The Capsicum baccatum-Specific Truncated NLR Protein CbCN Enhances the Innate Immunity against Colletotrichum acutatum
Abstract
:1. Introduction
2. Results
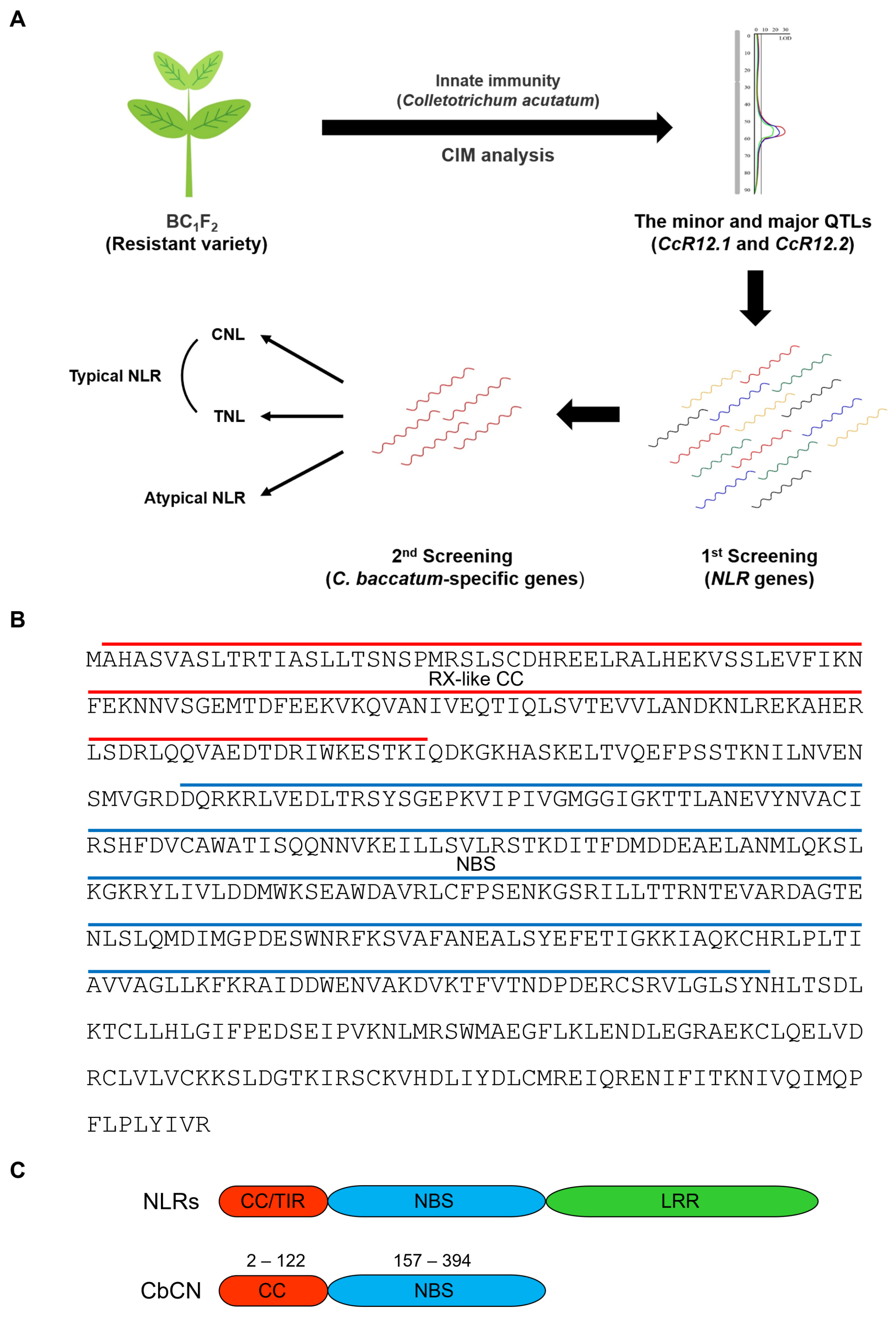
2.1. Identification of Truncated NLR Protein Specific of C. baccatum in the PBC80 Variety
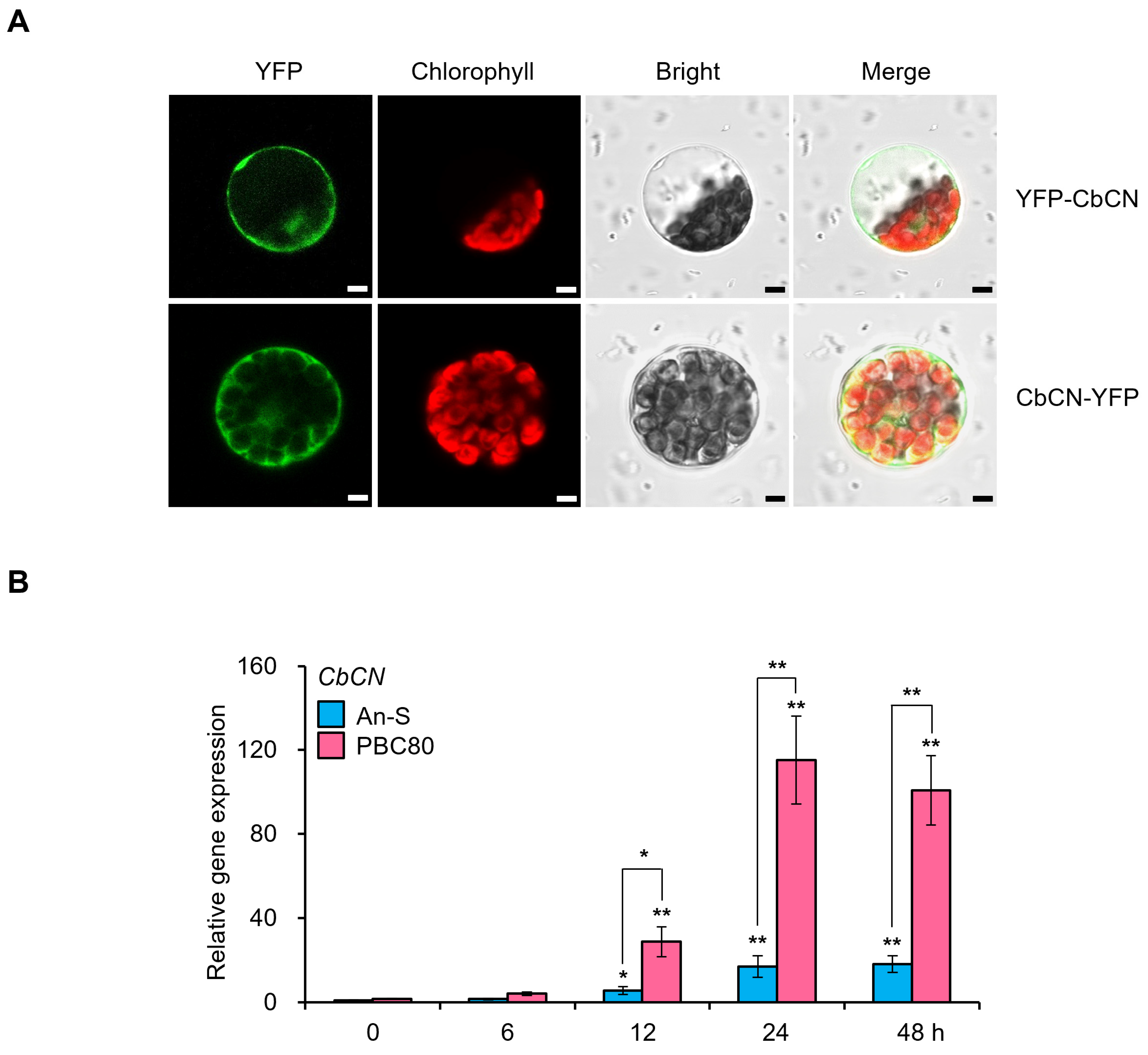
2.2. Upregulation of CbCN Transcription by C. acutatum
2.3. Enhanced Disease Resistance to C. acutatum in CbCN-Overexpressing Tobacco Plants
2.4. Attenuation of the Resistance to C. acutatum in Chili Pepper by CbCN Gene Silencing
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Condition
4.2. Cloning of CbCN and the Conserved Domain Analysis
4.3. Subcellular Localization in Chili Pepper Protoplasts
4.4. Gene Expression Analysis
4.5. Generation of Transgenic Tobacco Plants
4.6. Virus-Induced Gene Silencing in Chili Pepper Plants
4.7. Anthracnose Disease Resistance Assay
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.; Yeom, S.I.; Kim, Y.M.; Seo, E.; Kim, K.T.; Kim, M.S.; Lee, J.M.; Cheong, K.; Shin, H.S.; et al. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 2017, 18, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Moltke, J.; Ayres, J.S.; Kofoed, E.M.; Chavarria-Smith, J.; Vance, R.E. Recognition of bacteria by inflammasomes. Annu. Rev. Immunol. 2013, 31, 73–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Hu, M.; Wu, S.; Qi, J.; Wang, G.; Han, Z.; Qi, Y.; Gao, N.; Wang, H.W.; et al. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 2019, 364, eaav5868. [Google Scholar] [CrossRef]
- Hammond-Kosack, K.E.; Jones, J.D. Resistance gene-dependent plant defense responses. Plant Cell 1996, 8, 1773–1791. [Google Scholar] [CrossRef] [Green Version]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Mazzucotelli, E.; Marone, D.; Crosatti, C.; Michelotti, V.; Vale, G.; Mastrangelo, A.M. Regulation and Evolution of NLR Genes: A Close Interconnection for Plant Immunity. Int. J. Mol. Sci. 2018, 19, 1662. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, F.; Nishimura, M.T. Structural, Functional, and Genomic Diversity of Plant NLR Proteins: An Evolved Resource for Rational Engineering of Plant Immunity. Annu. Rev. Phytopathol. 2018, 56, 243–267. [Google Scholar] [CrossRef] [Green Version]
- Lolle, S.; Stevens, D.; Coaker, G. Plant NLR-triggered immunity: From receptor activation to downstream signaling. Curr. Opin. Immunol. 2020, 62, 99–105. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003, 15, 809–834. [Google Scholar] [CrossRef] [Green Version]
- Baggs, E.; Dagdas, G.; Krasileva, K.V. NLR diversity, helpers and integrated domains: Making sense of the NLR IDentity. Curr. Opin. Plant Biol. 2017, 38, 59–67. [Google Scholar] [CrossRef]
- Staal, J.; Kaliff, M.; Dewaele, E.; Persson, M.; Dixelius, C. RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 2008, 55, 188–200. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Wang, Z.; Zhang, X.; Yang, S. A missense mutation in CHS1, a TIR-NB protein, induces chilling sensitivity in Arabidopsis. Plant J. 2013, 75, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Rui, L.; Li, J.; Nishimura, M.T.; Vogel, J.P.; Liu, N.; Liu, S.; Zhao, Y.; Dangl, J.L.; Tang, D. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 2015, 11, e1004945. [Google Scholar] [CrossRef]
- Roth, C.; Ludke, D.; Klenke, M.; Quathamer, A.; Valerius, O.; Braus, G.H.; Wiermer, M. The truncated NLR protein TIR-NBS13 is a MOS6/IMPORTIN-alpha3 interaction partner required for plant immunity. Plant J. 2017, 92, 808–821. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; van Wersch, S.; Tong, M.; Li, X. TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol. 2019, 221, 2054–2066. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Tong, M.; Li, X. SUSA2 is an F-box protein required for autoimmunity mediated by paired NLRs SOC3-CHS1 and SOC3-TN2. Nat. Commun. 2020, 11, 5190. [Google Scholar] [CrossRef] [PubMed]
- Barragan, A.C.; Collenberg, M.; Wang, J.; Lee, R.R.Q.; Cher, W.Y.; Rabanal, F.A.; Ashkenazy, H.; Weigel, D.; Chae, E. A Truncated Singleton NLR Causes Hybrid Necrosis in Arabidopsis thaliana. Mol. Biol. Evol. 2021, 38, 557–574. [Google Scholar] [CrossRef] [PubMed]
- Nandety, R.S.; Caplan, J.L.; Cavanaugh, K.; Perroud, B.; Wroblewski, T.; Michelmore, R.W.; Meyers, B.C. The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 2013, 162, 1459–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Than, P.P.; Prihastuti, H.; Phoulivong, S.; Taylor, P.W.; Hyde, K.D. Chilli anthracnose disease caused by Colletotrichum species. J. Zhejiang Univ. Sci. B 2008, 9, 764–778. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.G.; Bae, J.H.; Jastrzebski, Z.; Cherkas, A.; Heo, B.G.; Gorinstein, S.; Ku, Y.G. Binding, Antioxidant and Anti-proliferative Properties of Bioactive Compounds of Sweet Paprika (Capsicum annuum L.). Plant Foods Hum. Nutr. 2016, 71, 129–136. [Google Scholar] [CrossRef]
- Surh, Y.J. More than spice: Capsaicin in hot chili peppers makes tumor cells commit suicide. J. Natl. Cancer Inst. 2002, 94, 1263–1265. [Google Scholar] [CrossRef] [PubMed]
- Ridzuan, R.; Rafii, M.Y.; Ismail, S.I.; Mohammad Yusoff, M.; Miah, G.; Usman, M. Breeding for Anthracnose Disease Resistance in Chili: Progress and Prospects. Int. J. Mol. Sci. 2018, 19, 3122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, E.; Zhang, D.; Mays, A.D.; Saftner, R.A.; Stommel, J.R. Genetic diversity in Capsicum baccatum is significantly influenced by its ecogeographical distribution. BMC Genet. 2012, 13, 68. [Google Scholar] [CrossRef] [Green Version]
- Manzur, J.P.; Fita, A.; Prohens, J.; Rodriguez-Burruezo, A. Successful Wide Hybridization and Introgression Breeding in a Diverse Set of Common Peppers (Capsicum annuum) Using Different Cultivated Aji (C. baccatum) Accessions as Donor Parents. PLoS ONE 2015, 10, e0144142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze-Lefert, P.; Panstruga, R. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 2011, 16, 117–125. [Google Scholar] [CrossRef]
- Lee, J.; Hong, J.-H.; Do, J.W.; Yoon, J.B. Identification of QTLs for resistance to anthracnose to two Colletotrichum species in pepper. J. Crop. Sci. Biotechnol. 2010, 13, 227–233. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.; Eulgem, T. Transcript-level expression control of plant NLR genes. Mol. Plant Pathol. 2018, 19, 1267–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, S.; Britton, M.; Martinez-Garcia, P.J.; Dandekar, A.M. Deep RNA-Seq profile reveals biodiversity, plant-microbe interactions and a large family of NBS-LRR resistance genes in walnut (Juglans regia) tissues. AMB Express 2016, 6, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eulgem, T.; Weigman, V.J.; Chang, H.S.; McDowell, J.M.; Holub, E.B.; Glazebrook, J.; Zhu, T.; Dangl, J.L. Gene expression signatures from three genetically separable resistance gene signaling pathways for downy mildew resistance. Plant Physiol. 2004, 135, 1129–1144. [Google Scholar] [CrossRef] [Green Version]
- Bartsch, M.; Gobbato, E.; Bednarek, P.; Debey, S.; Schultze, J.L.; Bautor, J.; Parker, J.E. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 2006, 18, 1038–1051. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Sohn, S.; Choi, M.S.; Kim, K.; Lomonossoff, G. The epigenetic phenotypes in transgenic Nicotiana benthamiana for CaMV 35S-GFP are mediated by spontaneous transgene silencing. Plant Biotechnol. Rep. 2011, 5, 273–281. [Google Scholar] [CrossRef]
- Tian, S.L.; Li, L.; Chai, W.G.; Shah, S.N.; Gong, Z.H. Effects of silencing key genes in the capsanthin biosynthetic pathway on fruit color of detached pepper fruits. BMC Plant Biol. 2014, 14, 314. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.B.; Park, H.G. Screening method for resistance to pepper fruit anthracnose: Pathogen sporulation, inoculation methods related to inoculum concentrations and post-inoculation environment. J. Korea Soc. Hort. Sci. 2001, 42, 389–393. [Google Scholar]
- Mutka, A.M.; Fentress, S.J.; Sher, J.W.; Berry, J.C.; Pretz, C.; Nusinow, D.A.; Bart, R. Quantitative, Image-Based Phenotyping Methods Provide Insight into Spatial and Temporal Dimensions of Plant Disease. Plant Physiol. 2016, 172, 650–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, S.; Kim, S.; Lee, K.S.; Oh, J.; Choi, I.; Do, J.W.; Yoon, J.B.; Han, J.; Park, S.R. The Capsicum baccatum-Specific Truncated NLR Protein CbCN Enhances the Innate Immunity against Colletotrichum acutatum. Int. J. Mol. Sci. 2021, 22, 7672. https://doi.org/10.3390/ijms22147672
Son S, Kim S, Lee KS, Oh J, Choi I, Do JW, Yoon JB, Han J, Park SR. The Capsicum baccatum-Specific Truncated NLR Protein CbCN Enhances the Innate Immunity against Colletotrichum acutatum. International Journal of Molecular Sciences. 2021; 22(14):7672. https://doi.org/10.3390/ijms22147672
Chicago/Turabian StyleSon, Seungmin, Soohong Kim, Kyong Sil Lee, Jun Oh, Inchan Choi, Jae Wahng Do, Jae Bok Yoon, Jungheon Han, and Sang Ryeol Park. 2021. "The Capsicum baccatum-Specific Truncated NLR Protein CbCN Enhances the Innate Immunity against Colletotrichum acutatum" International Journal of Molecular Sciences 22, no. 14: 7672. https://doi.org/10.3390/ijms22147672
APA StyleSon, S., Kim, S., Lee, K. S., Oh, J., Choi, I., Do, J. W., Yoon, J. B., Han, J., & Park, S. R. (2021). The Capsicum baccatum-Specific Truncated NLR Protein CbCN Enhances the Innate Immunity against Colletotrichum acutatum. International Journal of Molecular Sciences, 22(14), 7672. https://doi.org/10.3390/ijms22147672







