Antifungal Activity and Biosynthetic Potential of New Streptomyces sp. MW-W600-10 Strain Isolated from Coal Mine Water
Abstract
:1. Introduction
2. Results
2.1. Isolation of Streptomyces sp. MW-W600-10 Strain
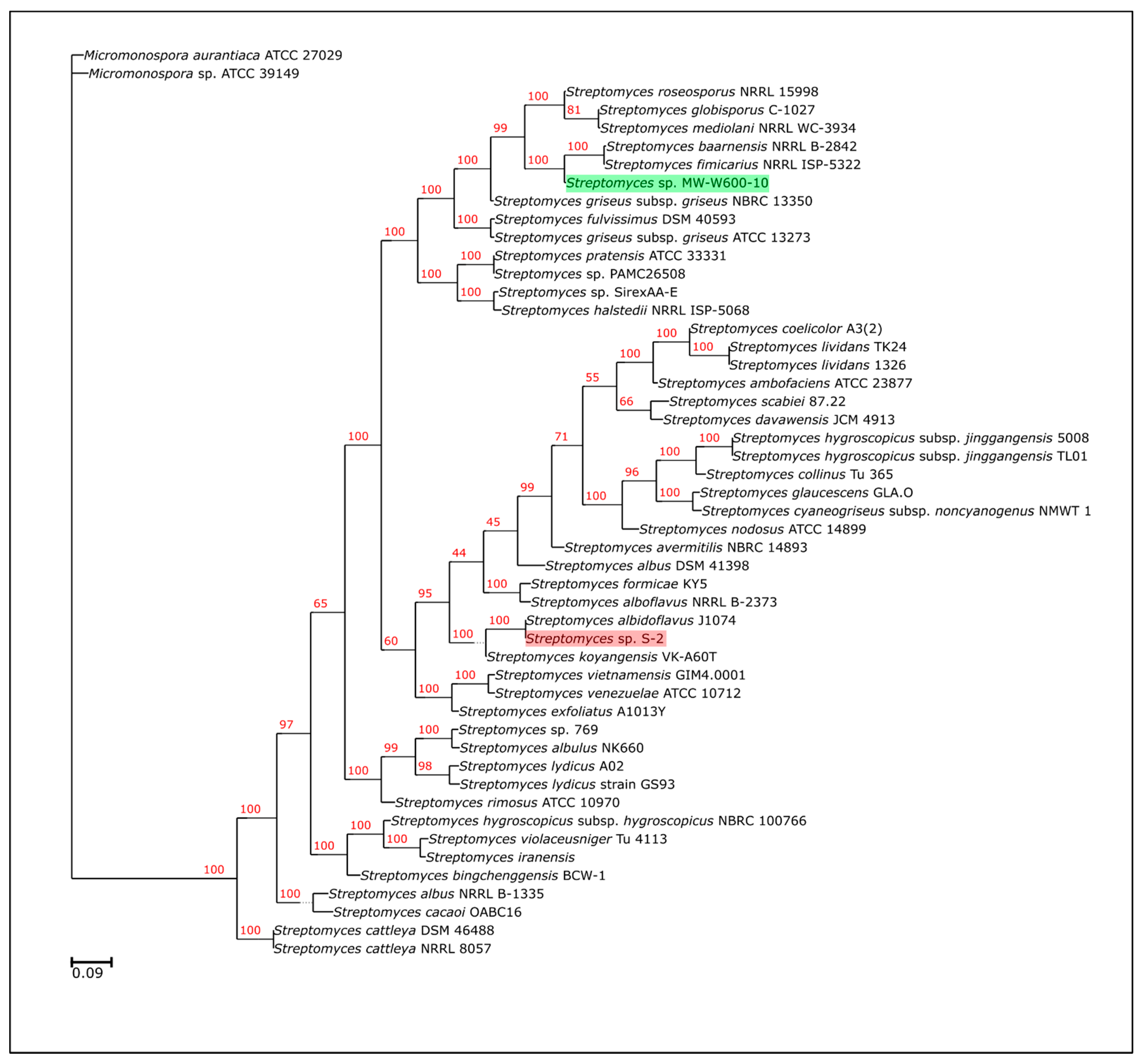
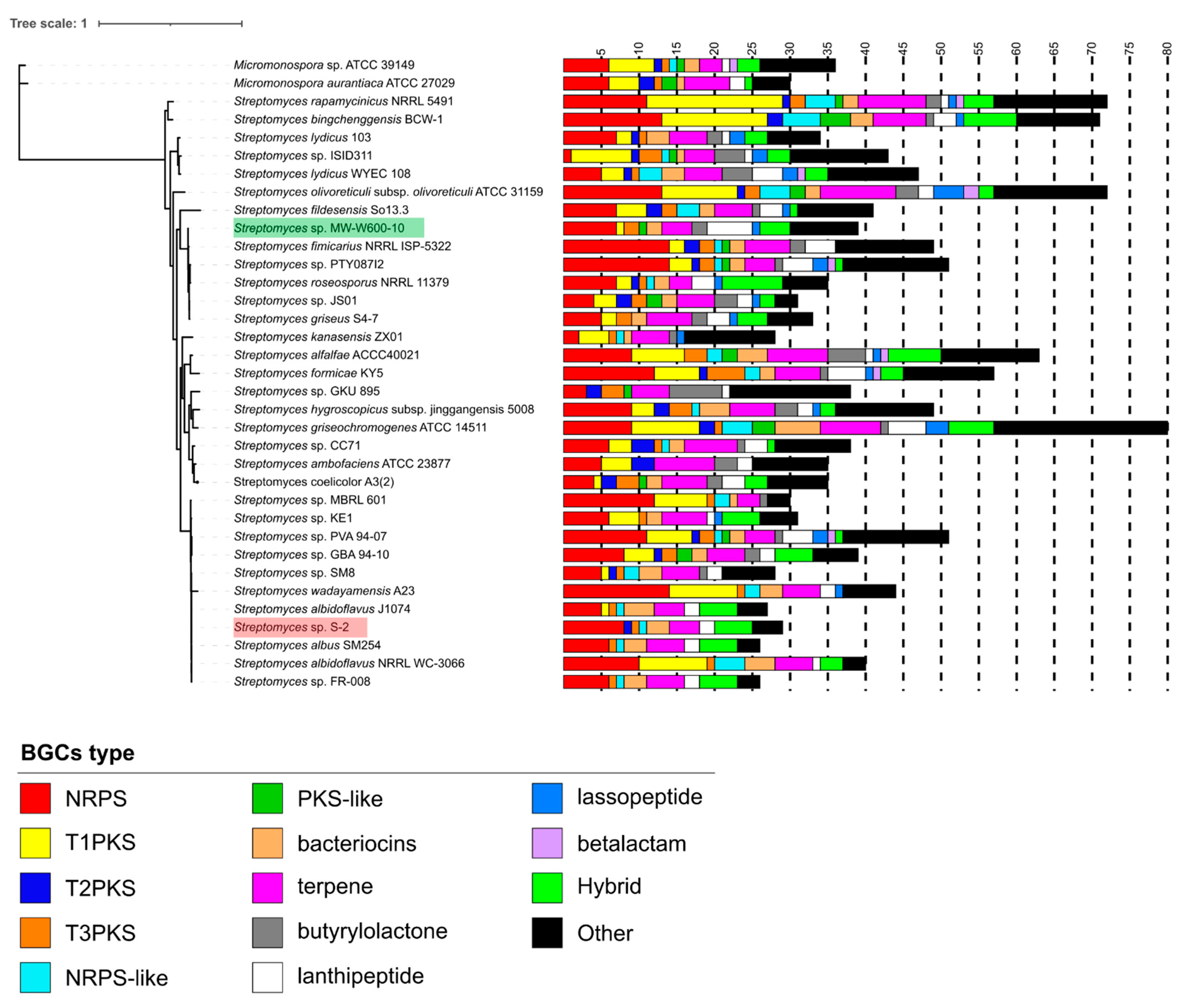
2.2. Genome Sequencing and Phylogeny of Streptomyces sp. MW-W600-10
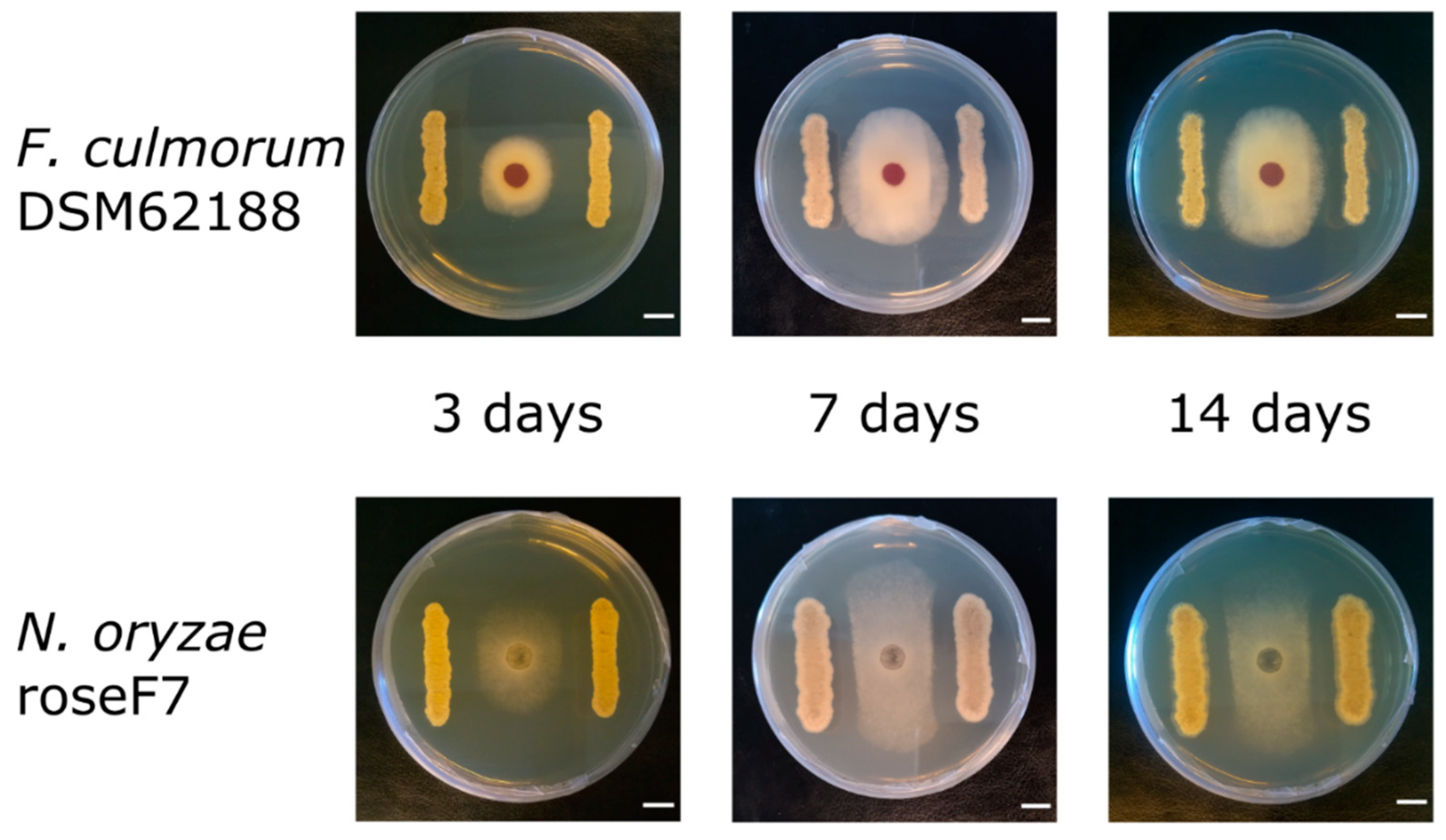
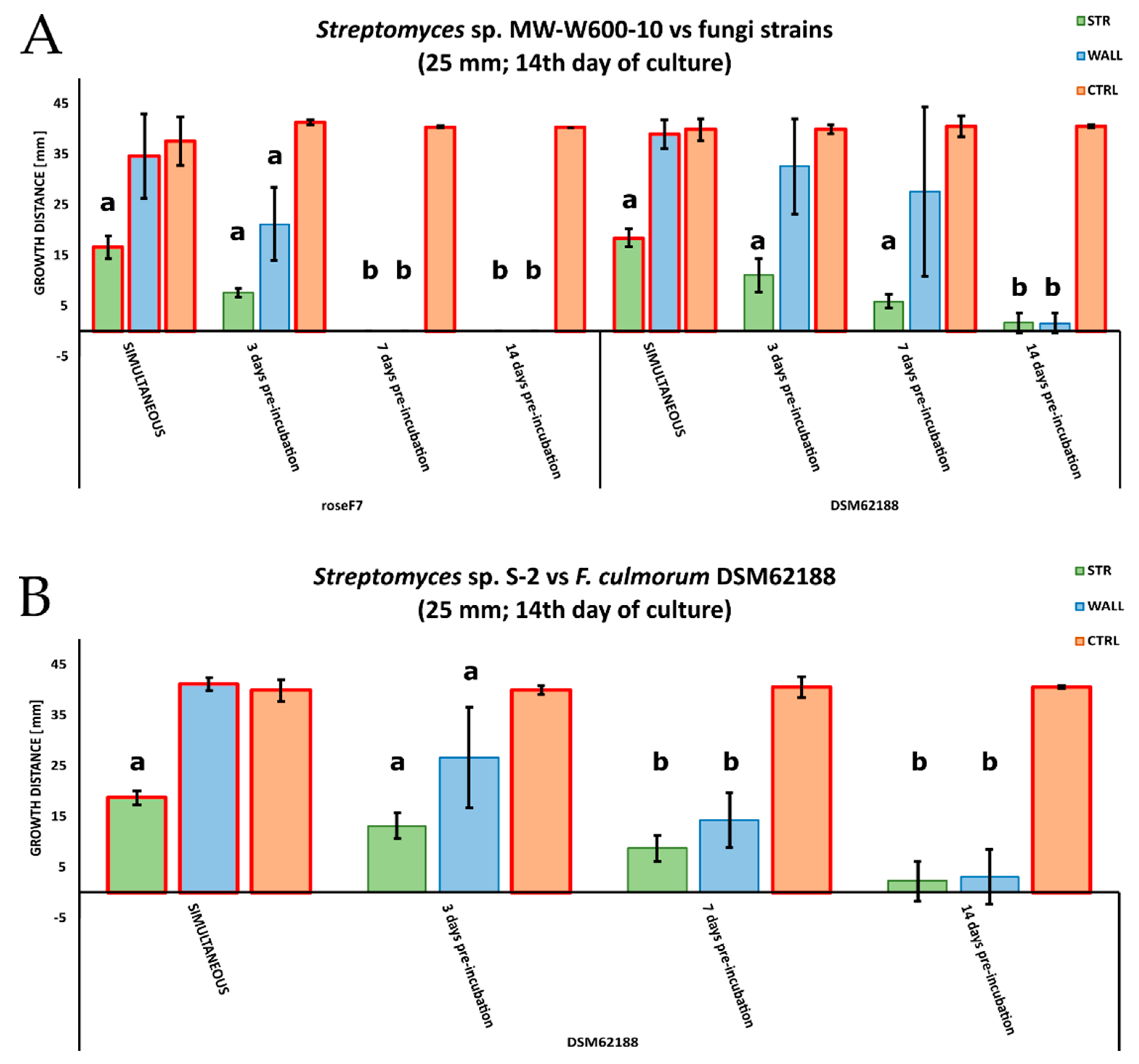
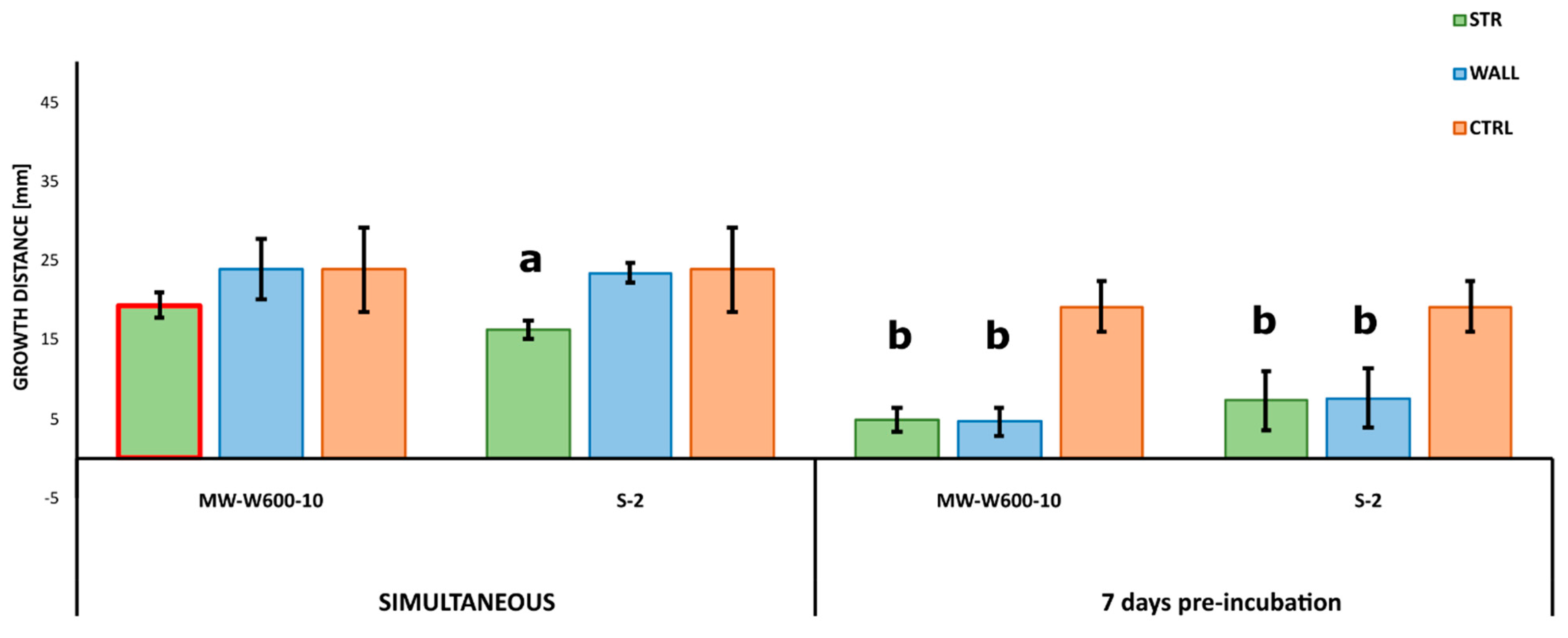
2.3. Antifungal Activities of Streptomyces from CRE
2.4. Putative Biosynthetic Gene Clusters Prediction in the Streptomyces sp. MW-W600-10 Strain
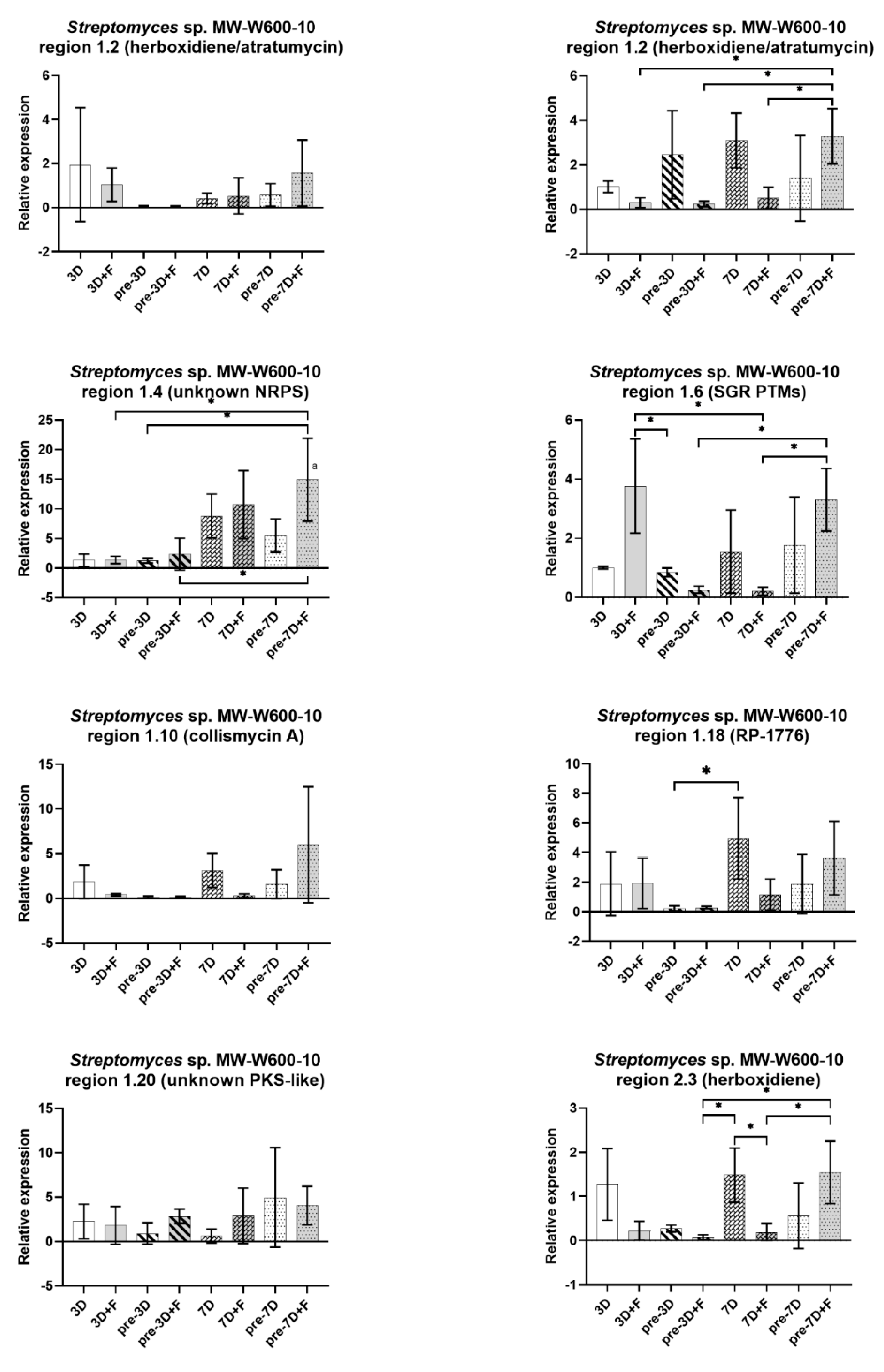
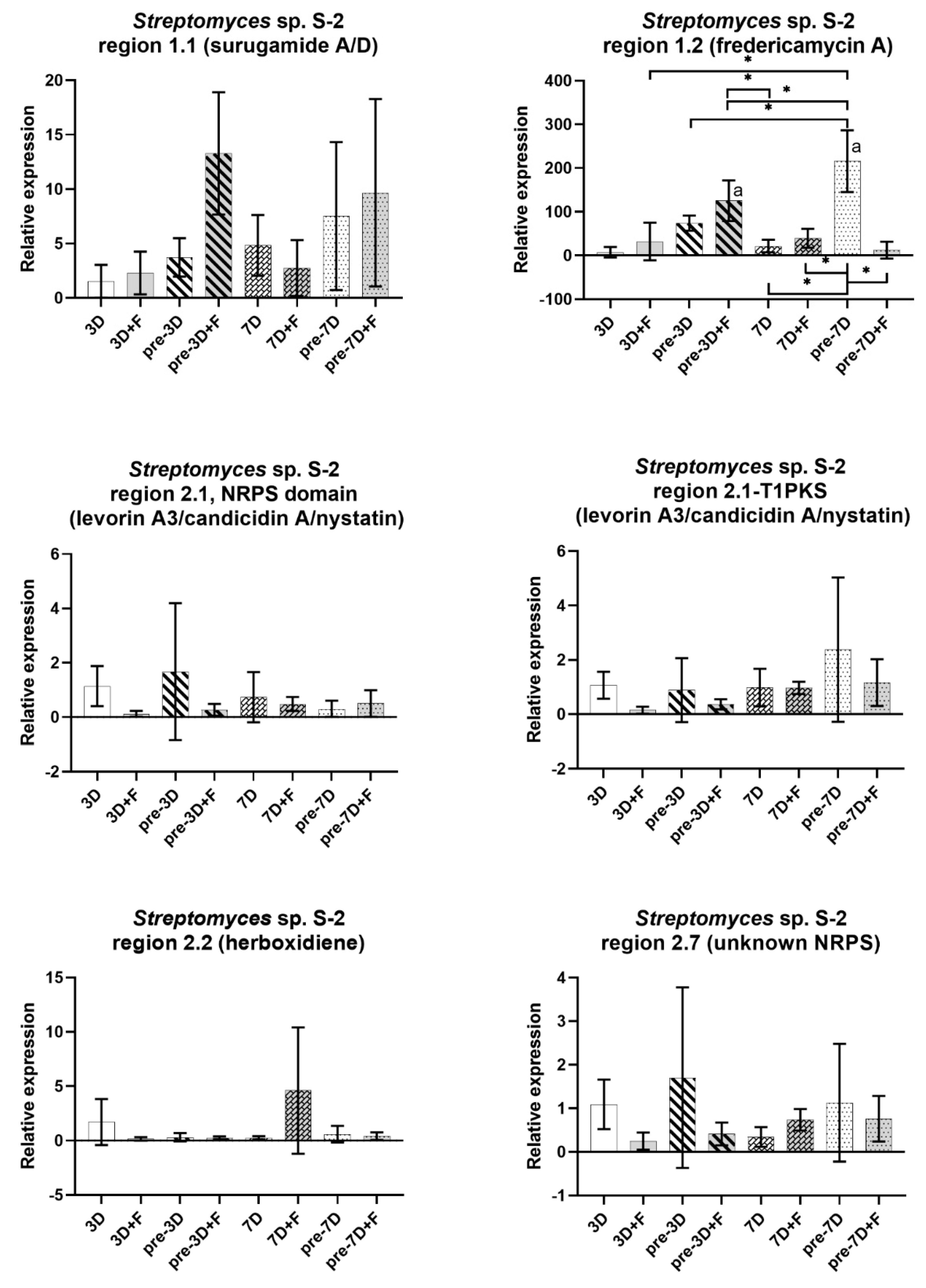
2.5. Expression of BGCs in Streptomyces from CRE
3. Discussion
4. Materials and Methods
4.1. Isolation and Culture of the Strains
4.2. Whole-Genome Sequencing of the MW-W600-10 Strain and Phylogeny
4.3. Putative Biosynthetic Gene Clusters Analysis
4.4. Antifungal Assay
4.5. RNA Isolation and RT-qPCR
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [Green Version]
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef]
- Feng, J.; Hwang, R.; Chang, K.F.; Hwang, S.F.; Strelkov, S.E.; Gossen, B.D.; Conner, R.L.; Turnbull, G.D. Genetic variation in Fusarium avenaceum causing root rot on field pea. Plant Pathol. 2010, 59, 845–852. [Google Scholar] [CrossRef]
- Scherm, B.; Balmas, V.; Spanu, F.; Pani, G.; Delogu, G.; Pasquali, M.; Migheli, Q. Fusarium culmorum: Causal agent of foot and root rot and head blight on wheat. Mol. Plant Pathol. 2013, 14, 323–341. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Bubici, G.; Kaushal, M.; Prigigallo, M.I.; Cabanás, C.G.L.; Mercado-Blanco, J. Biological control agents against Fusarium wilt of banana. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, T.; Zhang, M.; Zhou, D.; Jing, T.; Zang, X.; Qi, D.; Chen, Y.; Li, K.; Zhao, Y.; Tang, W.; et al. Anti-Foc RT4 Activity of a Newly Isolated Streptomyces sp. 5–10 From a Medicinal Plant (Curculigo capitulata). Front. Microbiol. 2021, 11, 5–10. [Google Scholar] [CrossRef]
- Pasqualotto, A.C. Differences in pathogenicity and clinical syndromes due to Aspergillus fumigatus and Aspergillus flavus. Med. Mycol. 2009, 47, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Nelson, P.E.; Dignani, M.C.; Anaissie, E.J. Taxonomy, biology, and clinical aspects of Fusarium species. Clin. Microbiol. Rev. 1994, 7, 479–504. [Google Scholar] [CrossRef]
- Castagnoli, E.; Marik, T.; Mikkola, R.; Kredics, L.; Andersson, M.A.; Salonen, H.; Kurnitski, J. Indoor Trichoderma strains emitting peptaibols in guttation droplets. J. Appl. Microbiol. 2018, 125, 1408–1422. [Google Scholar] [CrossRef] [Green Version]
- El-Nekeety, A.A.; Salman, A.S.; Hathout, A.S.; Sabry, B.A.; Abdel-Aziem, S.H.; Hassan, N.S.; Abdel-Wahhab, M.A. Evaluation of the bioactive extract of actinomyces isolated from the Egyptian environment against aflatoxin B1-induce cytotoxicity, genotoxicity and oxidative stress in the liver of rats. Food Chem. Toxicol. 2017, 105, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Maresca, M. From the gut to the brain: Journey and pathophysiological effects of the food-associated trichothecene mycotoxin deoxynivalenol. Toxins 2013, 5, 784–820. [Google Scholar] [CrossRef]
- Bardin, M.; Ajouz, S.; Comby, M.; Lopez-Ferber, M.; Graillot, B.; Siegwart, M.; Nicot, P.C. Is the efficacy of biological control against plant diseases likely to be more durable than that of chemical pesticides? Front. Plant Sci. 2015, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80. [Google Scholar] [CrossRef] [Green Version]
- Hasani, A.; Kariminik, A.; Isaazadeh, K. Streptomycetes: Characteristics and Their Antimicrobial Activities. Int. J. Adv. Biol. Biomed. Res. 2014, 2, 63–75. [Google Scholar]
- Berdi, J. Bioactive microbial metabolites. J. Antibiot. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stulberg, E.R.; Lozano, G.L.; Morin, J.B.; Park, H.; Baraban, E.G.; Mlot, C.; Heffelfinger, C.; Phillips, G.M.; Rush, J.S.; Phillips, A.J.; et al. Genomic and secondary metabolite analyses of Streptomyces sp. 2AW provide insight into the evolution of the cycloheximide pathway. Front. Microbiol. 2016, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Watve, M.G.; Tickoo, R.; Jog, M.M.; Bhole, B.D. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Natural products discovery and potential for new antibiotics. Curr. Opin. Microbiol. 2019, 51, 81–87. [Google Scholar] [CrossRef]
- Dewi, T.K.; Agustiani, D.; Antonius, S. Secondary Metabolites Production by Actinomycetes and their Antifungal Activity. KnE Life Sci. 2017, 3, 256. [Google Scholar] [CrossRef] [Green Version]
- Human, Z.R.; Moon, K.; Bae, M.; Wilhelm de Beer, Z.; Cha, S.; Wingfield, M.J.; Slippers, B.; Oh, D.C.; Venter, S.N. Antifungal Streptomyces spp. associated with the infructescences of Protea spp. in South Africa. Front. Microbiol. 2016, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.F.; Ser, H.L.; Khan, T.M.; Chuah, L.H.; Pusparajah, P.; Chan, K.G.; Goh, B.H.; Lee, L.H. The potential of Streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, D.; Zou, L.; Zhou, D.; Chen, Y.; Gao, Z.; Feng, R.; Zhang, M.; Li, K.; Xie, J.; Wang, W. Taxonomy and broad-spectrum antifungal activity of Streptomyces sp. SCA3-4 isolated from rhizosphere soil of opuntia stricta. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Koren, S.; Harhay, G.P.; Smith, T.P.L.; Bono, J.L.; Harhay, D.M.; Mcvey, S.D.; Radune, D.; Bergman, N.H.; Phillippy, A.M. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013, 14. [Google Scholar] [CrossRef]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef]
- Laver, T.; Harrison, J.; O’Neill, P.A.; Moore, K.; Farbos, A.; Paszkiewicz, K.; Studholme, D.J. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 2015, 3, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, C.; Miller, S. Next-Generation Sequencing. In Molecular Microbiology: Diagnostic Principles and Practice, 3rd ed.; Persing, D.H., Tenover, F.C., Hayden, R.T., Ieven, M., Miller, M.B., Nolte, F.S., Tang, Y.-W., van Belkum, A., Eds.; ASM Press: Washington, DC, USA, 2016; pp. 68–79. ISBN 9781555819071. [Google Scholar]
- Chen, Z.; Erickson, D.L.; Meng, J. Benchmarking long-read assemblers for genomic analyses of bacterial pathogens using oxford nanopore sequencing. Int. J. Mol. Sci. 2020, 21, 9161. [Google Scholar] [CrossRef]
- Pechtl, A.; Rückert, C.; Maus, I.; Koeck, D.E.; Trushina, N.; Kornberger, P.; Schwarz, W.H.; Schlüter, A.; Liebl, W.; Zverlov, V. Complete Genome Sequence of the Novel Cellulolytic, Established by Illumina and Nanopore MinION Sequencing. Genome Announc. 2018, 6, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Kranz, A.; Vogel, A.; Degner, U.; Kiefler, I.; Bott, M.; Usadel, B.; Polen, T. High precision genome sequencing of engineered Gluconobacter oxydans 621H by combining long nanopore and short accurate Illumina reads. J. Biotechnol. 2017, 258, 197–205. [Google Scholar] [CrossRef]
- Barajas, H.R.; Romero, M.F.; Martínez-Sánchez, S.; Alcaraz, L.D. Global genomic similarity and core genome sequence diversity of the Streptococcus genus as a toolkit to identify closely related bacterial species in complex environments. PeerJ 2019, 2019. [Google Scholar] [CrossRef] [Green Version]
- Terra, L.; Dyson, P.J.; Hitchings, M.D.; Thomas, L.; Abdelhameed, A.; Banat, I.M.; Gazze, S.A.; Vujaklija, D.; Facey, P.D.; Francis, L.W.; et al. A novel alkaliphilic Streptomyces inhibits ESKAPE pathogens. Front. Microbiol. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Almeida, E.L.; Carrillo Rincón, A.F.; Jackson, S.A.; Dobson, A.D.W. Comparative Genomics of Marine Sponge-Derived Streptomyces spp. Isolates SM17 and SM18 With Their Closest Terrestrial Relatives Provides Novel Insights Into Environmental Niche Adaptations and Secondary Metabolite Biosynthesis Potential. Front. Microbiol. 2019, 10, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Vicente, C.M.; Thibessard, A.; Lorenzi, J.N.; Benhadj, M.; Hôtel, L.; Gacemi-Kirane, D.; Lespinet, O.; Leblond, P.; Aigle, B. Comparative genomics among closely related Streptomyces strains revealed specialized metabolite biosynthetic gene cluster diversity. Antibiotics 2018, 7, 86. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Zhang, Z.; Yang, T.; Chen, M.; Li, J.; Chen, F.; Yang, J.; Li, W.; Zhang, B.; Zhang, Z.; et al. Comparative genomics analysis of Streptomyces species reveals their adaptation to the marine environment and their diversity at the genomic level. Front. Microbiol. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Takahashi, Y.; Ōmura, S. Search for new compounds from Kitasato microbial library by physicochemical screening. Biochem. Pharmacol. 2017, 134, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, S.; Onaka, H.; Abe, I. Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in Actinomycetes by co-culture with mycolic acid-containing bacteria. J. Ind. Microbiol. Biotechnol. 2019, 46, 363–374. [Google Scholar] [CrossRef]
- Alberti, F.; Leng, D.J.; Wilkening, I.; Song, L.; Tosin, M.; Corre, C. Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery. Chem. Sci. 2019, 10, 453–463. [Google Scholar] [CrossRef] [Green Version]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of Actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014, 25–64. [Google Scholar] [CrossRef] [Green Version]
- Siupka, P.; Piński, A.; Babicka, D.; Piotrowska-Seget, Z. Genome mining revealed a high biosynthetic potential for antifungal Streptomyces sp. S-2 isolated from black soot. Int. J. Mol. Sci. 2020, 21, 2558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Núñez-Montero, K.; Lamilla, C.; Abanto, M.; Maruyama, F.; Jorquera, M.A.; Santos, A.; Martinez-Urtaza, J.; Barrientos, L. Antarctic Streptomyces fildesensis So13.3 strain as a promising source for antimicrobials discovery. Sci. Rep. 2019, 9, 7488. [Google Scholar] [CrossRef] [PubMed]
- Gosse, J.T.; Ghosh, S.; Sproule, A.; Overy, D.; Cheeptham, N.; Boddy, C.N. Whole genome sequencing and metabolomic study of cave Streptomyces isolates ICC1 and ICC4. Front. Microbiol. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Carlson, C.M.; Ortega, H.E.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, J.; Baker, P.; Piper, C.; Cotter, P.D.; Walsh, M.; Mooij, M.J.; Bourke, M.B.; Rea, M.C.; O’Connor, P.M.; Ross, R.P.; et al. Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish waters. Mar. Biotechnol. 2009, 11, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Martinet, L.; Naômé, A.; Baiwir, D.; De Pauw, E.; Mazzucchelli, G.; Rigali, S. On the risks of phylogeny-based strain prioritization for drug discovery: Streptomyces lunaelactis as a case study. Biomolecules 2020, 10, 1027. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Trujilo, M.E.; Igarashi, Y.; Tamura, T. Diversity of PKS and NRPS gene clusters between Streptomyces abyssomicinicus sp. nov. and its taxonomic neighbor. J. Antibiot. 2020, 73, 141–151. [Google Scholar] [CrossRef]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Igarashi, Y.; Tamura, T. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci. Rep. 2018, 8, 6888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.O.; Shin, K.S.; Kim, M.N.; Shin, K.S.; Labeda, D.P.; Han, J.H.; Kim, S.B. Reassessment of the status of Streptomyces setonii and reclassification of Streptomyces fimicarius as a later synonym of Streptomyces setonii and Streptomyces albovinaceus as a later synonym of Streptomyces globisporus based on combined 16s rRNA/gyrB gene. Int. J. Syst. Evol. Microbiol. 2012, 62, 2978–2985. [Google Scholar] [CrossRef] [Green Version]
- Khulana, S.; Niranjan, B. Antimicrobial activity of thermotolerant bacterial isolate from coal mine spoil. Afr. J. Microbiol. Res. 2012, 6, 5459–5463. [Google Scholar] [CrossRef]
- Strandberg, G.W.; Lewis, S.N. Solubilization of coal by an extracellular product from Streptomyces setonii 75Vi2. J. Ind. Microbiol. 1987, 1, 371–375. [Google Scholar] [CrossRef]
- Maciejewska, M.; Adam, D.; Martinet, L.; Naômé, A.; Całusińska, M.; Delfosse, P.; Carnol, M.; Barton, H.A.; Hayette, M.P.; Smargiasso, N.; et al. A phenotypic and genotypic analysis of the antimicrobial potential of cultivable Streptomyces isolated from cave moonmilk deposits. Front. Microbiol. 2016, 7, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.K.; Kang, H.J.; Roh, S.G.; Park, J.S.; Kim, S.B. Streptomyces fodineus sp. nov., an actinobacterium with antifungal activity isolated from mine area soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Getha, K.; Vikineswary, S.; Wong, W.H.; Seki, T.; Ward, A.; Goodfellow, M. Evaluation of Streptomyces sp. strain g10 for suppression of Fusarium wilt and rhizosphere colonization in pot-grown banana plantlets. J. Ind. Microbiol. Biotechnol. 2005, 32, 24–32. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Babalola, O.O. Streptomyces: Implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019, 103, 1179–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Lu, L.; Pang, J.; Hu, Y.; Guo, Q.; Li, Z.; Wu, S.; Liu, H.; Wang, C. Biocontrol activity of volatile organic compounds from Streptomyces alboflavus TD-1 against Aspergillus flavus growth and aflatoxin production. J. Microbiol. 2019, 57, 396–404. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Q.; He, F.; Li, Y.; Xue, Q.; Lai, H. Biocontrol of Root Diseases and Growth Promotion of the Tuberous Plant Aconitum carmichaelii Induced by Actinomycetes Are Related to Shifts in the Rhizosphere Microbiota. Microb. Ecol. 2019. [Google Scholar] [CrossRef]
- Mahadevan, B.; Crawford, D.L. Properties of the chitinase of the antifungal biocontrol agent Streptomyces lydicus WYEC108. Enzyme Microb. Technol. 1997, 20, 489–493. [Google Scholar] [CrossRef]
- Hong, S.W.; Kim, D.R.; Kwon, Y.S.; Kwak, Y.S. Genome-wide screening antifungal genes in Streptomyces griseus S4-7, a Fusarium wilt disease suppressive microbial agent. FEMS Microbiol. Lett. 2019, 366, 1–8. [Google Scholar] [CrossRef]
- Remali, J.; Sarmin, N.I.M.; Ng, C.L.; Tiong, J.J.L.; Aizat, W.M.; Keong, L.K.; Zin, N.M. Genomic characterization of a new endophytic Streptomyces kebangsaanensis identifies biosynthetic pathway gene clusters for novel phenazine antibiotic production. PeerJ 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Sottorff, I.; Wiese, J.; Lipfert, M.; Preußke, N.; Sönnichsen, F.D.; Imhoff, J.F. Different secondary metabolite profiles of phylogenetically almost identical streptomyces griseus strains originating from geographically remote locations. Microorganisms 2019, 7, 166. [Google Scholar] [CrossRef] [Green Version]
- Chevrette, M.G.; Currie, C.R. Emerging evolutionary paradigms in antibiotic discovery. J. Ind. Microbiol. Biotechnol. 2019, 46, 257–271. [Google Scholar] [CrossRef]
- Li, N.; Overkleeft, H.S.; Florea, B.I. Activity-based protein profiling: An enabling technology in chemical biology research. Curr. Opin. Chem. Biol. 2012, 16, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Naz, A.; Bano, K.; Bano, F.; Ghafoor, N.A.; Akhtar, N. Conformational analysis (geometry optimization) of nucleosidic antitumor antibiotic showdomycin by ArgusLab 4 software. Pak. J. Pharm. Sci. 2009, 22, 78–82. [Google Scholar] [PubMed]
- Uehara, Y.I.; Fisher, J.M.; Rabinovitz, M. Showdomycin and its reactive moiety, maleimide. Biochem. Pharmacol. 1980, 29, 2199–2204. [Google Scholar] [CrossRef]
- Thirupathi, B.; Zilla, M.K. Syntheses and Biological Importance of Herboxidiene/GEX1A. ChemistrySelect 2019, 4, 11944–11958. [Google Scholar] [CrossRef]
- Shi, J.; Liu, C.L.; Zhang, B.; Guo, W.J.; Zhu, J.; Chang, C.Y.; Zhao, E.J.; Jiao, R.H.; Tan, R.X.; Ge, H.M. Genome mining and biosynthesis of kitacinnamycins as a STING activator. Chem. Sci. 2019, 10, 4839–4846. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a Cryptic Antifungal Compound from Streptomyces albus J1074 Using High-Throughput Elicitor Screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef] [Green Version]
- Latham, M.D.; King, C.K.; Gorycki, P.; Macdonald, T.L.; Ross, W.E. Inhibition of topoisomerases by fredericamycin A. Cancer Chemother. Pharmacol. 1989, 24, 167–171. [Google Scholar] [CrossRef]
- Hosotani, N.; Kumagai, K.; Nakagawa, H.; Shimatani, T.; Saji, I. Antimycins A10–A16, seven new antimycin antibiotics produced by Streptomyces spp. SPA-10191 and SPA-8893. J. Antibiot. 2005, 58, 460–467. [Google Scholar] [CrossRef]
- Kim, J.H.; Campbell, B.C.; Chan, K.L.; Mahoney, N.; Haff, R.P. Synergism of antifungal activity between mitochondrial respiration inhibitors and kojic acid. Molecules 2013, 18, 1564–1581. [Google Scholar] [CrossRef] [PubMed]
- Lechevalier, H.; Acker, R.F.; Corke, C.T.; Haenseler, C.M.; Waksman, S.A. Candicidin, a new antifungal antibiotic. Mycologia 1953, 45, 155–171. [Google Scholar] [CrossRef]
- Khalandi, H.; Masoori, L.; Farahyar, S.; Delbandi, A.A.; Raiesi, O.; Farzanegan, A.; Khalandi, G.; Mahmoudi, S.; Erfanirad, T.; Falahati, M. Antifungal Activity of Capric Acid, Nystatin, and Fluconazole and Their in Vitro Interactions against Candida Isolates from Neonatal Oral Thrush. Assay Drug Dev. Technol. 2020, 18, 195–201. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, H.; Liang, J.; Wang, M.; Lu, L.; Shao, Z.; Cobb, R.E.; Zhao, H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 2013, 4, 2894. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Ye, K.X.; Dai, W.H.; Sun, C.; Xu, L.H.; Han, B.N. Comparative genomic insights into secondary metabolism biosynthetic gene cluster distributions of marine Streptomyces. Mar. Drugs 2019, 17, 498. [Google Scholar] [CrossRef] [Green Version]
- Makarasen, A.; Reukngam, N.; Khlaychan, P.; Chuysinuan, P.; Isobe, M.; Techasakul, S. Mode of action and synergistic effect of valinomycin and cereulide with amphotericin B against Candida albicans and Cryptococcus albidus. J. Mycol. Med. 2018, 28, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Park, C.N.; Lee, J.M.; Lee, D.; Kim, B.S. Antifungal activity of valinomycin, a peptide antibiotic produced by Streptomyces sp. strain M10 antagonistic to Botrytis cinerea. J. Microbiol. Biotechnol. 2008, 18, 880–884. [Google Scholar] [PubMed]
- Huang, S.; Liu, Y.; Liu, W.; Neubauer, P.; Li, J. The nonribosomal peptide valinomycin: From discovery to bioactivity and biosynthesis. Microorganisms 2021, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Gomi, S.; Amano, S.; Sato, E.; Miyadoh, S.; Kodama, Y. Novel antibiotics SF2738A, B and C, and their analogs produced by Streptomyces sp. J. Antibiot. 1994, 47, 1385–1394. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, M.; Zhang, Q.; Zhang, X.; Yang, P.; Liu, Z.; Deng, Y.; Zhu, Y.; Huang, X.; Han, L.; et al. Diisonitrile Natural Product SF2768 Functions As a Chalkophore That Mediates Copper Acquisition in Streptomyces thioluteus. ACS Chem. Biol. 2017, 12, 3067–3075. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- Nicault, M.; Zaiter, A.; Dumarcay, S.; Chaimbault, P.; Gelhaye, E.; Leblond, P.; Bontemps, C. Elicitation of antimicrobial active compounds by Streptomyces-fungus co-cultures. Microorganisms 2021, 9, 178. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, M.; Tian, J.; Liu, J.; Guo, Z.; Tang, W.; Chen, Y. Activation of paulomycin production by exogenous γ-butyrolactone signaling molecules in Streptomyces albidoflavus J1074. Appl. Microbiol. Biotechnol. 2020, 104, 1695–1705. [Google Scholar] [CrossRef]
- Tsigkinopoulou, A.; Takano, E.; Breitling, R. Unravelling the γ-butyrolactone network in Streptomyces coelicolor by computational ensemble modelling. PLoS Comput. Biol. 2020, 16, 1–17. [Google Scholar] [CrossRef]
- Colombo, E.M.; Pizzatti, C.; Kunova, A.; Gardana, C.; Saracchi, M.; Cortesi, P.; Pasquali, M. Evaluation of in-vitro methods to select effective Streptomycetes against toxigenic Fusaria. PeerJ 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Rózkowski, A.; Chmura, A.; Gajowiec, B.; Rózkowski, J.; Wagner, J. Impact of the underground coal exploitation on the surface and groundwater chemistry in the upper silesian coal basin (Poland). Mine Water Environ. 1993, 12, 95–105. [Google Scholar] [CrossRef]
- Parzentny, H.R. Spatial Macroscale Variability of the Role of Mineral Matter in Concentrating Some Trace Elements in Bituminous Coal in a Coal Basin—A Case Study from the Upper Silesian Coal Basin in Poland. Minerals 2020, 10, 422. [Google Scholar] [CrossRef]
- Malik, A.Y.; Ali, M.; Jamal, A.; Ali, M.I. Isolation and Characterization of Coal Solubilizing Aerobic Microorganisms from Salt Range Coal Mines, Pakistan. Geomicrobiol. J. 2017, 34, 109–118. [Google Scholar] [CrossRef]
- Westphal, K.R.; Nielsen, K.A.H.; Wollenberg, R.D.; Møllehøj, M.B.; Bachleitner, S.; Studt, L.; Lysøe, E.; Giese, H.; Wimmer, R.; Sørensen, J.L.; et al. Fusaoctaxin a, an example of a two-step mechanism for non-ribosomal peptide assembly and maturation in Fungi. Toxins 2019, 11, 277. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics 2016, 32, 2103–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Avram, O.; Rapoport, D.; Portugez, S.; Pupko, T. M1CR0B1AL1Z3R-a user-friendly web server for the analysis of large-scale microbial genomics data. Nucleic Acids Res. 2019, 47, W88–W92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, 339–346. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Pascal Andreu, V.; De Los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2019, 47, D625–D630. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Medema, M.H.; Kottmann, R.; Lee, S.Y.; Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, D555–D559. [Google Scholar] [CrossRef] [Green Version]
- Tsypik, O.; Makitrynskyy, R.; Yan, X.; Koch, H.-G.; Paululat, T.; Bechthold, A. Regulatory Control of Rishirilide(s) Biosynthesis in Streptomyces bottropensis. Microorganisms 2021, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Kerepesi, C.; Bánky, D.; Grolmusz, V. AmphoraNet: The webserver implementation of the AMPHORA2 metagenomic workflow suite. Gene 2014, 533, 538–540. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Varlamov, A.; Vaskin, Y.; Efremov, I.; German Grehov, O.G.; Kandrov, D.; Rasputin, K.; Syabro, M.; et al. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| General Genome Statistics | ||
| Streptomyces sp. S-2 | Streptomyces sp. MW-W600-10 | |
| Number of contigs | 3 | 4 |
| N50 | 4,185,286 bp | 5,533,251 bp |
| Genome Length | 7,243,498 bp | 8,432,369 bp |
| G + C content | 73.28% | 71.70% |
| Completeness | 100% | 99.8% |
| CDS | 6635 | 7738 |
| tRNA | 67 | 66 |
| rRNA | 21 | 18 |
| Protein Features | ||
| Hypothetical proteins | 2269 | 2603 |
| Proteins with functional assignments | 4366 | 5135 |
| Proteins with EC number assignments | 1185 | 1246 |
| Proteins with GO assignments | 1032 | 1083 |
| Proteins with pathway assignments | 925 | 970 |
| Speciality Genes | ||
| Antibiotic resistance [CARD] | 2 | 5 |
| Antibiotic resistance [NDARO] | 2 | 5 |
| Antibiotic resistance [PATRIC] | 40 | 60 |
| Drug target [DrugBank] | 7 | 7 |
| Drug target [TTD] | - | 1 |
| Transporters [TCBD] | 30 | 40 |
| Virulence factors genes [PATRIC_VF] | 2 | 2 |
| Virulence factor genes [Victors] | 1 | 1 |
| BGCs Type | Number of Clusters | |||
|---|---|---|---|---|
| Streptomyces sp. S-2 | S. Albidoflavus J1074 | Streptomyces sp. MW-W600-10 | S. Fimicarius NRRL ISP-5322 | |
| NRPS | 8 (2) | 5 (2) | 7 (1) | 14 (5) |
| NRPS-like | 1 | 1 | 0 | 1 |
| T1PKS | 0 | 1 (1) | 1 (1) | 2 (1) |
| T2PKS | 1 | 0 | 0 | 2 |
| T3PKS | 1 | 1 | 2 | 2 |
| PKS-like | 0 | 0 | 1 (1) | 1 |
| Bacteriocin | 3 (3) | 4 (3) | 2 (1) | 2 (2) |
| Siderophore | 2 | 2 | 2 | 2 |
| Lanthipeptide | 2 (1) | 2 | 6 (4) | 4 (2) |
| Terpene | 4 (1) | 4 | 4 (1) | 6 (2) |
| Ectoine | 1 | 1 | 3 | 2 |
| HgIE-KS | 0 | 0 | 1 | 2 (2) |
| Butyrolactone | 0 | 0 | 2 (1) | 2 |
| Melanin | 0 | 0 | 1 | 0 |
| Lassopeptide | 0 | 0 | 1 | 0 |
| Hybrid | 5 (1) | 5 | 4 (1) | 0 |
| Other | 0 | 1 | 2 (2) | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siupka, P.; Hansen, F.T.; Schier, A.; Rocco, S.; Sørensen, T.; Piotrowska-Seget, Z. Antifungal Activity and Biosynthetic Potential of New Streptomyces sp. MW-W600-10 Strain Isolated from Coal Mine Water. Int. J. Mol. Sci. 2021, 22, 7441. https://doi.org/10.3390/ijms22147441
Siupka P, Hansen FT, Schier A, Rocco S, Sørensen T, Piotrowska-Seget Z. Antifungal Activity and Biosynthetic Potential of New Streptomyces sp. MW-W600-10 Strain Isolated from Coal Mine Water. International Journal of Molecular Sciences. 2021; 22(14):7441. https://doi.org/10.3390/ijms22147441
Chicago/Turabian StyleSiupka, Piotr, Frederik Teilfeldt Hansen, Aleksandra Schier, Simone Rocco, Trine Sørensen, and Zofia Piotrowska-Seget. 2021. "Antifungal Activity and Biosynthetic Potential of New Streptomyces sp. MW-W600-10 Strain Isolated from Coal Mine Water" International Journal of Molecular Sciences 22, no. 14: 7441. https://doi.org/10.3390/ijms22147441
APA StyleSiupka, P., Hansen, F. T., Schier, A., Rocco, S., Sørensen, T., & Piotrowska-Seget, Z. (2021). Antifungal Activity and Biosynthetic Potential of New Streptomyces sp. MW-W600-10 Strain Isolated from Coal Mine Water. International Journal of Molecular Sciences, 22(14), 7441. https://doi.org/10.3390/ijms22147441







