Hypertrophy of Rat Skeletal Muscle Is Associated with Increased SIRT1/Akt/mTOR/S6 and Suppressed Sestrin2/SIRT3/FOXO1 Levels
Abstract
:1. Introduction
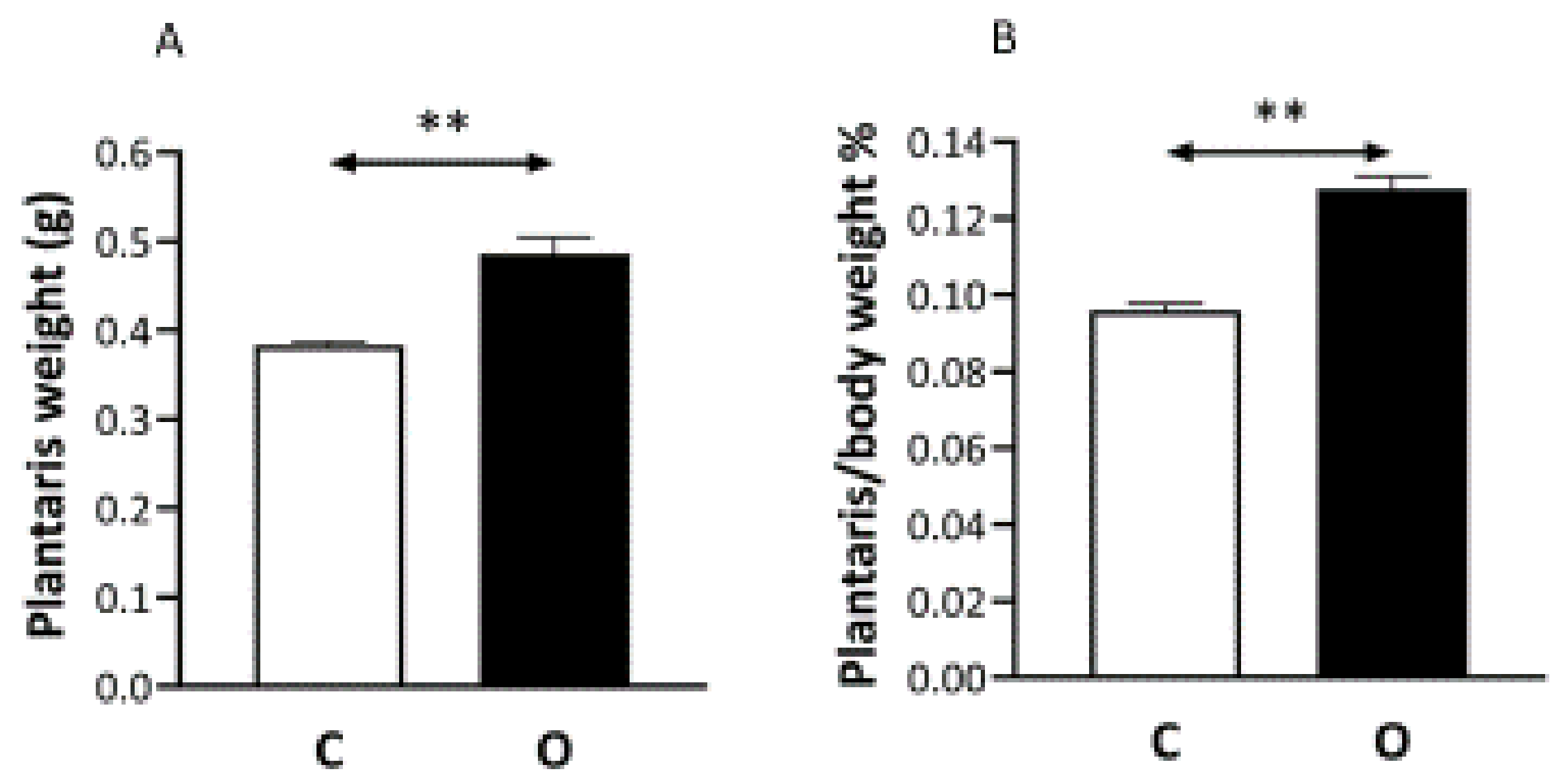
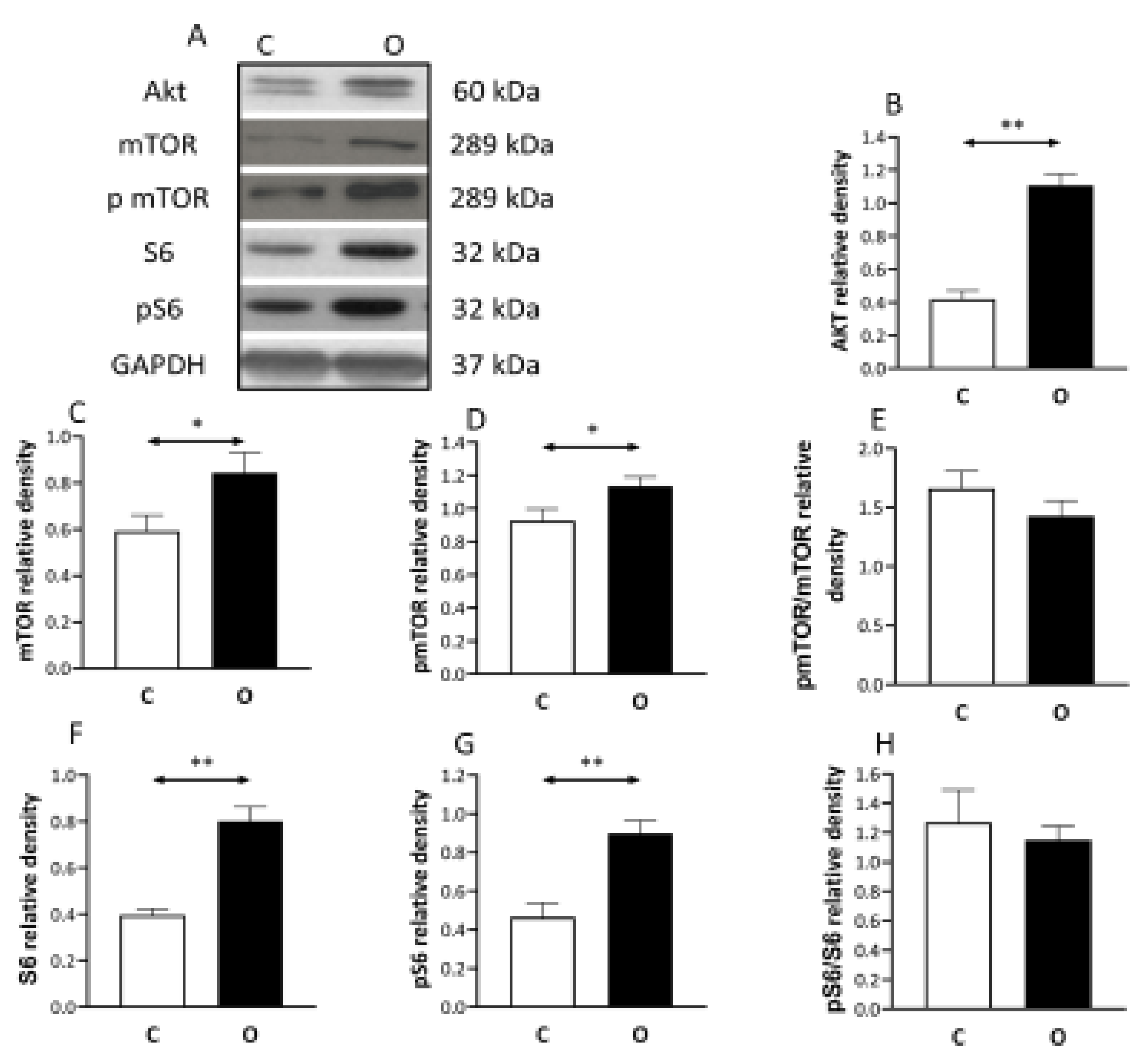
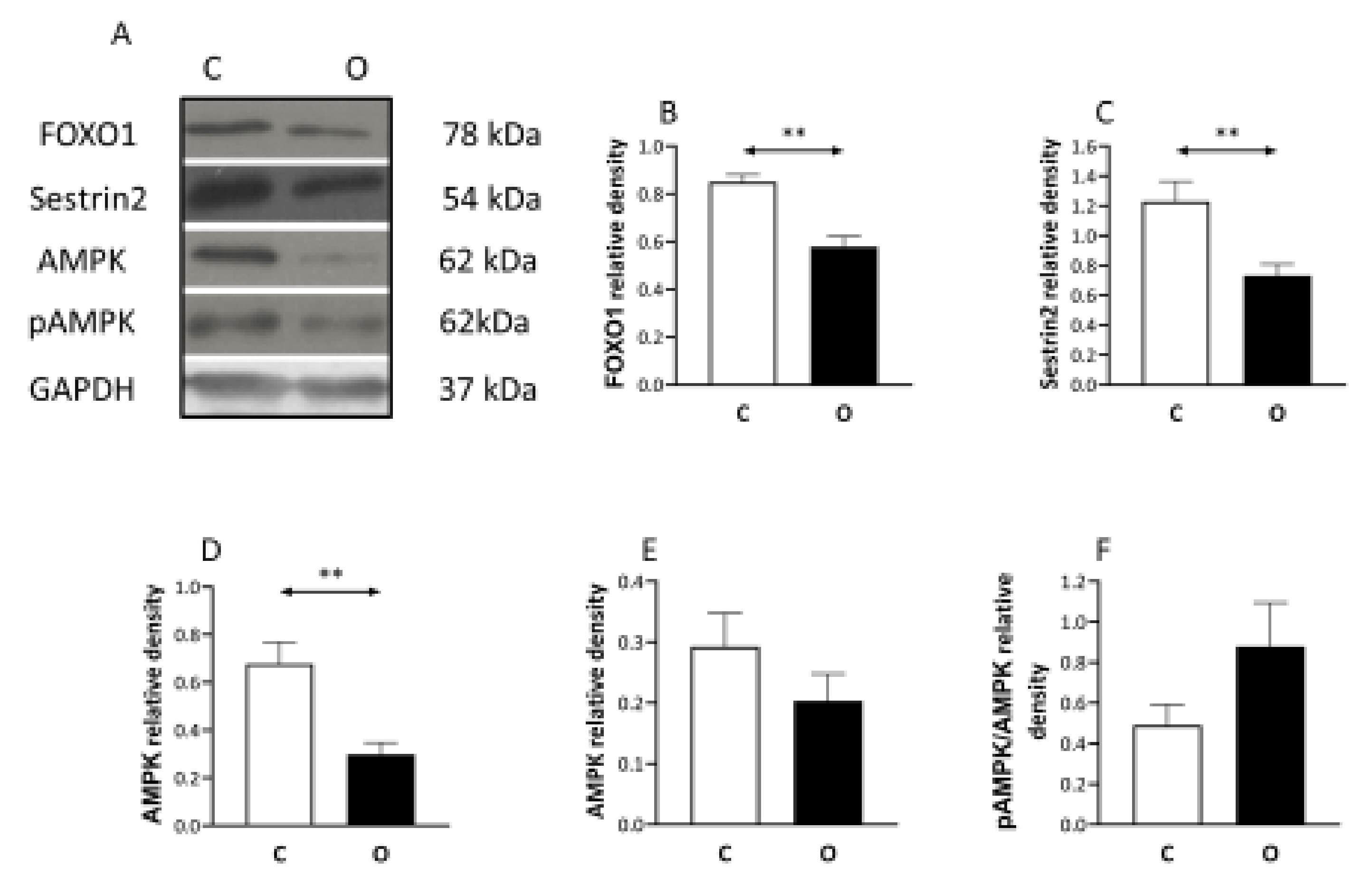
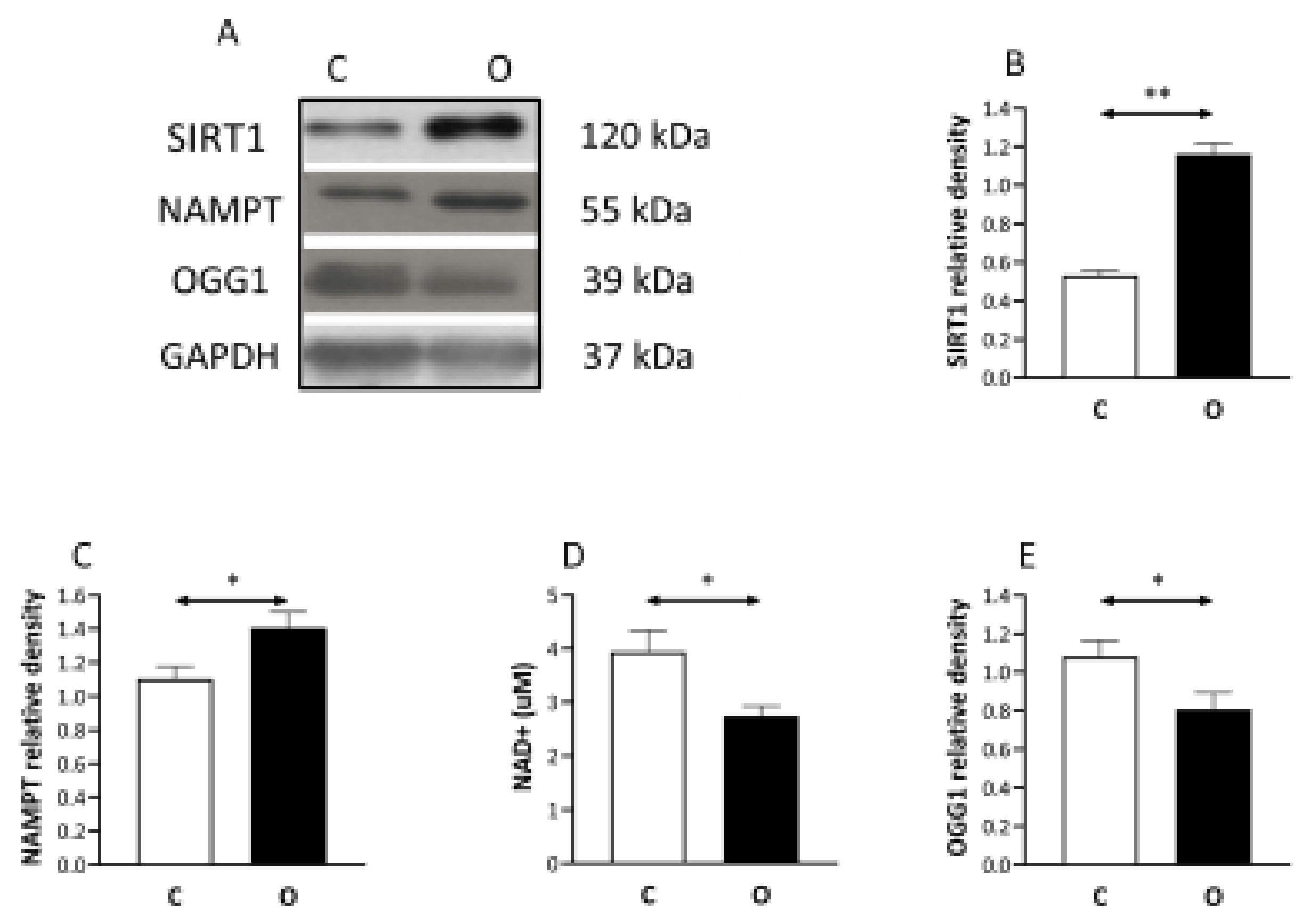
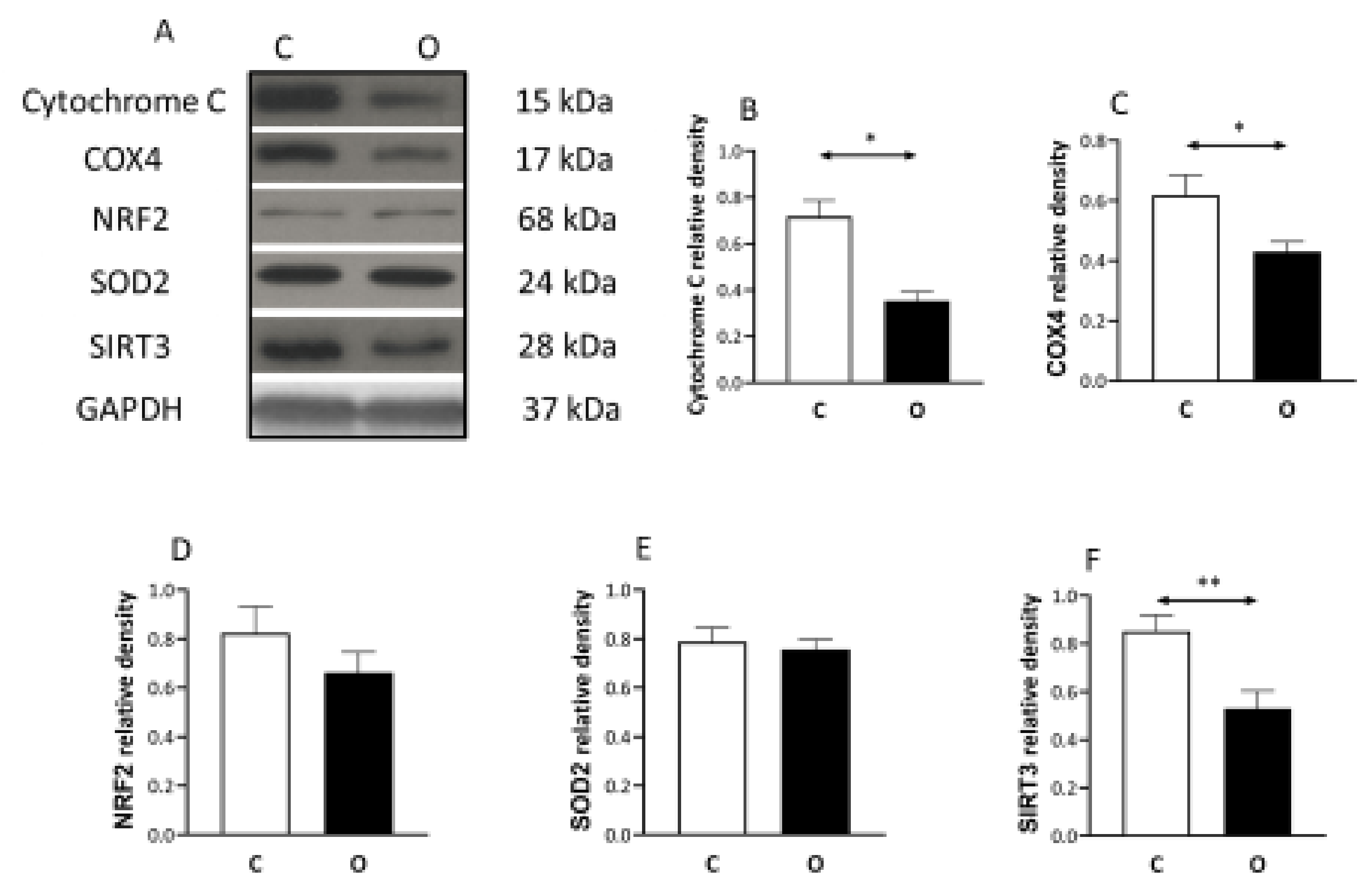
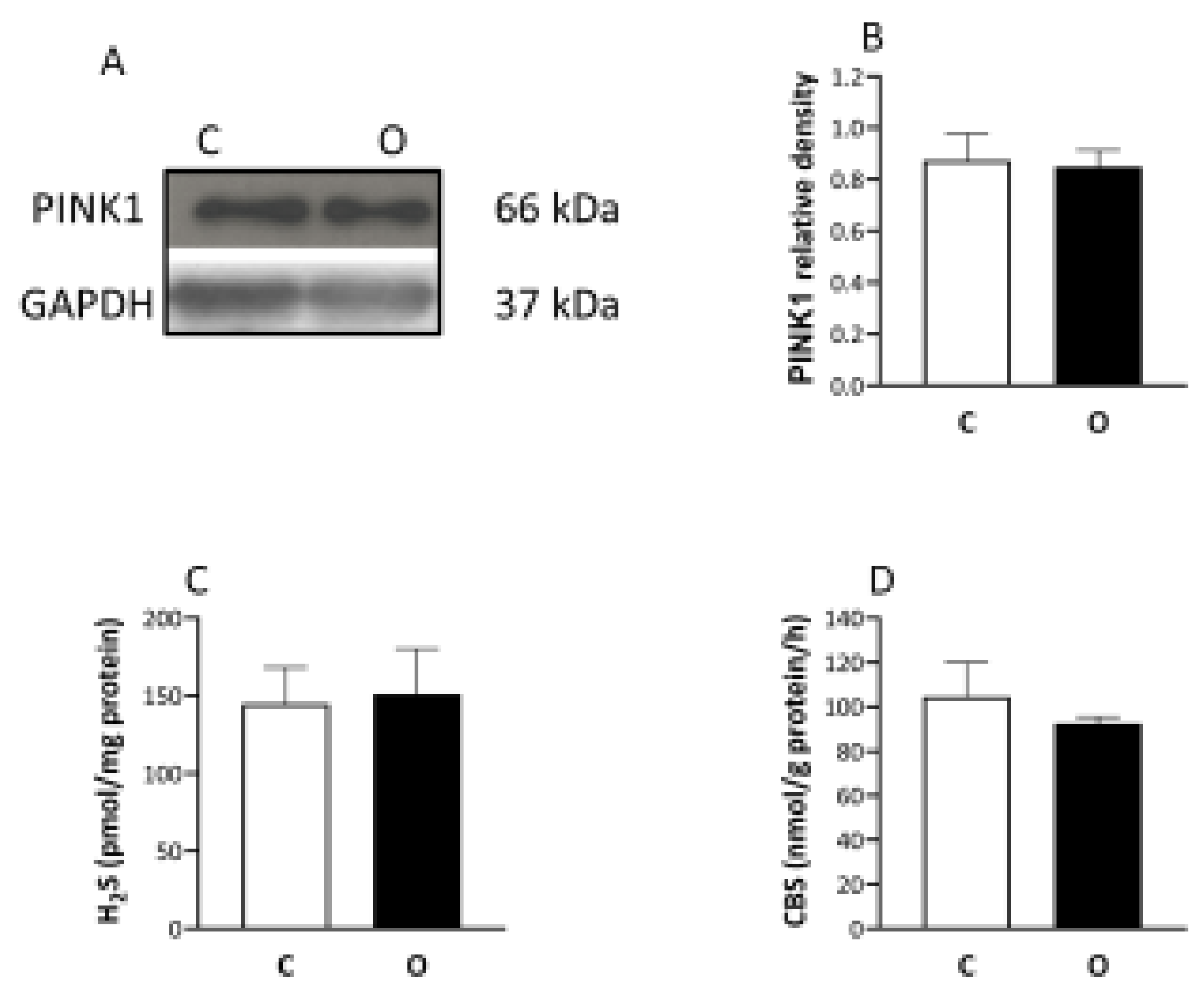
2. Results
3. Discussion
4. Methods
4.1. Animals
4.2. Synergist Muscle Ablation
4.3. NAD Measurement
4.4. Western Blots
4.5. Measurement of H2S with the Monobromobimane Method
4.6. Measurement of CBS Activity
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atherton, P.J.; Greenhaff, P.; Phillips, S.; Bodine, S.; Adams, C.; Lang, C.H. Control of skeletal muscle atrophy in response to disuse: Clinical/preclinical contentions and fallacies of evidence. Am. J. Physiol. Metab. 2016, 311, 594–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyatt, H.; Deminice, R.; Yoshihara, T.; Powers, S.K. Mitochondrial dysfunction induces muscle atrophy during prolonged inactivity: A review of the causes and effects. Arch. Biochem. Biophys. 2019, 662, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.L.; Yeo, D. Mitochondrial dysregulation and muscle disuse atrophy. F1000 Res. 2019, 8, 1621. [Google Scholar] [CrossRef]
- Ebert, S.M.; Al-Zougbi, A.; Bodine, S.C.; Adams, C.M. Skeletal Muscle Atrophy: Discovery of Mechanisms and Potential Therapies. Physiology 2019, 34, 232–239. [Google Scholar] [CrossRef]
- Cheema, B.S.; Chan, D.; Fahey, P.; Atlantis, E. Effect of progressive resistance training on measures of skeletal muscle hypertrophy, muscular strength and health-related quality of life in patients with chronic kidney disease: A systematic review and meta-analysis. Sports Med. 2014, 44, 1125–1138. [Google Scholar] [CrossRef]
- Thomson, D.M. The role of AMPK in the regulation of skeletal muscle size, hypertrophy, and regeneration. Int. J. Mol. Sci. 2018, 19, 3125. [Google Scholar] [CrossRef] [Green Version]
- Kjøbsted, R.; Hingst, J.R.; Fentz, J.; Foretz, M.; Sanz, M.; Pehmøller, C.; Shum, M.; Marette, A.; Mounier, R.; Treebak, J.T.; et al. AMPK in skeletal muscle function and metabolism. FASEB J. 2018, 32, 1741–1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koltai, E.; Bori, Z.; Chabert, C.; Dubouchaud, H.; Naito, H.; Machida, S.; Davies, K.J.; Murlasits, Z.; Fry, A.C.; Boldogh, I.; et al. SIRT1 may play a crucial role in overload-induced hypertrophy of skeletal muscle. J. Physiol. 2017, 595, 3361–3376. [Google Scholar] [CrossRef]
- Makanae, Y.; Kawada, S.; Sasaki, K.; Nakazato, K.; Ishii, N. Vitamin C administration attenuates overload-induced skeletal muscle hypertrophy in rats. Acta Physiol. 2012, 208, 57–65. [Google Scholar] [CrossRef]
- Novak, M.L.; Billich, W.; Smith, S.M.; Sukhija, K.B.; McLoughlin, T.J.; Hornberger, T.A.; Koh, T.J. COX-2 inhibitor reduces skeletal muscle hypertrophy in mice. Am. J. Physiol. Integr. Comp. Physiol. 2009, 296, 1132–1139. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.S.L.; Cui, W. Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development 2016, 143, 3050–3060. [Google Scholar] [CrossRef] [Green Version]
- Timmer, L.T.; Hoogaars, W.M.H.; Jaspers, R.T. The role of IGF-1 signaling in skeletal muscle atrophy. Adv. Exp. Med. Biol. 2018, 1088, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Budanov, A.V.; Park, E.J.; Birse, R.; Kim, T.E.; Perkins, G.A.; Ocorr, K.; Ellisman, M.H.; Bodmer, R.; Bier, E.; et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 2010, 327, 1223–1228. [Google Scholar] [CrossRef] [Green Version]
- Budanov, A.V.; Sablina, A.A.; Feinstein, E.; Koonin, E.V.; Chumakov, P. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 2004, 304, 596–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budanov, A.V.; Karin, M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Tang, Z.-H.; Ren, Z.; Qu, S.-L.; Liu, M.-H.; Liu, L.-S.; Jiang, Z.-S. Hydrogen sulfide, the next potent preventive and therapeutic agent in aging and age-associated diseases. Mol. Cell. Biol. 2013, 33, 1104–1113. [Google Scholar] [CrossRef] [Green Version]
- Nagy, P. Mechanistic chemical perspective of hydrogen sulfide signaling. Methods Enzymol. 2015, 554, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Dóka, É.; Ida, T.; Dagnell, M.; Abiko, Y.; Luong, N.C.; Balog, N.; Takata, T.; Espinosa, B.; Nishimura, A.; Cheng, Q.; et al. Control of protein function through oxidation and reduction of persulfidated states. Sci. Adv. 2020, 6, 8358. [Google Scholar] [CrossRef] [Green Version]
- Shang, Z.; Lu, C.; Chen, S.; Hua, L.; Qian, R. Effect of H2S on the circadian rhythm of mouse hepatocytes. Lipids Health Dis. 2012, 11, 23. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Goldberg, A.L. SIRT1 protein, by blocking the activities of transcription factors FoxO1 and FoxO3, inhibits muscle atrophy and promotes muscle growth. J. Biol. Chem. 2013, 288, 30515–30526. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.H.; Sung, S.H.; Oh, S.Y.; Lim, J.M.; Lee, S.K.; Park, Y.N.; Lee, H.E.; Kang, D.; Rhee, S.G. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013, 17, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.-J.; Kim, J.W.; Chung, H.S.; Seo, J.A.; Kim, S.G.; Kim, N.H.; Choi, K.M.; Baik, S.H.; Yoo, H.J. Knockdown of sestrin2 increases lipopolysaccharide-induced oxidative stress, apoptosis, and fibrotic reactions in H9c2 cells and heart tissues of mice via an AMPK-dependent mechanism. Mediat. Inflamm. 2018, 2018, 6209140. [Google Scholar] [CrossRef] [Green Version]
- Koltai, E.; Bori, Z.; Osvath, P.; Ihasz, F.; Peter, S.; Toth, G.; Degens, H.; Rittweger, J.; Boldogh, I.; Radak, Z. Master athletes have higher miR-7, SIRT3 and SOD2 expression in skeletal muscle than age-matched sedentary controls. Redox Biol. 2018, 19, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Sujkowski, A.; Namkoong, S.; Gu, B.; Cobb, T.; Kim, B.; Kowalsky, A.H.; Cho, C.-S.; Semple, I.; Ro, S.-H.; et al. Sestrins are evolutionarily conserved mediators of exercise benefits. Nat. Commun. 2020, 11, 190. [Google Scholar] [CrossRef]
- Uemichi, K.; Shirai, T.; Hanakita, H.; Takemasa, T. Effect of mechanistic/mammalian target of rapamycin complex 1 on mitochondrial dynamics during skeletal muscle hypertrophy. Physiol. Rep. 2021, 9, 14789. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.H.; Yang, J.H.; Shin, B.Y.; Seo, K.; Shin, S.M.; Cho, I.J.; Ki, S.H. Resveratrol inhibits LXRα-dependent hepatic lipogenesis through novel antioxidant Sestrin2 gene induction. Toxicol. Appl. Pharmacol. 2013, 271, 95–105. [Google Scholar] [CrossRef]
- Çelik, H.; Karahan, H.; Kelicen-Uğur, P. Effect of atorvastatin on Aβ1–42-induced alteration of SESN2, SIRT1, LC3II and TPP1 protein expressions in neuronal cell cultures. J. Pharm. Pharmacol. 2020, 72, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Tsilogianni, Z.; Baker, J.R.; Papaporfyriou, A.; Papaioannou, A.I.; Papathanasiou, E.; Koulouris, N.G.; Daly, L.; Ito, K.; Hillas, G.; Papiris, S.; et al. Sirtuin 1: Endocan and sestrin 2 in different biological samples in patients with asthma. Does severity make the difference? J. Clin. Med. 2020, 9, 473. [Google Scholar] [CrossRef] [Green Version]
- Zeng, N.; D’Souza, R.F.; Mitchell, C.J.; Cameron-Smith, D. Sestrins are differentially expressed with age in the skeletal muscle of men: A cross-sectional analysis. Exp. Gerontol. 2018, 110, 23–34. [Google Scholar] [CrossRef]
- Zeng, N.; D’Souza, R.F.; MacRae, C.L.; Figueiredo, V.C.; Pileggi, C.A.; Markworth, J.F.; Merry, T.L.; Cameron-Smith, D.; Mitchell, C.J. Daily protein supplementation attenuates immobilization-induced blunting of postabsorptive muscle mTORC1 activation in middle-aged men. Am. J. Physiol. Physiol. 2021, 320, 591–601. [Google Scholar] [CrossRef]
- Radak, Z.; Koltai, E.; Taylor, A.W.; Higuchi, M.; Kumagai, S.; Ohno, H.; Goto, S.; Boldogh, I. Redox-regulating sirtuins in aging, caloric restriction, and exercise. Free Radic. Biol. Med. 2013, 58, 87–97. [Google Scholar] [CrossRef]
- Cohen, H.Y.; Miller, C.; Bitterman, K.J.; Wall, N.; Hekking, B.; Kessler, B.; Howitz, K.T.; Gorospe, M.; de Cabo, R.; Sinclair, D. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 2004, 305, 390–392. [Google Scholar] [CrossRef] [Green Version]
- Hine, C.; Harputlugil, E.; Zhang, Y.; Ruckenstuhl, C.; Lee, B.C.; Brace, L.; Longchamp, A.; Treviño-Villarreal, J.H.; Mejia, P.; Ozaki, C.K.; et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 2015, 160, 132–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabo, C.; Papapetropoulos, A. International union of basic and clinical pharmacology. CII: Pharmacological modulation of H2S Levels: H2S donors and H2S biosynthesis inhibitors. Pharmacol. Rev. 2017, 69, 497–564. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Li, K.; Wang, H.; Jiao, H.; Wang, X.; Zhao, J.; Lin, H. Endogenous CSE/hydrogen sulfide system regulates the effects of glucocorticoids and insulin on muscle protein synthesis. Oxidative Med. Cell. Longev. 2019, 2019, 9752698. [Google Scholar] [CrossRef]
- Du, C.; Lin, X.; Xu, W.; Zheng, F.; Cai, J.; Yang, J.; Cui, Q.; Tang, C.; Cai, J.; Xu, G.; et al. Sulfhydrated sirtuin-1 increasing its deacetylation activity is an essential epigenetics mechanism of anti-atherogenesis by hydrogen sulfide. Antioxid. Redox Signal. 2019, 30, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Weichhart, T. mTOR as regulator of lifespan, aging, and cellular senescence: A mini-review. Gerontology 2018, 64, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.J.; Candau, R.; Bernardi, H. FoxO transcription factors: Their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci. 2013, 71, 1657–1671. [Google Scholar] [CrossRef] [PubMed]
- Seabright, A.P.; Lai, Y.-C. Regulatory roles of PINK1-parkin and AMPK in ubiquitin-dependent skeletal muscle mitophagy. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Qiao, A.; Wang, K.; Yuan, Y.; Guan, Y.; Ren, X.; Li, L.; Chen, X.; Li, F.; Chen, A.F.; Zhou, J.; et al. Correction: Sirt3-mediated mitophagy protects tumor cells against apoptosis under hypoxia. Oncotarget 2018, 9, 27318. [Google Scholar] [CrossRef] [PubMed]
- Ditrói, T.; Nagy, A.; Martinelli, D.; Rosta, A.; Kožich, V.; Nagy, P. Comprehensive analysis of how experimental parameters affect H2S measurements by the monobromobimane method. Free Radic. Biol. Med. 2019, 136, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Krijt, J.; Kopecká, J.; Hnizda, A.; Moat, S.; Kluijtmans, L.A.J.; Mayne, P.; Kožich, V. Determination of cystathionine beta-synthase activity in human plasma by LC-MS/MS: Potential use in diagnosis of CBS deficiency. J. Inherit. Metab. Dis. 2011, 34, 49–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Time (min) | Solvent B% |
|---|---|
| 0 | 15 |
| 4762 | 35 |
| 14,178 | 35 |
| 17,328 | 90 |
| 18,904 | 90 |
| 20,479 | 15 |
| 23,629 | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gombos, Z.; Koltai, E.; Torma, F.; Bakonyi, P.; Kolonics, A.; Aczel, D.; Ditroi, T.; Nagy, P.; Kawamura, T.; Radak, Z. Hypertrophy of Rat Skeletal Muscle Is Associated with Increased SIRT1/Akt/mTOR/S6 and Suppressed Sestrin2/SIRT3/FOXO1 Levels. Int. J. Mol. Sci. 2021, 22, 7588. https://doi.org/10.3390/ijms22147588
Gombos Z, Koltai E, Torma F, Bakonyi P, Kolonics A, Aczel D, Ditroi T, Nagy P, Kawamura T, Radak Z. Hypertrophy of Rat Skeletal Muscle Is Associated with Increased SIRT1/Akt/mTOR/S6 and Suppressed Sestrin2/SIRT3/FOXO1 Levels. International Journal of Molecular Sciences. 2021; 22(14):7588. https://doi.org/10.3390/ijms22147588
Chicago/Turabian StyleGombos, Zoltan, Erika Koltai, Ferenc Torma, Peter Bakonyi, Attila Kolonics, Dora Aczel, Tamas Ditroi, Peter Nagy, Takuji Kawamura, and Zsolt Radak. 2021. "Hypertrophy of Rat Skeletal Muscle Is Associated with Increased SIRT1/Akt/mTOR/S6 and Suppressed Sestrin2/SIRT3/FOXO1 Levels" International Journal of Molecular Sciences 22, no. 14: 7588. https://doi.org/10.3390/ijms22147588
APA StyleGombos, Z., Koltai, E., Torma, F., Bakonyi, P., Kolonics, A., Aczel, D., Ditroi, T., Nagy, P., Kawamura, T., & Radak, Z. (2021). Hypertrophy of Rat Skeletal Muscle Is Associated with Increased SIRT1/Akt/mTOR/S6 and Suppressed Sestrin2/SIRT3/FOXO1 Levels. International Journal of Molecular Sciences, 22(14), 7588. https://doi.org/10.3390/ijms22147588







