C.; Ni, J.J.; Cui, W.Y.; Wang, B.Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef]
- Léveillé, N.; Melo, C.A.; Rooijers, K.; Díaz-Lagares, A.; Melo, S.A.; Korkmaz, G.; Lopes, R.; Moqadam, F.A.; Maia, A.R.; Wijchers, P.J.; et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun. 2015, 6, 6520. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef]
- Kim, T.; Jeon, Y.J.; Cui, R.; Lee, J.H.; Peng, Y.; Kim, S.H.; Tili, E.; Alder, H.; Croce, C.M. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J. Natl. Cancer Inst. 2015, 107, 4. [Google Scholar] [CrossRef] [PubMed]
- Hart, J.R.; Roberts, T.C.; Weinberg, M.S.; Morris, K.V.; Vogt, P.K. MYC regulates the non-coding transcriptome. Oncotarget 2014, 5, 12543–12554. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, Y.; Segura, V.; Marín-Béjar, O.; Athie, A.; Marchese, F.P.; González, J.; Bujanda, L.; Guo, S.; Matheu, A.; Huarte, M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat. Commun. 2014, 5, 5812. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef]
- Marchese, F.P.; Huarte, M. Long non-coding RNAs and chromatin modifiers: Their place in the epigenetic code. Epigenetics 2014, 9, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, P.; Wang, L.; Piao, H.L.; Ma, L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim. Biophys. Sin. 2014, 46, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer me-tastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Heward, J.A.; Lindsay, M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014, 35, 408–419. [Google Scholar] [CrossRef]
- Jiang, R.; Tang, J.; Chen, Y.; Deng, L.; Ji, J.; Xie, Y.; Wang, K.; Jia, W.; Chu, W.M.; Sun, B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellu-lar carcinoma immune evasion. Nat. Commun. 2017, 8, 15129. [Google Scholar] [CrossRef]
- Serghiou, S.; Kyriakopoulou, A.; Ioannidis, J.P. Long noncoding RNAs as novel predictors of survival in human cancer: A systematic review and meta-analysis. Mol. Cancer 2016, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, M.; Lin, S.; Guo, Y.; Dai, Z.; Liu, K.; Yang, P.; Dai, C.; Zhu, Y.; Zheng, Y.; et al. The Impact of lncRNA Dysregulation on Clinicopathology and Survival of Breast Cancer: A Systematic Review and Meta-analysis. Molecular therapy. Nucleic Acids 2018, 12, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Jendrzejewski, J.; He, H.; Radomska, H.S.; Li, W.; Tomsic, J.; Liyanarachchi, S.; Davuluri, R.V.; Nagy, R.; de la Chapelle, A. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc. Natl. Acad. Sci. USA 2012, 109, 8646–8651. [Google Scholar] [CrossRef] [PubMed]
- Pasmant, E.; Sabbagh, A.; Vidaud, M.; Bièche, I. ANRIL, a long, noncoding RNA, is an unex-pected major hotspot in GWAS. FASEB J. 2011, 25, 444–448. [Google Scholar] [CrossRef]
- Betts, J.A.; Marjaneh, M.M.; Al-Ejeh, F.; Lim, Y.C.; Shi, W.; Sivakumaran, H.; Tropée, R.; Patch, A.M.; Clark, M.B.; Bartonicek, N.; et al. Long Noncoding RNAs CUPID1 and CUPID2 Mediate Breast Cancer Risk at 11q13 by Modulating the Response to DNA Damage. Am. J. Hum. Genet. 2017, 101, 255–266. [Google Scholar] [CrossRef]
- Daley, J.M.; Sung, P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 2014, 34, 1380–1388. [Google Scholar] [CrossRef]
- Fachal, L.; Aschard, A.; Beesley, J.; Barnes, D.R.; Allen, J.; Kar, S.; Pooley, K.A.; Dennis, J.; Michailidou, K.; Turman, C.; et al. Fine mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat. Genet. 2020, 52, 56–73. [Google Scholar] [CrossRef]
- Marjaneh, M.M.; Beesley, J.; O’Mara, T.A.; Mukhopadhyay, P.; Koufariotis, L.T.; Kazakoff, S.; Hussein, N.; Fachal, L.; Bartonicek, N.; Hillman, K.M. French Non-coding RNAs underlie genetic predis-position to breast cancer. Genome Biol. 2020, 21, 7. [Google Scholar] [CrossRef]
- Mathias, C.; Pedroso, G.A.; Pabst, F.R.; de Lima, R.S.; Kuroda, F.; Cavalli, I.J.; de Oliveira, J.C.; de Souza Fonseca Ribeiro, E.M.; Gradia, D.F. So alike yet so different. Differential expres-sion of the long non-coding RNAs NORAD and HCG11 in breast cancer subtypes. Genet. Mol. Biol. 2021, 44, e20200153. [Google Scholar] [CrossRef] [PubMed]
- Mathias, C.; Muzzi, J.C.D.; Antunes, B.B.; Gradia, D.F.; Castro, M.A.A.; de Oliveira, J.C. Un-raveling immune-related lncRNAs in breast cancer molecular subtypes. Front. Oncol. 2021, 11, 692170. [Google Scholar] [CrossRef]
- Chehade, M.; Bullock, M.; Moore, K.; Hutvagner, G.; Sidhu, S.B. The long non-coding RNA psoriasis susceptibility-associated RNA induced by stress (PRINS) as a tumour suppressor and potential biomarker in invasive breast cancer. Abstr. J. Breast Surg. 2020, 90, 21. [Google Scholar]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef] [PubMed]
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultracon-served elements in the human genome. Science 2004, 304, 1321–1325. [Google Scholar] [CrossRef]
- Calin, G.A.; Liu, C.G.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Terracciano, D.; Terreri, S.; de Nigris, F.; Costa, V.; Calin, G.A.; Cimmino, A. The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochimica et biophysica acta. Rev. Cancer 2017, 1868, 449–455. [Google Scholar]
- Ferdin, J.; Nishida, N.; Wu, X.; Nicoloso, M.S.; Shah, M.Y.; Devlin, C.; Ling, H.; Shimizu, M.; Kumar, K.; Cortez, M.A.; et al. HINCUTs in cancer: Hypoxia-induced noncoding ultraconserved tran-scripts. Cell Death Differ. 2013, 20, 1675–1687. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Fredlund, E.; Pattyn, F.; Rihani, A.; Van Maerken, T.; Vermeulen, J.; Kumps, C.; Menten, B.; De Preter, K.; Schramm, A.; et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene 2010, 29, 3583–3592. [Google Scholar] [CrossRef]
- Sana, J.; Hankeova, S.; Svoboda, M.; Kiss, I.; Vyzula, R.; Slaby, O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology 2012, 82, 114–118. [Google Scholar] [CrossRef]
- Hudson, R.S.; Yi, M.; Volfovsky, N.; Prueitt, R.L.; Esposito, D.; Volinia, S.; Liu, C.G.; Schetter, A.J.; Van Roosbroeck, K.; Stephens, R.M.; et al. Transcription signatures en-coded by ultraconserved genomic regions in human prostate cancer. Mol. Cancer 2013, 12, 13. [Google Scholar] [CrossRef]
- Jiang, J.; Azevedo-Pouly, A.C.; Redis, R.S.; Lee, E.J.; Gusev, Y.; Allard, D.; Sutaria, D.S.; Badawi, M.; Elgamal, O.A.; Lerner, M.R.; et al. Globally increased ultraconserved noncoding RNA expression in pancreatic adenocarcinoma. Oncotarget 2016, 7, 53165–53177. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Wang, C. Uc.206 regulates cell proliferation and apoptosis by targeting P53 in cervical cancer cells. Neoplasma 2016, 63, 411–418. [Google Scholar] [CrossRef]
- Olivieri, M.; Ferro, M.; Terreri, S.; Durso, M.; Romanelli, A.; Avitabile, C.; De Cobelli, O.; Messere, A.; Bruzzese, D.; Vannini, I.; et al. Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget 2016, 7, 20636–20654. [Google Scholar] [CrossRef]
- Sekino, Y.; Sakamoto, N.; Goto, K.; Honma, R.; Shigematsu, Y.; Sentani, K.; Oue, N.; Teishima, J.; Matsubara, A.; Yasui, W. Transcribed ultraconserved region Uc.63+ promotes resistance to docet-axel through regulation of androgen receptor signaling in prostate cancer. Oncotarget 2017, 8, 94259–94270. [Google Scholar] [CrossRef] [PubMed]
- Honma, R.; Goto, K.; Sakamoto, N.; Sekino, Y.; Sentani, K.; Oue, N.; Yasui, W. Expression and function of Uc.160+, a transcribed ultraconserved region, in gastric cancer. Gastric Cancer 2017, 20, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Li, Q.; Zhang, Y.; Deng, B.; Wu, F.; Wang, J.; Wu, K.; Ding, Y.; Yu, D. Transcribed ultraconserved noncoding RNA uc.160 acts as a negative regulator in gastric cancer. Am. J. Transl. Res. 2018, 10, 2822–2833. [Google Scholar]
- Zhou, J.; Wang, C.; Huang, C.; Ding, Z.; Shi, M. TUCR.454 inhabits metastasis in lung cancer cells. Int. J. Clin. Exp. Pathol. 2018, 11, 1289–1296. [Google Scholar]
- Marini, A.; Lena, A.M.; Panatta, E.; Ivan, C.; Han, L.; Liang, H.; Annicchiarico-Petruzzelli, M.; Di Daniele, N.; Calin, G.A.; Candi, E.; et al. Ultraconserved long non-coding RNA uc.63 in breast cancer. Oncotarget 2017, 8, 35669–35680. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, L.X.; Xu, L.; Zhang, C.H.; Lu, Y.H.; Ji, T.H.; Ling, L.J. uc.38 induces breast cancer cell apoptosis via PBX1. Am. J. Cancer Res. 2017, 7, 2438–2451. [Google Scholar]
- Magnani, L.; Ballantyne, E.B.; Zhang, X.; Lupien, M. PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet. 2011, 7, e1002368. [Google Scholar] [CrossRef]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.J.; Rothnagel, J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014, 15, 193–204. [Google Scholar] [CrossRef]
- Wu, P.; Mo, Y.; Peng, M.; Tang, T.; Zhong, Y.; Deng, X.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 2020, 19, 22. [Google Scholar] [CrossRef]
| lncRNA | Genomic Location | Expression in Patients | Function in Tumorigenesis |
|---|---|---|---|
| PCGEM1 | 2q32.2 | Increased in prostate cancer | oncogene |
| MALAT1 | 11q13.1 | Increased in colon, lung, and liver cancers | oncogene |
| MEG3 | 14q32.2 | Down-regulated in multiple cancers | tumor suppressor |
| HOTAIR | 12q13.13 | Increased in primary breast tumors and metastases, GIST, and pancreatic cancers | oncogene |
| Clinicopathological Features | lncRNA |
|---|---|
| PR status | MALAT1 |
| HER status | TUSC7 |
| Histological grade | MEG3 |
| TNM stage | NEAT1, TP73-AS1 |
| survival | - |
| shorter overall survival | CCAT2, MALAT1, NEAT1 |
| longer overall survival | MEG3 |
| overall survival (better prognosis) | FGF14-AS2, AFAP1-AS1, EPB41L4A-AS2, BC040587, EGOT, GAS6-AS1, FENDRR |
| overall survival (worse prognosis) | BCAR4, HOTTIP, CCAT1, Z38, TUNAR, CRNDE, HULC, MVIH, TP73-AS1, linc-ITGB1, PVT1, UCA1, OR3A4, DANCR, LINP1, SNHG15, SUMO1P3 |
| metastasis-free survival | CCAT2, HOTAIR, BCAR4 |
| disease-free survival | MALAT1, HOTTIP, MVIH, LINC00978, linc-ITGB1, MEG3, GAS6-AS1, HOTAIR, LINP1 |
| progression-free survival | MALAT1, MEG3, HOTAIR, RFS, CCAT1, MEG3, FENDRR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolarz, B.; Zadrożna-Nowak, A.; Romanowicz, H. The Role of lncRNA in the Development of Tumors, including Breast Cancer. Int. J. Mol. Sci. 2021, 22, 8427. https://doi.org/10.3390/ijms22168427
Smolarz B, Zadrożna-Nowak A, Romanowicz H. The Role of lncRNA in the Development of Tumors, including Breast Cancer. International Journal of Molecular Sciences. 2021; 22(16):8427. https://doi.org/10.3390/ijms22168427
Chicago/Turabian StyleSmolarz, Beata, Anna Zadrożna-Nowak, and Hanna Romanowicz. 2021. "The Role of lncRNA in the Development of Tumors, including Breast Cancer" International Journal of Molecular Sciences 22, no. 16: 8427. https://doi.org/10.3390/ijms22168427
APA StyleSmolarz, B., Zadrożna-Nowak, A., & Romanowicz, H. (2021). The Role of lncRNA in the Development of Tumors, including Breast Cancer. International Journal of Molecular Sciences, 22(16), 8427. https://doi.org/10.3390/ijms22168427






