Molecular Responses to Cadmium Exposure in Two Contrasting Durum Wheat Genotypes
Abstract
1. Introduction
2. Results and Discussion
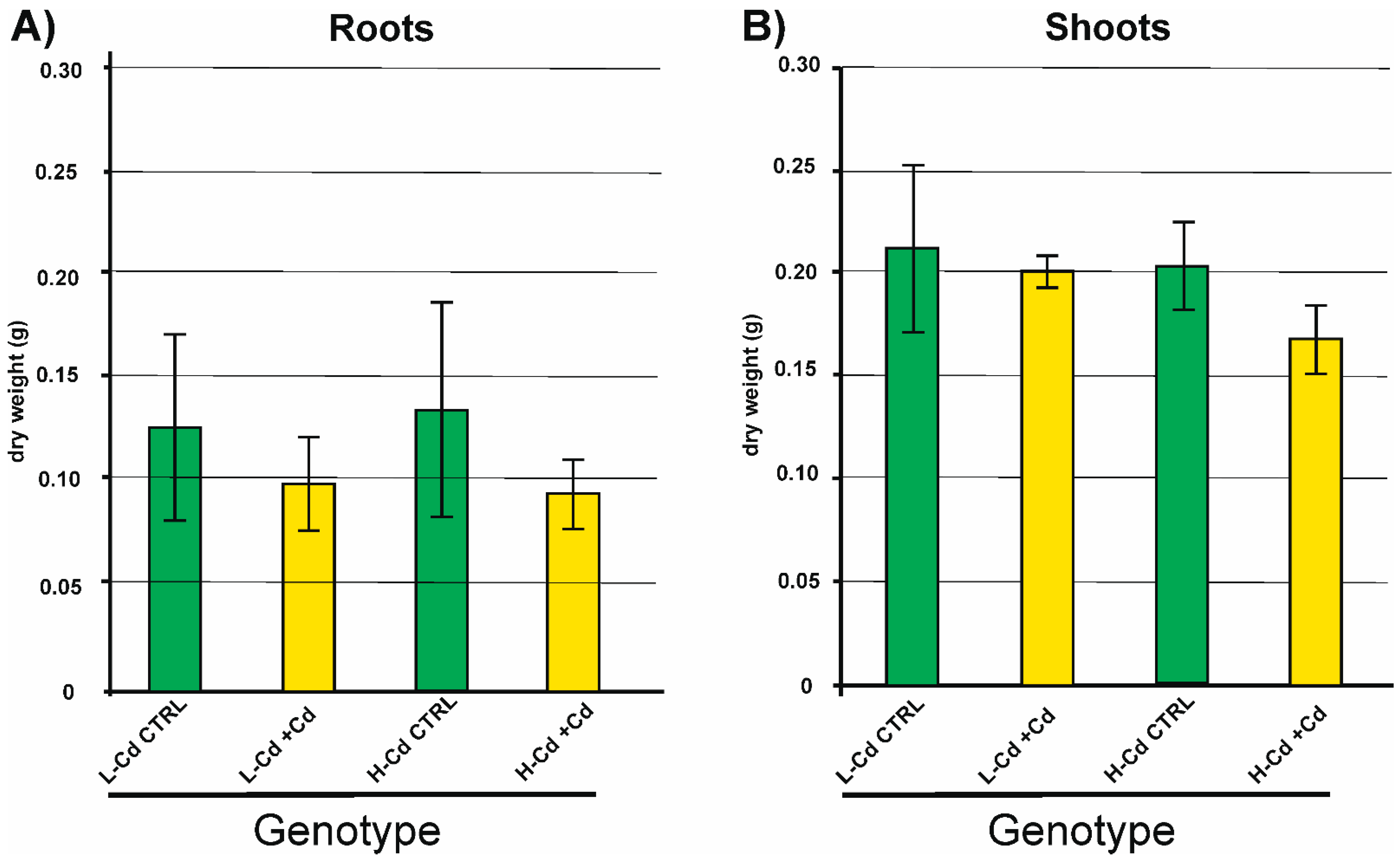
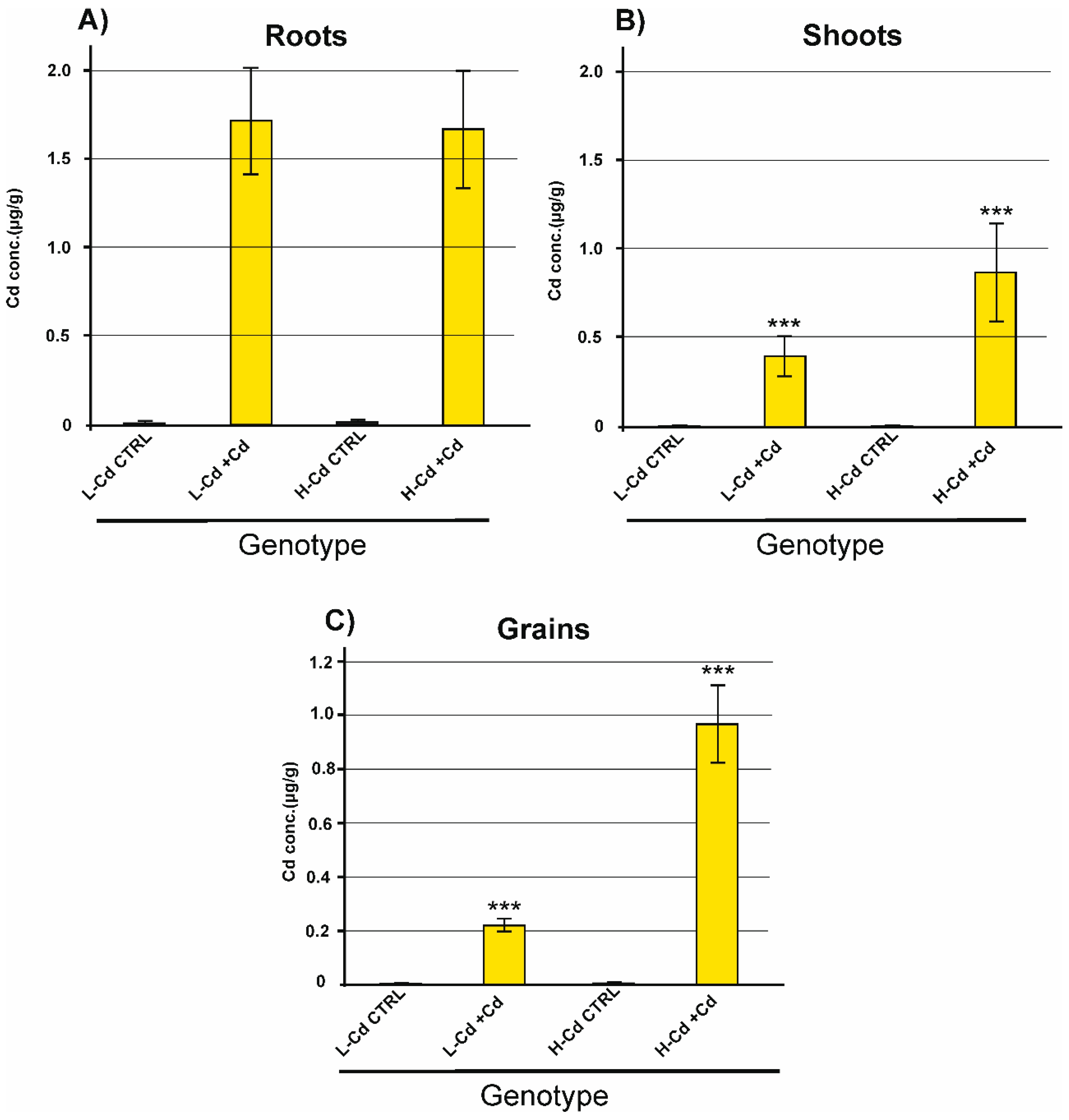
2.1. Biomass and Cd Concentration in Plant Tissues
2.2. RNA Sequencing and Quality of Data
2.3. Hypothesis-Based Analysis of the Transcriptome Modulation in NILs Treated with Cadmium
3. Materials and Methods
3.1. Plant Materials and Treatment
3.2. Biomass Analysis and Determination of Cd Concentration
3.3. RNA Isolation and mRNA Sequencing
3.4. Identification of Differentially Expressed Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huybrechts, M.; Cuypers, A.; Deckers, J.; Iven, V.; Vandionant, S.; Jozefczak, M.; Hendrix, S. Cadmium and Plant Development: An Agony from Seed to Seed. Int. J. Mol. Sci. 2019, 20, 3971. [Google Scholar] [CrossRef]
- DalCorso, G.; Farinati, S.; Furini, A. Regulatory networks of cadmium stress in plants. Plant Sign. Behav. 2010, 6, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Satarug, S.; Garrett, S.H.; Sens, M.A.; Sens, D.A. Cadmium, environmental exposure, and health outcomes. Environ. Health Perspect. 2010, 118, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Haider, F.H.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 2021, 211, 111887. [Google Scholar] [CrossRef] [PubMed]
- El Rasafi, T.; Oukarroum, A.; Haddioui, A.; Song, H.; Kwon, E.E.; Bolan, N.; Tack, F.M.G.; Sebastian, A.; Prasad, M.N.V.; Rinklebe, J. Cadmium stress in plants: A critical review of the effects, mechanisms, and tolerance strategies. Crit. Rev. Environ. Sci. Technol. 2020, 1–52. [Google Scholar] [CrossRef]
- Eller, F.; Brix, H. Influence of low calcium availability on cadmium uptake and translocation in a fast-growing shrub and a metal-accumulating herb. AoB Plants 2016, 8, plv143. [Google Scholar] [CrossRef]
- Viehweger, K. How plants cope with heavy metals. Bot. Stud. 2014, 55, 35. [Google Scholar] [CrossRef]
- Chen, H.; Li, Y.; Ma, X.; Guo, L.; He, Y.; Ren, Z.; Kuang, Z.; Zhang, X.; Zhang, Z. Analysis of potential strategies for cadmium stress tolerance revealed by transcriptome analysis of upland cotton. Sci. Rep. 2019, 9, 86. [Google Scholar] [CrossRef]
- Xiao, Y.; Du, Z.; Qi, X.; Wu, H.; Guo, W.; Zhao, Z. RNA-sequencing analysis reveals transcriptional changes in the roots of low-cadmium-accumulating winter wheat under cadmium stress. Acta Physiol. Plant 2019, 41, 13. [Google Scholar] [CrossRef]
- Qiao, K.; Liang, S.; Wang, F.; Wang, H.; Hu, Z.; Chai, T. Effects of cadmium toxicity on diploid wheat (Triticum urartu) and the molecular mechanism of the cadmium response. J. Hazard. Mater. 2019, 374, 1–10. [Google Scholar] [CrossRef]
- Herbette, S.; Taconnat, L.; Hugouvieux, V.; Piette, L.; Magniette, M.-L.M.; Cuine, S.; Auroy, P.; Richaud, P.; Forestier, C.; Bourguignon, J.; et al. Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoot. Biochimie 2006, 88, 1751–1765. [Google Scholar] [CrossRef]
- Gutsch, A.; Hendrix, S.; Guerriero, G.; Renaut, J.; Lutts, S.; Alseekh, S.; Fernie, A.R.; Hausman, J.F.; Vangronsveld, J.; Cuypers, A.; et al. Long-Term Cd Exposure Alters the Metabolite Profile in Stem Tissue of Medicago sativa. Cells 2020, 9, 2707. [Google Scholar] [CrossRef]
- He, J.; Li, H.; Luo, J.; Ma, C.; Li, S.; Qu, L.; Gai, Y.; Jiang, X.; Janz, D.; Polle, A.; et al. A Transcriptomic Network Underlies Microstructural and Physiological Responses to Cadmium in Populus x canescens. Plant Physiol. 2013, 162, 424–439. [Google Scholar] [CrossRef]
- Jamers, A.; Blust, R.; De Coen, W.; Griffin, J.L.; Jones, O.A.-H. An omics-based assessment of cadmium toxicity in the green alga Chlamydomonas reinhardtii. Aquat. Toxicol. 2013, 126, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-Y.; Trinh, N.N.; Fu, S.-F.; Hsiung, Y.-C.; Chia, L.-C.; Lin, C.-W.; Huang, H.-J. Comparison of early transcriptome responses to copper and cadmium in rice roots. Plant Mol. Biol. 2013, 81, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Harris, N.S.; Taylor, G.J. Cadmium uptake and translocation in seedlings of near isogenic lines of durum wheat that differ in grain cadmium accumulation. BMC Plant Biol. 2004, 4, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vergine, M.; Aprile, A.; Sabella, E.; Genga, A.; Siciliano, M.; Rampino, P.; Lenucci, M.S.; Luvisi, A.; De Bellis, L. Cadmium concentration in grains of durum wheat (Triticum turgidum L. subsp. durum). J. Agric. Food Chem. 2017, 65, 6240–6246. [Google Scholar] [CrossRef]
- Aprile, A.; Sabella, E.; Vergine, M.; Genga, A.; Siciliano, M.; Nutricati, E.; Rampino, P.; De Pascali, M.; Luvisi, A.; Miceli, A.; et al. Activation of a gene network in durum wheat roots exposed to cadmium. BMC Plant Biol. 2018, 18, 238. [Google Scholar] [CrossRef]
- Knox, R.; Pozniak, C.; Clarke, F.; Clarke, J.; Houshmand, S.; Singh, A.K. Chromosomal location of the cadmium uptake gene (Cdu1-B) in durum wheat. Genome 2009, 52, 741–747. [Google Scholar] [CrossRef]
- Penner, G.A.; Bezte, L.J.; Leisle, D.; Clarke, J. Identification of RAPD markers linked to a gene governing cadmium uptake in durum wheat. Genome 1995, 38, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, K.; Harris, N.; Faris, J.; Clarke, J.; Knox, R.; Taylor, G.J.; Pozniak, C.J. Targeted mapping of Cdu1-B, a major locus regulating grain cadmium concentration in durum wheat (Triticum turgidum L. var durum). Theor. Appl. Genet. 2010, 121, 1047–1058. [Google Scholar] [CrossRef]
- Wiebe, K. Molecular Characterization of Cdu-B1, a Major Locus Controlling Cadmium Accumulation in Durum Wheat (Triticum turgidum L. var Durum) Grain. Master’s Thesis, University of Saskatchewan, Saskatoon, SK, Canada, 2012. [Google Scholar]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannag, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [PubMed]
- AbuHammad, W.A.; Mamidi, S.; Kumar, A.; Pirseyedi, S.; Manthey, F.A.; Kianian, S.F.; Alamri, M.S.; Mergoum, M.; Elias, E.M. Identification and validation of a major cadmium accumulation locus and closely associated SNP markers in North Dakota durum wheat cultivars. Mol. Breed. 2016, 36, 112. [Google Scholar] [CrossRef]
- Oladzad-Abbasabadi, A.; Kumar, A.; Pirseyedi, S.; Salsman, E.; Dobrydina, M.; Poudel, R.S.; AbuHammad, W.A.; Chao, S.; Faris, J.D.; Elias, E.M. Identification and Validation of a New Source of Low Grain Cadmium Accumulation in Durum Wheat. G3 Genes Genomes Genet. 2018, 8, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, J.; Singh, R.; Garg, T.; Chhuneja, P.; Balyan, H.S.; Gupta, P.K. QTL analysis for grain color and preharvest sprouting in bread wheat. Plant Sci. 2009, 177, 114–122. [Google Scholar] [CrossRef]
- Harris, N.S.; Taylor, G.J. Cadmium uptake and partitioning in durum wheat during grain filling. BMC Plant Biol. 2013, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.M.; Leisle, D.; DePauw, R.M.; Thiessen, L.L. The registration of genetic stocks: Registration of five pairs of near-isogenic lines for cadmium concentration. Crop Sci. 1997, 37, 297. [Google Scholar] [CrossRef]
- Ortiz, D.F.; Ruscitti, T.; Kent, F.; McCue, K.F.; Ow, D.W. Transport of Metal-binding Peptides by HMT1, A Fission Yeast ABC-type Vacuolar Membrane Protein. J. Biol. Chem. 1995, 270, 4721–4728. [Google Scholar] [CrossRef]
- Kim, S.; Selote, D.S.; Vatamaniuk, O.K. The N-Terminal Extension Domain of the C. elegans Half-Molecule ABC Transporter, HMT-1, Is Required for Protein-Protein Interactions and Function. PLoS ONE 2010, 5, e12938. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, Y.; Peng, J.-S.; Zhong, C.; Yi, H.-Y.; Ow, D.W.; Gong, J.M. Fission Yeast HMT1 Lowers Seed Cadmium through Phytochelatin-Dependent Vacuolar Sequestration in Arabidopsis. Plant Physiol. 2012, 158, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Fukuoka, H.; Arao, T.; Ohyama, A.; Nunome, T.; Miyatake, K.; Negoro, S. Gene expression analysis in cadmium-stressed roots of a low cadmium-accumulating solanaceous plant, Solanum torvum. J. Exp. Bot. 2010, 61, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler—A web server for functional interpretation of gene lists. Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Loix, C.; Huybrechts, M.; Vangronsveld, J.; Gielen, M.; Keunen, E.; Cuypers, A. Reciprocal Interactions between Cadmium-Induced Cell Wall Responses and Oxidative Stress in Plants. Front. Plant Sci. 2017, 8, 1867. [Google Scholar] [CrossRef]
- Haydon, M.J.; Cobbett, C.S. A Novel Major Facilitator Superfamily Protein at the Tonoplast Influences Zinc Tolerance and Accumulation in Arabidopsis. Plant Physiol. 2007, 143, 1705–1719. [Google Scholar] [CrossRef]
- Yao, X.; Cai, Y.; Yu, D.; Liang, G. bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J. Integr. Plant Biol. 2018, 60, 691–702. [Google Scholar] [CrossRef]
- Wu, H.; Chen, C.; Du, J.; Liu, H.; Cui, Y.; Zhang, Y.; He, Y.; Wang, Y.; Chu, C.; Feng, Z.; et al. Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 2012, 158, 790–800. [Google Scholar] [CrossRef]
- Gravot, A.; Lieutaud, A.; Verret, F.; Auroy, P.; Vavasseur, A.; Richaud, P. AtHMA3, a plant P-1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 2004, 561, 22–28. [Google Scholar] [CrossRef]
- Verret, F.; Gravot, A.; Auroy, P.; Preveral, S.; Forestier, C.; Vavasseur, A.; Richaud, P. Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His11 stretch. FEBS Lett. 2005, 579, 1515–1522. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, G.; Honsbein, A.; Meda, A.R.; Kirchner, S.; Wipf, D.; von Wirén, N. AtIREG2 Encodes a Tonoplast Transport Protein Involved in Iron-dependent Nickel Detoxification in Arabidopsis thaliana Roots. J. Biol. Chem. 2006, 281, 25532–25540. [Google Scholar] [CrossRef]
- Higuchi, K.; Kanazawa, K.; Nishizawa, N.K.; Chino, M.; Mori, S. Purification and characterization of nicotianamine synthase from Fe-deficient barley roots. Plant Soil 1994, 165, 173–179. [Google Scholar] [CrossRef]
- Less, H.; Galili, G. Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 2008, 147, 316–330. [Google Scholar] [CrossRef]
- Bonneau, J.; Baumann, U.; Beasley, J.; Li, Y.; Johnson, A.A.T. Identification and molecular characterization of the nicotianamine synthase gene family in bread wheat. Plant Biotech. J. 2016, 14, 2228–2239. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, J.S.; Lamb, A.L. Opine Metallophore Biosynthesis. Compr. Nat. Prod. III 2020, 5, 395–414. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high-resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH transcription factor POPEYE regulates response to Iron deficiency in Arabidopsis roots. Plant Cell. 2010, 22, 2219–2236. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The Basic Helix–Loop–Helix Transcription Factor Family in Plants: A Genome-Wide Study of Protein Structure and Functional Diversity. Mol. Biol. Evol. 2003, 5, 735–747. [Google Scholar] [CrossRef]
- Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004, 5, 226. [Google Scholar] [CrossRef]
- Parker, D.R.; Norvell, W.A. Advances in solution culture methods for plant mineral nutrition research. Adv. Agron. 1999, 65, 151–213. [Google Scholar] [CrossRef]
- Massadeh, A.M.; Snook, R.D. Determination of Pb and cd in road dusts over the period in which Pb was removed from petrol in the UK. J. Environ. Monit. 2002, 4, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Aprile, A.; Havlickova, L.; Panna, R.; Marè, C.; Borrelli, G.M.; Marone, D.; Perrotta, C.; Rampino, P.; De Bellis, L.; Curn, V.; et al. Different stress responsive strategies to drought and heat in two durum wheat cultivars with contrasting water use efficiency. BMC Genom. 2013, 14, 821. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene expression analysis of RNAseq experiments with TopHat and cufflinks. Nat. Prot. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Patcher, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotech. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
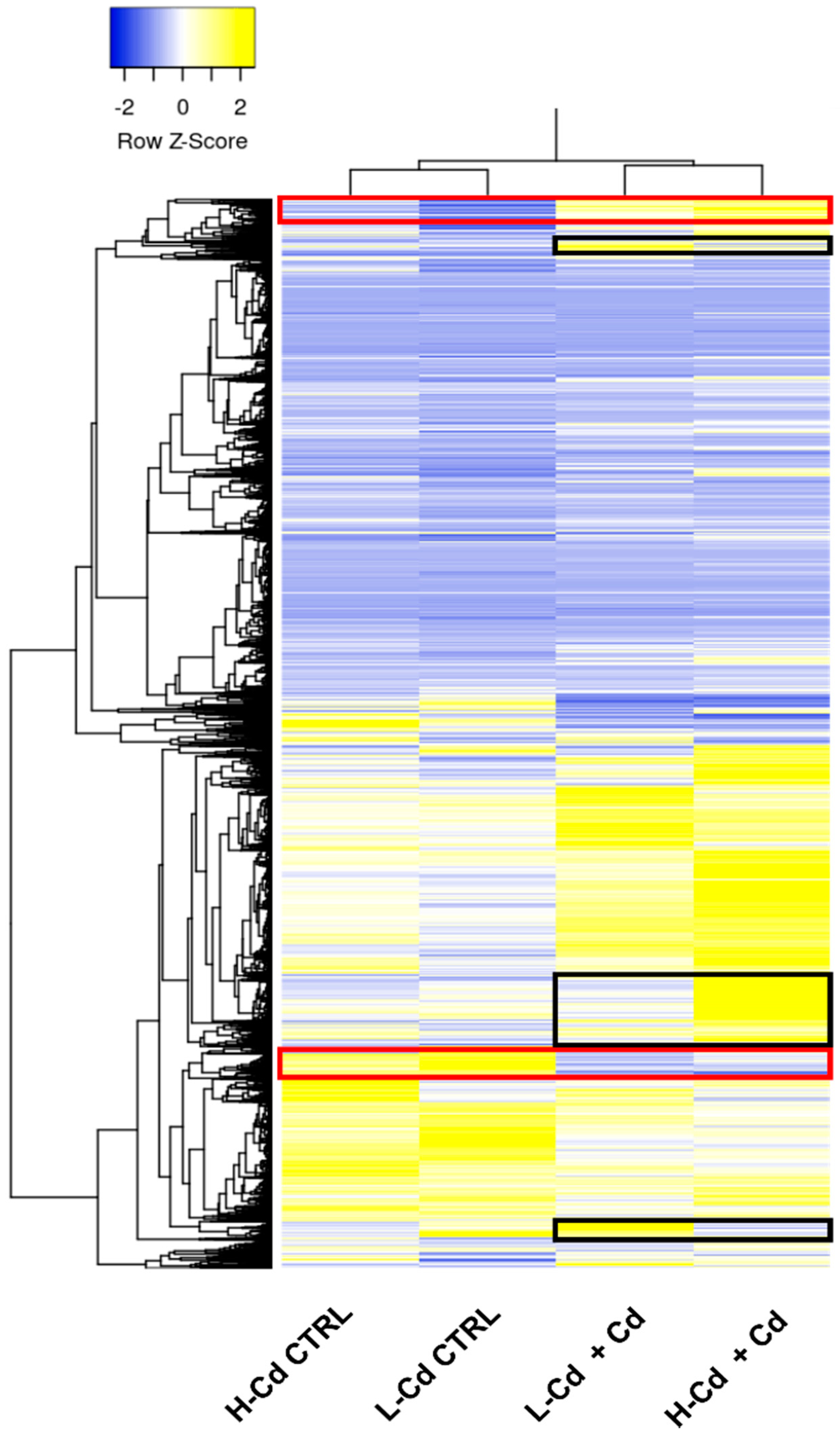
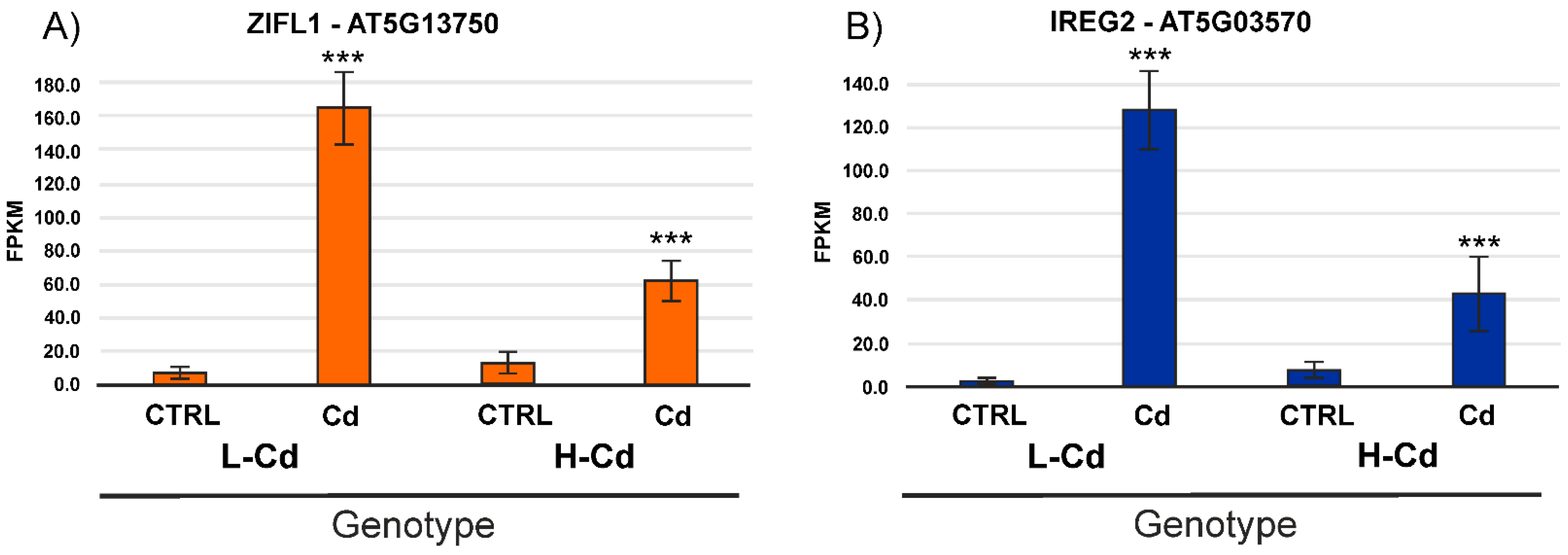


| Contig ID | L-Cd CTRL | L-Cd +Cd | H-Cd CTRL | H-Cd +Cd | Annotation |
|---|---|---|---|---|---|
| contig19712 | 27.4 | 31.2 | 2.4 | 2.1 | Cyclophilin (AT3G66654) |
| contig23984 | 76.5 | 49.8 | 0.1 | 0.1 | no hits—unknown |
| contig5761 | 36.4 | 25.7 | 4.9 | 4.0 | 3-mercaptopyruvate sulfurtransferase (AT1G79230) |
| contig90107 | 57.1 | 30.7 | 0.0 | 0.0 | no hits—unknown |
| contig9490 | 53.2 | 43.9 | 2.8 | 0.2 | ABC transporter 3-like (AT1G64550) |
| Contig ID | L-Cd CTRL | L-Cd +Cd | H-Cd CTRL | H-Cd +Cd | Annotation |
|---|---|---|---|---|---|
| contig13191 | 0.1 | 0.3 | 97.8 | 34.2 | aquaporin pip1-2 (AT1G01620) |
| contig17660 | 0.3 | 0.2 | 148.0 | 65.1 | aquaporin pip1-5 (AT4G00430) |
| contig17681 | 0.0 | 0.1 | 33.2 | 27.0 | no hits—unknown |
| contig20402 | 0.1 | 0.0 | 62.1 | 58.1 | no hits—unknown |
| contig35423 | 0.0 | 0.1 | 70.6 | 43.7 | Ribosomal protein (AT1G33140) |
| contig3675 | 0.4 | 0.4 | 104.1 | 84.4 | no hits—unknown |
| contig48835 | 4.4 | 4.4 | 62.9 | 103.4 | no hits—unknown |
| contig5342 | 3.3 | 3.9 | 52.2 | 67.4 | RNA ligase (AT4G18930) |
| contig1280 | 3.1 | 1.9 | 30.9 | 32.6 | transcription elongation factor (AT1G32130) |
| L-Cd | H-Cd | |||||||
|---|---|---|---|---|---|---|---|---|
| Contig ID | CTRL | 0.5 µM | FC | CTRL | 0.5 µM | FC | AGI Code | Annotation |
| contig26634 | 7.8 | 604.5 | 77.1 | 24.3 | 240.7 | 9.9 | NAS 1 | |
| contig55580 | 3.6 | 707.5 | 195.8 | 20.5 | 383.4 | 18.7 | NAS 1 | |
| contig44671 | 5.4 | 774.2 | 143.6 | 49.4 | 437.2 | 8.8 | NAS 2 | |
| contig18293 | 1.8 | 502.3 | 278.2 | 11.1 | 149.3 | 13.5 | AT5G56080 | NAS 2 |
| contig18375 | 23.5 | 653.3 | 27.8 | 64.6 | 386.0 | 6.0 | AT5G56080 | NAS 2 |
| contig18424 | 10.6 | 1178.3 | 111.5 | 41.5 | 522.9 | 12.6 | AT1G09240 | NAS 3 |
| contig33892 | 1.1 | 623.2 | 572.5 | 10.0 | 262.0 | 26.3 | AT1G09240 | NAS 3 |
| contig40805 | 9.8 | 629.4 | 64.2 | 55.9 | 348.5 | 6.2 | AT1G09240 | NAS 3 |
| contig5670 | 1.1 | 179.2 | 158.9 | 5.2 | 23.2 | 4.5 | AT1G09240 | NAS 3 |
| contig6866 | 7.6 | 663.0 | 87.8 | 51.9 | 281.9 | 5.4 | AT1G09240 | NAS 3 |
| contig16879 | 0.8 | 155.6 | 204.0 | 5.2 | 68.2 | 13.0 | NAS 4 | |
| contig36649 | 7.8 | 482.7 | 61.8 | 19.6 | 183.5 | 9.4 | NAS 4 | |
| contig13016 | 2.1 | 391.5 | 186.2 | 30.2 | 114.6 | 3.8 | AT1G56430 | NAS 4 |
| contig18348 | 2.4 | 626.8 | 264.5 | 25.2 | 257.9 | 10.2 | AT1G56430 | NAS 4 |
| contig27606 | 7.4 | 321.2 | 43.7 | 31.3 | 106.5 | 3.4 | AT1G56430 | NAS 4 |
| contig46013 | 7.5 | 1488.7 | 197.4 | 64.4 | 306.9 | 4.8 | AT1G56430 | NAS 4 |
| contig51008 | 1.2 | 369.2 | 318.0 | 19.5 | 85.3 | 4.4 | AT1G56430 | NAS 4 |
| contig5669 | 12.5 | 845.6 | 67.6 | 44.2 | 248.1 | 5.6 | AT1G56430 | NAS 4 |
| contig56695 | 1.7 | 169.2 | 102.4 | 12.5 | 75.1 | 6.0 | AT1G56430 | NAS 4 |
| contig62096 | 6.4 | 778.0 | 122.2 | 39.4 | 270.5 | 6.9 | AT1G56430 | NAS 4 |
| contig6432 | 5.3 | 1071.6 | 200.7 | 81.6 | 218.6 | 2.7 | AT1G56430 | NAS 4 |
| contig35044 | 10.3 | 691.1 | 67.3 | 49.2 | 419.8 | 8.5 | NAS 5B | |
| CTRL | 0.5 µM | FC | CTRL | 0.5 µM | FC | |||
| Mean | 6.3 | 632.1 | 161.5 | 34.4 | 245.0 | 8.7 | ||
| L-Cd | H-Cd | |||||||
|---|---|---|---|---|---|---|---|---|
| Contig ID | CTRL | 0.5 µM | FC | CTRL | 0.5 µM | FC | AGI Code | Annotation |
| contig24852 | 0.7 | 246.9 | 356.6 | 23.8 | 103.0 | 4.3 | AT2G20610 | NAAT |
| contig7573 | 1.0 | 209.9 | 209.6 | 10.9 | 117.3 | 10.7 | AT5G53970 | NAAT |
| contig20039 | 3.2 | 462.9 | 145.4 | 49.6 | 287.9 | 5.8 | AT5G53970 | NAAT |
| contig40067 | 21.9 | 274.7 | 12.6 | 33.9 | 175.8 | 5.2 | AT1G59960 | DMAS |
| contig10233 | 11.4 | 253.8 | 22.3 | 22.9 | 139.9 | 6.1 | AT1G59960 | DMAS |
| CTRL | 0.5 µM | FC | CTRL | 0.5 µM | FC | |||
| Mean | 7.6 | 289.6 | 149.3 | 28.2 | 164.8 | 6.4 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabella, E.; Luvisi, A.; Genga, A.; De Bellis, L.; Aprile, A. Molecular Responses to Cadmium Exposure in Two Contrasting Durum Wheat Genotypes. Int. J. Mol. Sci. 2021, 22, 7343. https://doi.org/10.3390/ijms22147343
Sabella E, Luvisi A, Genga A, De Bellis L, Aprile A. Molecular Responses to Cadmium Exposure in Two Contrasting Durum Wheat Genotypes. International Journal of Molecular Sciences. 2021; 22(14):7343. https://doi.org/10.3390/ijms22147343
Chicago/Turabian StyleSabella, Erika, Andrea Luvisi, Alessandra Genga, Luigi De Bellis, and Alessio Aprile. 2021. "Molecular Responses to Cadmium Exposure in Two Contrasting Durum Wheat Genotypes" International Journal of Molecular Sciences 22, no. 14: 7343. https://doi.org/10.3390/ijms22147343
APA StyleSabella, E., Luvisi, A., Genga, A., De Bellis, L., & Aprile, A. (2021). Molecular Responses to Cadmium Exposure in Two Contrasting Durum Wheat Genotypes. International Journal of Molecular Sciences, 22(14), 7343. https://doi.org/10.3390/ijms22147343










