Transcriptional Modulation of the Hippo Signaling Pathway by Drugs Used to Treat Bipolar Disorder and Schizophrenia
Abstract
:1. Introduction
2. Results
2.1. GSEA
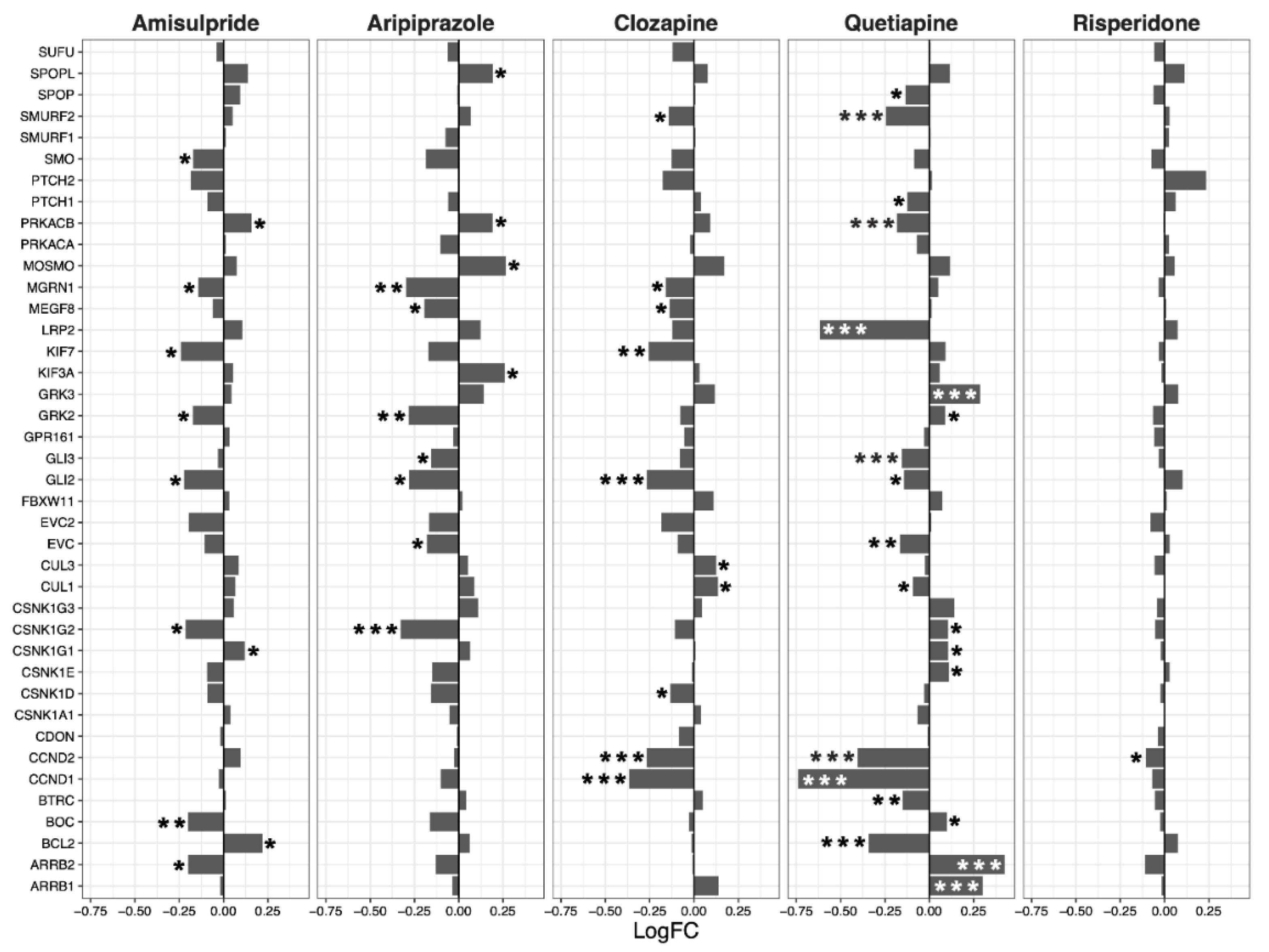
2.2. Hippo Core Genes and Transcription Factors
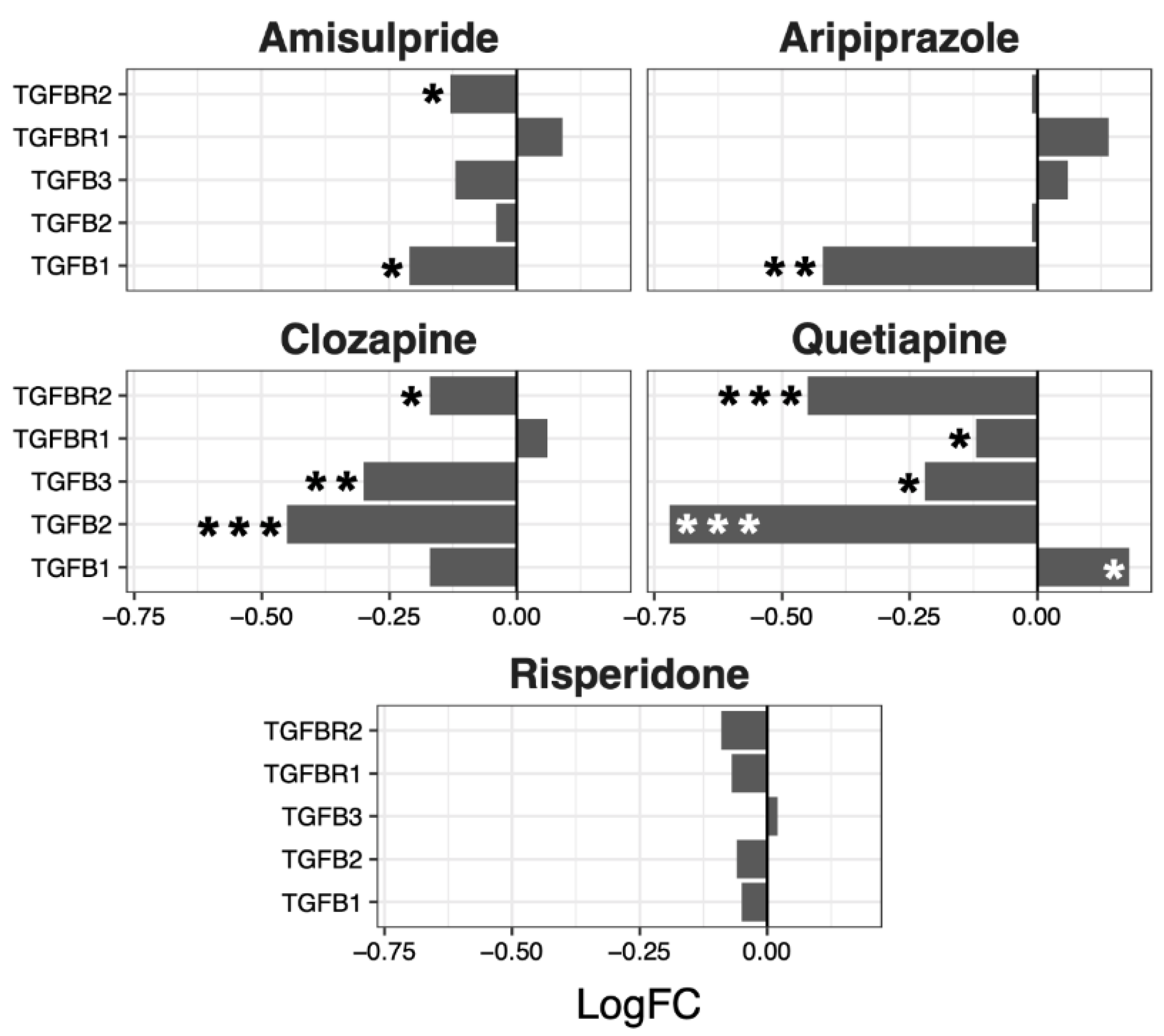
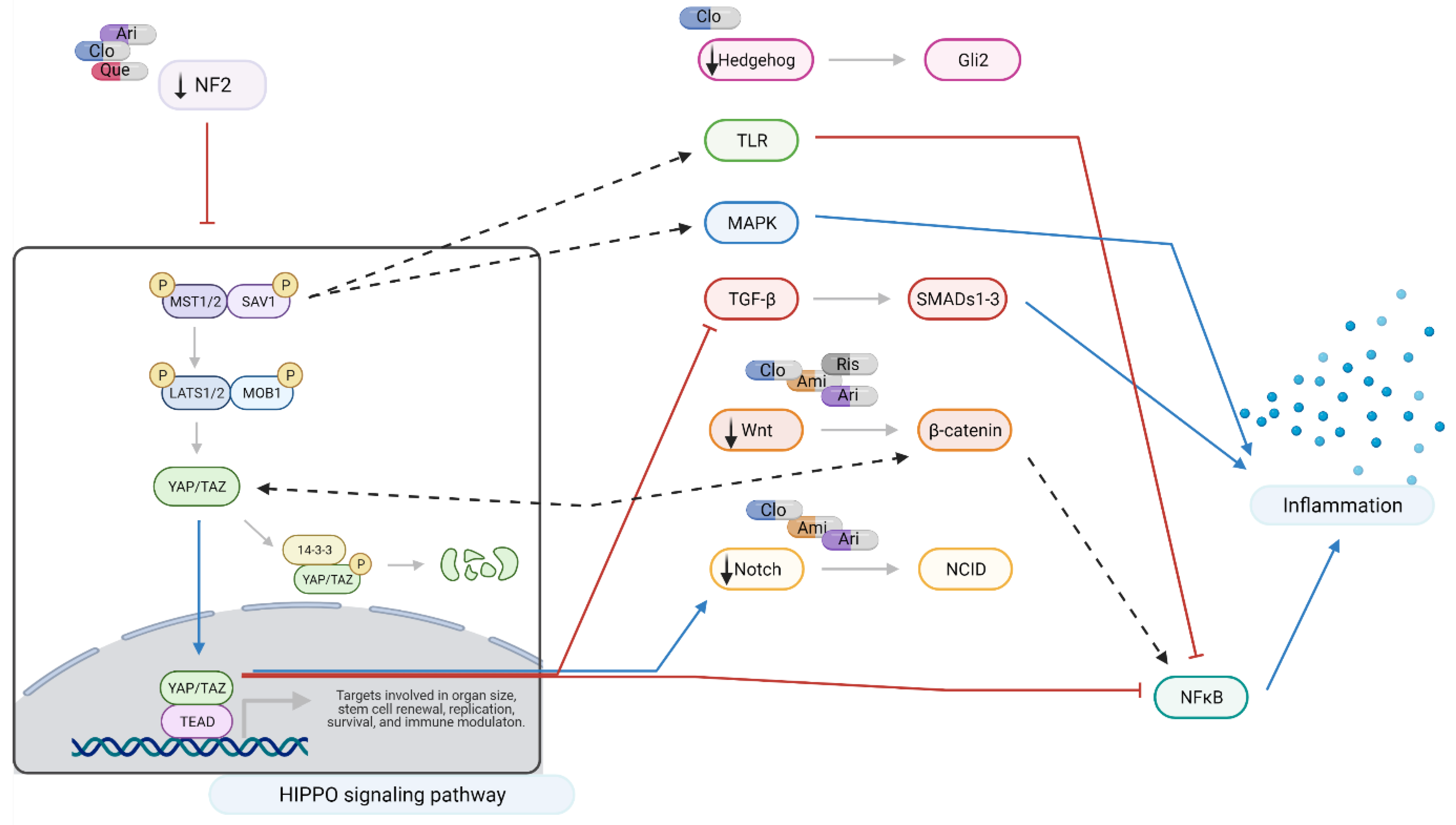
2.3. Interacting Pathways
2.4. CMap
3. Discussion
3.1. Aripiprazole and Clozapine
3.2. Amisulpride
3.3. Quetiapine and Risperidone
3.4. Summary
3.5. Drug Repurposing
4. Materials and Methods
4.1. Cell Culture
4.2. Gene Expression
4.3. Genome-Wide Gene Expression Analysis
4.4. Gene Set Enrichment Analysis (GSEA)
4.5. Interacting Pathways
4.6. Connectivity Map (CMap)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.-L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Li, L.; Guan, K.-L. Hippo Signaling at a Glance. J. Cell Sci. 2010, 123, 4001–4006. [Google Scholar] [CrossRef] [Green Version]
- Fu, V.; Plouffe, S.W.; Guan, K.-L. The Hippo Pathway in Organ Development, Homeostasis, and Regeneration. Curr. Opin. cell Biol. 2017, 49, 99–107. [Google Scholar] [CrossRef]
- Hong, L.; Li, X.; Zhou, D.; Geng, J.; Chen, L. Role of Hippo Signaling in Regulating Immunity. Cell. Mol. Immunol. 2018, 15, 1003–1009. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wang, L.; Yin, F.; Yu, Y.; Wang, Y.; Shepard, M.J.; Zhuang, Z.; Qin, J. Probing Impaired Neurogenesis in Human Brain Organoids Exposed to Alcohol. Integr. Biol. 2017, 9, 968–978. [Google Scholar] [CrossRef]
- Emoto, K. The Growing Role of the Hippo–NDR Kinase Signalling in Neuronal Development and Disease. J. Biochem. 2011, 150, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Rojek, K.O.; Krzemień, J.; Doleżyczek, H.; Boguszewski, P.M.; Kaczmarek, L.; Konopka, W.; Rylski, M.; Jaworski, J.; Holmgren, L.; Prószyński, T.J. Amot and Yap1 Regulate Neuronal Dendritic Tree Complexity and Locomotor Coordination in Mice. PLoS Biol. 2019, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emoto, K. Signaling Mechanisms That Coordinate the Development and Maintenance of Dendritic Fields. Curr. Opin. Neurobiol. 2012, 22, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wang, S.; Dong, Y.; Yuan, Z. The Role and Regulatory Mechanism of Hippo Signaling Components in the Neuronal System. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Huang, T.; Zhang, J.; Cheng, A.S.L.; Yu, J.; Kang, W.; To, K.F. Emerging Roles of Hippo Signaling in Inflammation and YAP-Driven Tumor Immunity. Cancer Lett. 2018, 426, 73–79. [Google Scholar] [CrossRef]
- Yang, Y.; Gong, Z.; Wang, Z. Yes-Associated Protein Reduces Neuroinflammation through Upregulation of Sirt3 and Inhibition of JNK Signaling Pathway. J. Recept. Signal Transduct. 2019, 39, 479–487. [Google Scholar] [CrossRef]
- Lavado, A.; Park, J.Y.; Paré, J.; Finkelstein, D.; Pan, H.; Xu, B.; Fan, Y.; Kumar, R.P.; Neale, G.; Kwak, Y.D.; et al. The Hippo Pathway Prevents YAP/TAZ–Driven Hypertranscription and Controls Neural Progenitor Number. Dev. Cell 2018, 47, 576–591.e8. [Google Scholar] [CrossRef] [Green Version]
- Rivas, S.; Antón, I.M.; Wandosell, F. WIP-YAP/TAZ as A New Pro-Oncogenic Pathway in Glioma. Cancers 2018, 10, 191. [Google Scholar] [CrossRef] [Green Version]
- Mueller, K.A.; Glajch, K.E.; Huizenga, M.N.; Wilson, R.A.; Granucci, E.J.; Dios, A.M.; Tousley, A.R.; Iuliano, M.; Weisman, E.; LaQuaglia, M.J.; et al. Hippo Signaling Pathway Dysregulation in Human Huntington’s Disease Brain and Neuronal Stem Cells. Sci. Rep. 2018, 8, 11355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, H.; Homma, H.; Fujita, K.; Kondo, K.; Yamada, S.; Jin, X.; Waragai, M.; Ohtomo, G.; Iwata, A.; Tagawa, K.; et al. YAP-Dependent Necrosis Occurs in Early Stages of Alzheimer’s Disease and Regulates Mouse Model Pathology. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Melka, M.G.; Castellani, C.A.; O’Reilly, R.; Singh, S.M. Insights into the Origin of DNA Methylation Differences between Monozygotic Twins Discordant for Schizophrenia. J. Mol. Psychiatry 2015, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, K.S.; McGregor, N.W.; Lochner, C.; Emsley, R.; Warnich, L. The Genetic Architecture of Schizophrenia, Bipolar Disorder, Obsessive-Compulsive Disorder and Autism Spectrum Disorder. Mol. Cell. Neurosci. 2018, 88, 300–307. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, H.-Y.; Zhu, J.; Niu, Y.-M.; Zhang, C.; Guo, G.-L. Identification of Hub Genes and Key Pathways Associated With Bipolar Disorder Based on Weighted Gene Co-Expression Network Analysis. Front. Physiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bame, M.; McInnis, M.; O’Shea, K.S. MicroRNA Alterations in Induced Pluripotent Stem Cell-Derived Neurons from Bipolar Disorder Patients: Pathways Involved in Neuronal Differentiation, Axon Guidance and Plasticity. Stem Cells Dev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-E.; Lee, W.-Y.; Cheng, H.-W.; Chung, C.-H.; Mi, F.-L.; Lin, C.-W. The Antipsychotic Chlorpromazine Suppresses YAP Signaling, Stemness Properties, and Drug Resistance in Breast Cancer Cells. Chem. Biol. Interact. 2019, 302, 28–35. [Google Scholar] [CrossRef]
- Davood, Z.-A.; Shamsi, S.; Ghaedi, H.; Sahand, R.-I.; Mojtaba, G.; Mahdi, T.; Reza, M.; Ebrahimi, M.J.; Miri-Moosavi, R.S.; Boosaliki, S.; et al. Valproic Acid May Exerts Its Cytotoxic Effect through Rassf1a Expression Induction in Acute Myeloid Leukemia. Tumour Biol. 2016, 37, 11001–11006. [Google Scholar] [CrossRef]
- Horacek, J.; Bubenikova-Valesova, V.; Kopecek, M.; Palenicek, T.; Dockery, C.; Mohr, P.; Höschl, C. Mechanism of Action of Atypical Antipsychotic Drugs and the Neurobiology of Schizophrenia. CNS Drugs 2006, 20, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo Pathway and Human Cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Yu, J.; Zheng, Y.; Chen, Q.; Zhang, N.; Pan, D. Spatial Organization of Hippo Signaling at the Plasma Membrane Mediated by the Tumor Suppressor Merlin/NF2. Cell 2013, 154, 1342–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, J.; Giancotti, F.G. Molecular Insights into NF2/Merlin Tumor Suppressor Function. FEBS Lett. 2014, 588, 2743–2752. [Google Scholar] [CrossRef] [Green Version]
- Sourbier, C.; Liao, P.-J.; Ricketts, C.J.; Wei, D.; Yang, Y.; Baranes, S.M.; Gibbs, B.K.; Ohanjanian, L.; Spencer Krane, L.; Scroggins, B.T.; et al. Targeting Loss of the Hippo Signaling Pathway in NF2-Deficient Papillary Kidney Cancers. Oncotarget 2018, 9, 10723–10733. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Li, S.; Mo, J.; Hawley, E.; Wang, Y.; He, Y.; Brosseau, J.-P.; Shipman, T.; Clapp, D.W.; Carroll, T.J.; et al. Schwannoma Development Is Mediated by Hippo Pathway Dysregulation and Modified by RAS/MAPK Signaling. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Höffken, V.; Hermann, A.; Pavenstädt, H.; Kremerskothen, J. WWC Proteins: Important Regulators of Hippo Signaling in Cancer. Cancers 2021, 13, 306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, S.; Chen, J.; Gan, C.; Chen, D.; Li, Y.; Wen, J.; Kremerskothen, J.; Chen, S.; Zhang, J.; et al. WWC2 Is an Independent Prognostic Factor and Prevents Invasion via Hippo Signalling in Hepatocellular Carcinoma. J. Cell. Mol. Med. 2017, 21, 3718–3729. [Google Scholar] [CrossRef] [PubMed]
- Park, H.W.; Kim, Y.C.; Yu, B.; Moroishi, T.; Mo, J.-S.; Plouffe, S.W.; Meng, Z.; Lin, K.C.; Yu, F.-X.; Alexander, C.M.; et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell 2015, 162, 780–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Xia, W.; Fisher, G.J.; Voorhees, J.J.; Quan, T. YAP/TAZ Regulates TGF-β/Smad3 Signaling by Induction of Smad7 via AP-1 in Human Skin Dermal Fibroblasts. Cell Commun. Signal. 2018, 16, 18. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Peng, L.; Li, Z.; Tan, G.; Liang, E.; Chen, S.; Zhao, X.; Zhi, F. YAP Triggers the Wnt/β-Catenin Signalling Pathway and Promotes Enterocyte Self-Renewal, Regeneration and Tumorigenesis after DSS-Induced Injury. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Ouyang, T.; Meng, W.; Li, M.; Hong, T.; Zhang, N. Recent Advances of the Hippo/YAP Signaling Pathway in Brain Development and Glioma. Cell Mol. Neurobiol. 2020, 40, 495–510. [Google Scholar] [CrossRef]
- Lv, Y.; Kim, K.; Sheng, Y.; Cho, J.; Qian, Z.; Zhao, Y.-Y.; Hu, G.; Pan, D.; Malik, A.B.; Hu, G. YAP Controls Endothelial Activation and Vascular Inflammation Through TRAF6. Circ. Res. 2018, 123, 43–56. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, J.; Li, W.; Wu, A.; Zhang, X.; Tong, W.; Ho, K.K.; Qin, L.; Song, H.; Mak, K.K. Reciprocal Inhibition of YAP/TAZ and NF-ΚB Regulates Osteoarthritic Cartilage Degradation. Nat. Commun. 2018, 9, 4564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Zhou, L.; Ling, L.; Meng, X.; Chu, F.; Zhang, S.; Zhou, F. The Crosstalk Between Hippo-YAP Pathway and Innate Immunity. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Palma, V.; Lim, D.A.; Dahmane, N.; Sánchez, P.; Brionne, T.C.; Herzberg, C.D.; Gitton, Y.; Carleton, A.; Alvarez-Buylla, A.; Ruiz i Altaba, A. Sonic Hedgehog Controls Stem Cell Behavior in the Postnatal and Adult Brain. Development 2005, 132, 335–344. [Google Scholar] [CrossRef] [Green Version]
- Galvin, K.E.; Ye, H.; Erstad, D.J.; Feddersen, R.; Wetmore, C. Gli1 Induces G2/M Arrest and Apoptosis in Hippocampal but Not Tumor-Derived Neural Stem Cells. Stem Cells 2008, 26, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Lauth, M.; Rohnalter, V.; Bergström, A.; Kooshesh, M.; Svenningsson, P.; Toftgård, R. Antipsychotic Drugs Regulate Hedgehog Signaling by Modulation of 7-Dehydrocholesterol Reductase Levels. Mol. Pharmacol. 2010, 78, 486–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyd, P.J.; Cunliffe, V.T.; Roy, S.; Wood, J.D. Sonic Hedgehog Functions Upstream of Disrupted-in-Schizophrenia 1 (Disc1): Implications for Mental Illness. Biol. Open 2015, 4, 1336–1343. [Google Scholar] [CrossRef] [Green Version]
- Hrckulak, D.; Kolar, M.; Strnad, H.; Korinek, V. TCF/LEF Transcription Factors: An Update from the Internet Resources. Cancers 2016, 8, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imajo, M.; Miyatake, K.; Iimura, A.; Miyamoto, A.; Nishida, E. A Molecular Mechanism That Links Hippo Signalling to the Inhibition of Wnt/β-Catenin Signalling. EMBO J. 2012, 31, 1109–1122. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.C.; Moroishi, T.; Meng, Z.; Jeong, H.-S.; Plouffe, S.W.; Sekido, Y.; Han, J.; Park, H.W.; Guan, K.-L. Regulation of Hippo Pathway Transcription Factor TEAD by P38 MAPK-Induced Cytoplasmic Translocation. Nat. Cell Biol. 2017, 19, 996–1002. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.C.; Park, H.W.; Guan, K.-L. Regulation of the Hippo Pathway Transcription Factor TEAD. Trends Biochem. Sci. 2017, 42, 862–872. [Google Scholar] [CrossRef]
- Lee, D.-H.; Park, J.O.; Kim, T.-S.; Kim, S.-K.; Kim, T.-H.; Kim, M.-C.; Park, G.S.; Kim, J.-H.; Kuninaka, S.; Olson, E.N.; et al. LATS-YAP/TAZ Controls Lineage Specification by Regulating TGFβ Signaling and Hnf4α Expression during Liver Development. Nat. Commun. 2016, 7, 11961. [Google Scholar] [CrossRef]
- Varelas, X.; Samavarchi-Tehrani, P.; Narimatsu, M.; Weiss, A.; Cockburn, K.; Larsen, B.G.; Rossant, J.; Wrana, J.L. The Crumbs Complex Couples Cell Density Sensing to Hippo-Dependent Control of the TGF-β-SMAD Pathway. Dev. Cell 2010, 19, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liu, H.; Shen, S.; Guo, X.; Yan, H.; Ji, X.; Li, L.; Huang, J.; Feng, X.-H.; Zhao, B. YAP Activates the Hippo Pathway in a Negative Feedback Loop. Cell Res. 2015, 25, 1175–1178. [Google Scholar] [CrossRef] [Green Version]
- Moroishi, T.; Park, H.W.; Qin, B.; Chen, Q.; Meng, Z.; Plouffe, S.W.; Taniguchi, K.; Yu, F.-X.; Karin, M.; Pan, D.; et al. A YAP/TAZ-Induced Feedback Mechanism Regulates Hippo Pathway Homeostasis. Genes Dev. 2015, 29, 1271–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, H.D.; Kim, D.H.; Jeong, H.-S.; Park, H.W. Regulation of TEAD Transcription Factors in Cancer Biology. Cells 2019, 8, 600. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, R.D.; Angelo, L.S.; Kurzrock, R. NF2/Merlin in Hereditary Neurofibromatosis 2 versus Cancer: Biologic Mechanisms and Clinical Associations. Oncotarget 2013, 5, 67–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shy, B.R.; Wu, C.-I.; Khramtsova, G.F.; Zhang, J.Y.; Olopade, O.I.; Goss, K.H.; Merrill, B.J. Regulation of Tcf7l1 DNA Binding and Protein Stability as Principal Mechanisms of Wnt/β-Catenin Signaling. Cell Rep. 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Medoff, B.D.; Chen, L.; Li, L.; Zhang, X.; Praskova, M.; Liu, M.; Landry, A.; Blumberg, R.S.; Boussiotis, V.A.; et al. The Nore1B/Mst1 Complex Restrains Antigen Receptor-Induced Proliferation of Naïve T Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 20321–20326. [Google Scholar] [CrossRef] [Green Version]
- Mou, F.; Praskova, M.; Xia, F.; Van Buren, D.; Hock, H.; Avruch, J.; Zhou, D. The Mst1 and Mst2 Kinases Control Activation of Rho Family GTPases and Thymic Egress of Mature Thymocytes. J. Exp. Med. 2012, 209, 741–759. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Sun, X.; Wang, P.; Zhang, S.; Wang, X.; Wu, H.; Hong, L.; Xie, C.; Li, X.; Zhao, H.; et al. Kinases Mst1 and Mst2 Positively Regulate Phagocytic Induction of Reactive Oxygen Species and Bactericidal Activity. Nat. Immunol. 2015, 16, 1142–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boro, M.; Singh, V.; Balaji, K.N. Mycobacterium Tuberculosis -Triggered Hippo Pathway Orchestrates CXCL1/2 Expression to Modulate Host Immune Responses. Sci. Rep. 2016, 6, 37695. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, Y.; Hu, G.; Zhou, J.; Mei, L.; Xiong, W.-C. YAP Is a Critical Inducer of SOCS3, Preventing Reactive Astrogliosis. Cereb. Cortex 2016, 26, 2299–2310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalilzadeh, M.; Hassanzadeh, F.; Aghamiri, H.; Dehpour, A.R.; Shafaroodi, H. Aripiprazole Prevents from Development of Vincristine-Induced Neuropathic Nociception by Limiting Neural NOS Overexpression and NF-KB Hyperactivation. Cancer Chemothe. Pharm. 2020, 86, 393–404. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Tian, Y.; Wu, X.; He, Y.; Li, C.; Namaka, M.; Kong, J.; Li, H.; Xiao, L. Quetiapine Inhibits Microglial Activation by Neutralizing Abnormal STIM1-Mediated Intercellular Calcium Homeostasis and Promotes Myelin Repair in a Cuprizone-Induced Mouse Model of Demyelination. Front. Cell. Neurosci. 2015, 9. [Google Scholar] [CrossRef]
- Jeon, S.; Kim, S.H.; Shin, S.Y.; Lee, Y.H. Clozapine Reduces Toll-like Receptor 4/NF-ΚB-Mediated Inflammatory Responses through Inhibition of Calcium/Calmodulin-Dependent Akt Activation in Microglia. Prog. Neuro Psychopharmacol. Biol. Psychiatry 2018, 81, 477–487. [Google Scholar] [CrossRef]
- Troib, A.; Azab, A.N. Effects of Psychotropic Drugs on Nuclear Factor Kappa B. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 1198–1208. [Google Scholar]
- Shishodia, S.; Majumdar, S.; Banerjee, S.; Aggarwal, B.B. Ursolic Acid Inhibits Nuclear Factor-KappaB Activation Induced by Carcinogenic Agents through Suppression of IkappaBalpha Kinase and P65 Phosphorylation: Correlation with down-Regulation of Cyclooxygenase 2, Matrix Metalloproteinase 9, and Cyclin D1. Cancer Res. 2003, 63, 4375–4383. [Google Scholar] [PubMed]
- Humar, M.; Dohrmann, H.; Stein, P.; Andriopoulos, N.; Goebel, U.; Roesslein, M.; Schmidt, R.; Schwer, C.I.; Loop, T.; Geiger, K.K.; et al. Thionamides Inhibit the Transcription Factor Nuclear Factor-KappaB by Suppression of Rac1 and Inhibitor of KappaB Kinase Alpha. J. Pharmacol. Exp. Ther. 2008, 324, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Mack Strong, V.E.; Mackrell, P.J.; Concannon, E.M.; Mestre, J.R.; Smyth, G.P.; Schaefer, P.A.; Stapleton, P.P.; Daly, J.M. NS-398 Treatment after Trauma Modifies NF-KappaB Activation and Improves Survival. J. Surg. Res. 2001, 98, 40–46. [Google Scholar] [CrossRef]
- Roy, A.; Ghosh, A.; Jana, A.; Liu, X.; Brahmachari, S.; Gendelman, H.E.; Pahan, K. Sodium Phenylbutyrate Controls Neuroinflammatory and Antioxidant Activities and Protects Dopaminergic Neurons in Mouse Models of Parkinson’s Disease. PLoS ONE 2012, 7, e38113. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.-G.; Ji, C.-H.; Shi, M.-H. The Anti-Infection Drug Furazolidone Inhibits NF-ΚB Signaling and Induces Cell Apoptosis in Small Cell Lung Cancer. Kaohsiung J. Med. Sci. 2020, 36, 998–1003. [Google Scholar] [CrossRef]
- Birukova, A.A.; Wu, T.; Tian, Y.; Meliton, A.; Sarich, N.; Tian, X.; Leff, A.; Birukov, K.G. Iloprost Improves Endothelial Barrier Function in Lipopolysaccharide-Induced Lung Injury. Eur. Respir. J. 2013, 41, 165–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen-Lahav, M.; Shany, S.; Tobvin, D.; Chaimovitz, C.; Douvdevani, A. Vitamin D Decreases NFkappaB Activity by Increasing IkappaBalpha Levels. Nephrol. Dial. Transpl. 2006, 21, 889–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Xia, Y.; Gao, T.; Xu, F.; Lei, Q.; Peng, C.; Yang, Y.; Xue, Q.; Hu, X.; Wang, Q.; et al. The Antipsychotic Agent Trifluoperazine Hydrochloride Suppresses Triple-Negative Breast Cancer Tumor Growth and Brain Metastasis by Inducing G0/G1 Arrest and Apoptosis. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreau, R.A.; Whitaker, B.D.; Hicks, K.B. Phytosterols, Phytostanols, and Their Conjugates in Foods: Structural Diversity, Quantitative Analysis, and Health-Promoting Uses. Prog. Lipid Res. 2002, 41, 457–500. [Google Scholar] [CrossRef]
- Ikeda, Y.; Murakami, A.; Ohigashi, H. Ursolic Acid: An Anti- and pro-Inflammatory Triterpenoid. Mol. Nutr. Food Res. 2008, 52, 26–42. [Google Scholar] [CrossRef]
- Checker, R.; Sandur, S.K.; Sharma, D.; Patwardhan, R.S.; Jayakumar, S.; Kohli, V.; Sethi, G.; Aggarwal, B.B.; Sainis, K.B. Potent Anti-Inflammatory Activity of Ursolic Acid, a Triterpenoid Antioxidant, Is Mediated through Suppression of NF-ΚB, AP-1 and NF-AT. PLoS ONE 2012, 7, e31318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, D.G.; Neis, V.B.; Balen, G.O.; Colla, A.; Cunha, M.P.; Dalmarco, J.B.; Pizzolatti, M.G.; Prediger, R.D.; Rodrigues, A.L.S. Antidepressant-like Effect of Ursolic Acid Isolated from Rosmarinus officinalis L. in Mice: Evidence for the Involvement of the Dopaminergic System. Pharmacol. Biochem. Behav. 2012, 103, 204–211. [Google Scholar] [CrossRef] [Green Version]
- Habtemariam, S. Iridoids and Other Monoterpenes in the Alzheimer’s Brain: Recent Development and Future Prospects. Molecules 2018, 23, 117. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Hryb, A.B.; Pazini, F.L.; Kaster, M.P.; Rodrigues, A.L.S. Therapeutic Potential of Ursolic Acid to Manage Neurodegenerative and Psychiatric Diseases. CNS Drugs 2017, 31, 1029–1041. [Google Scholar] [CrossRef]
- Gudoityte, E.; Arandarcikaite, O.; Mazeikiene, I.; Bendokas, V.; Liobikas, J. Ursolic and Oleanolic Acids: Plant Metabolites with Neuroprotective Potential. Int. J. Mol. Sci. 2021, 22, 4599. [Google Scholar] [CrossRef]
- Bortolasci, C.C.; Spolding, B.; Kidnapillai, S.; Richardson, M.F.; Vasilijevic, N.; Martin, S.D.; Gray, L.J.; McGee, S.L.; Berk, M.; Walder, K. Effects of Psychoactive Drugs on Cellular Bioenergetic Pathways. World J. Biol. Psychiatry Off. J. World Fed. Soc. Biol. Psychiatry 2020, 1–15. [Google Scholar] [CrossRef]
- Kidnapillai, S.; Bortolasci, C.C.; Udawela, M.; Panizzutti, B.; Spolding, B.; Connor, T.; Sanigorski, A.; Dean, O.M.; Crowley, T.; Jamain, S.; et al. The Use of a Gene Expression Signature and Connectivity Map to Repurpose Drugs for Bipolar Disorder. World J. Biol. Psychiatry 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bortolasci, C.C.; Spolding, B.; Callaly, E.; Martin, S.; Panizzutti, B.; Kidnapillai, S.; Connor, T.; Hasebe, K.; Mohebbi, M.; Dean, O.M.; et al. Mechanisms Underpinning the Polypharmacy Effects of Medications in Psychiatry. Int. J. Neuropsychopharmacol. 2018, 21, 582–591. [Google Scholar] [CrossRef]
- Melbourne, J.K.; Pang, Y.; Park, M.R.; Sudhalkar, N.; Rosen, C.; Sharma, R.P. Treatment with the Antipsychotic Risperidone Is Associated with Increased M1-like JAK-STAT1 Signature Gene Expression in PBMCs from Participants with Psychosis and THP-1 Monocytes and Macrophages. Int. Immunopharmacol. 2020, 79, 106093. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Gwag, T.; Sui, Y.; Park, S.-H.; Zhou, X.; Zhou, C. The Atypical Antipsychotic Quetiapine Induces Hyperlipidemia by Activating Intestinal PXR Signaling. JCI Insight 2019, 4, e125657. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- STAR: Ultrafast Universal RNA-Seq Aligner Bioinformatics Oxford Academic. Available online: https://Academic.Oup.com/Bioinformatics/Article/29/1/15/272537 (accessed on 2 June 2021).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/tool/81287/R-A-Language-and-Environment-for-Statistical-Computing (accessed on 2 June 2021).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiser, T.; Feng, G. Modeling Psychiatric Disorders for Developing Effective Treatments. Nat. Med. 2015, 21, 979–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
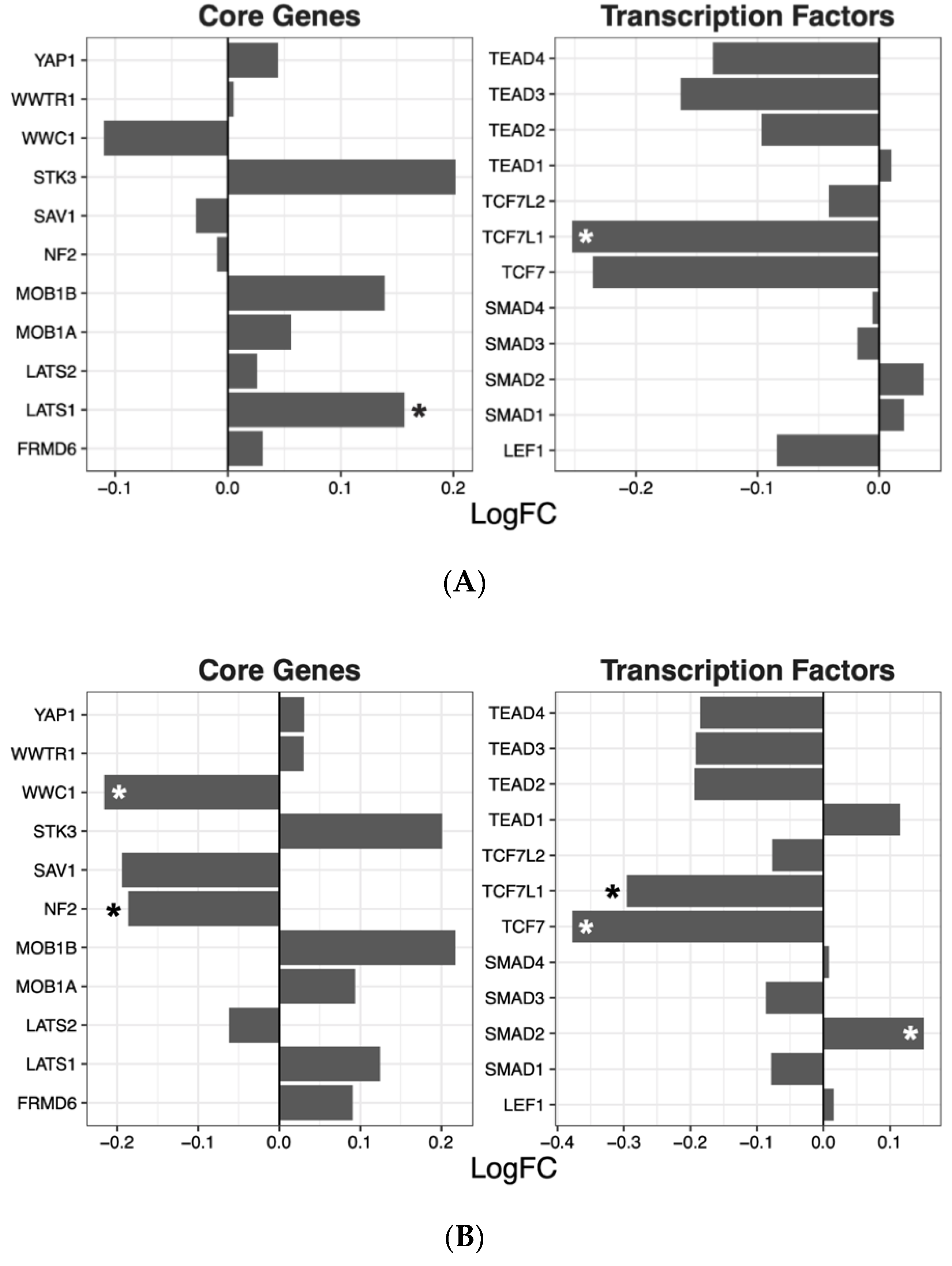
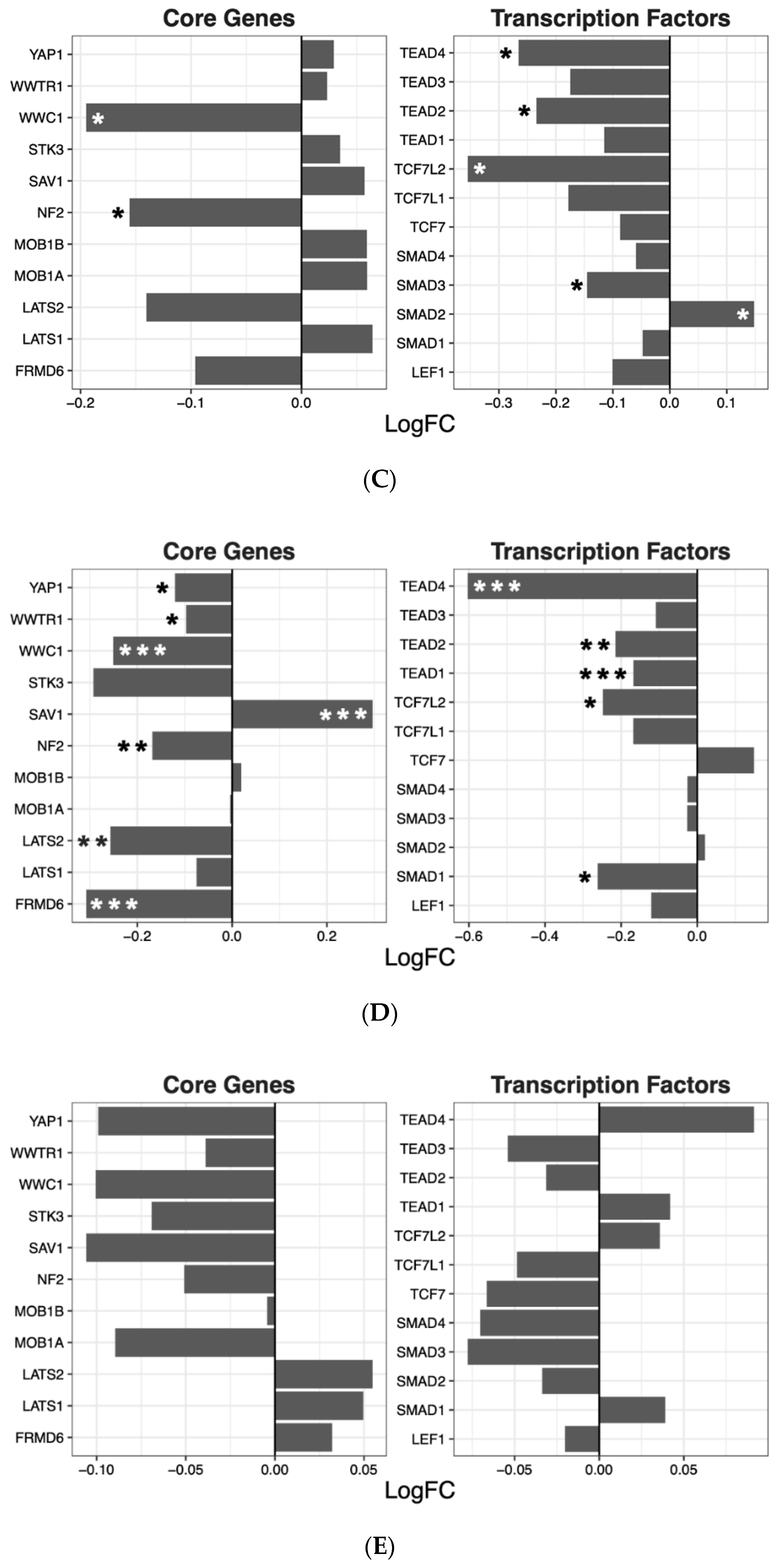
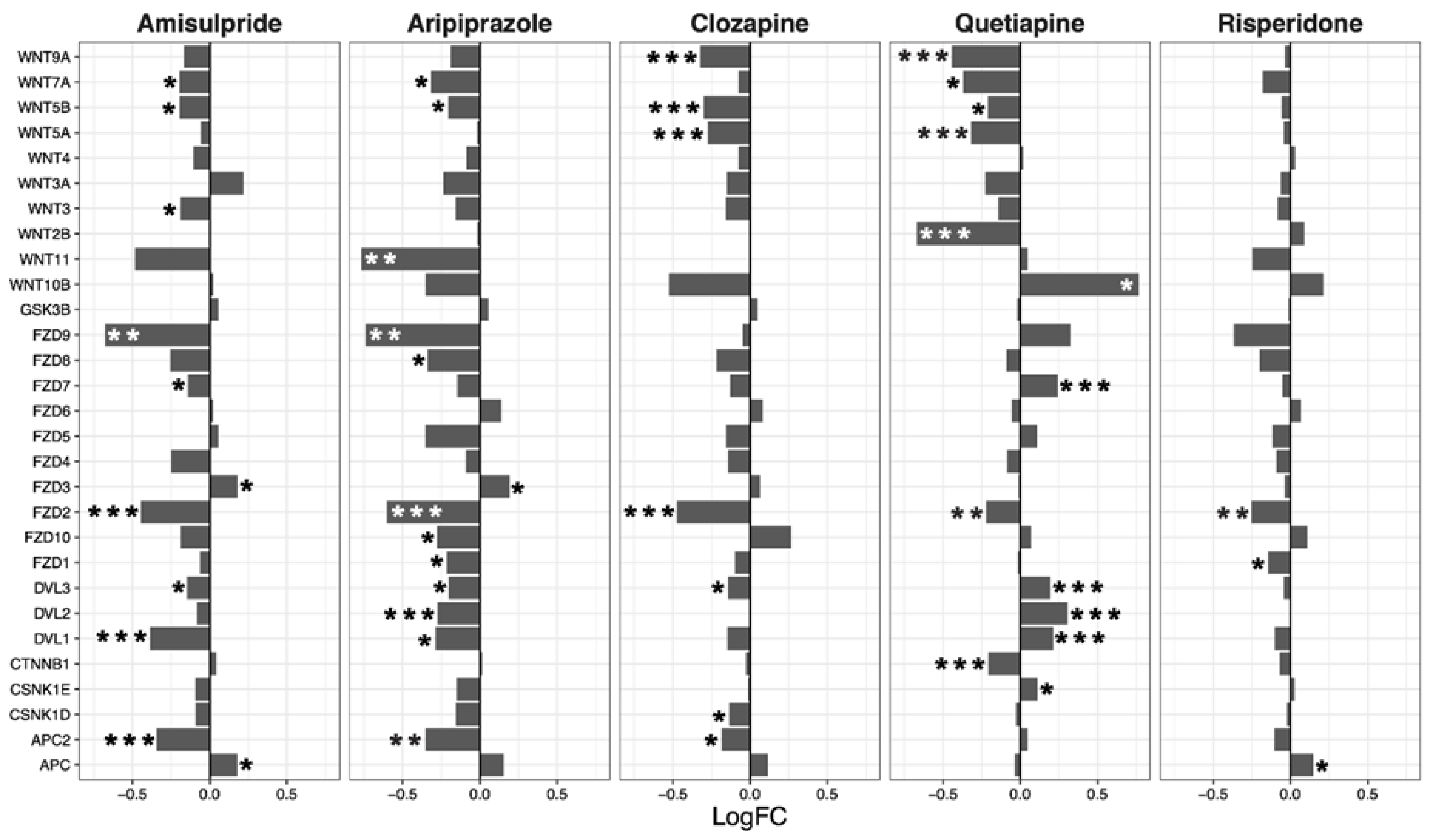
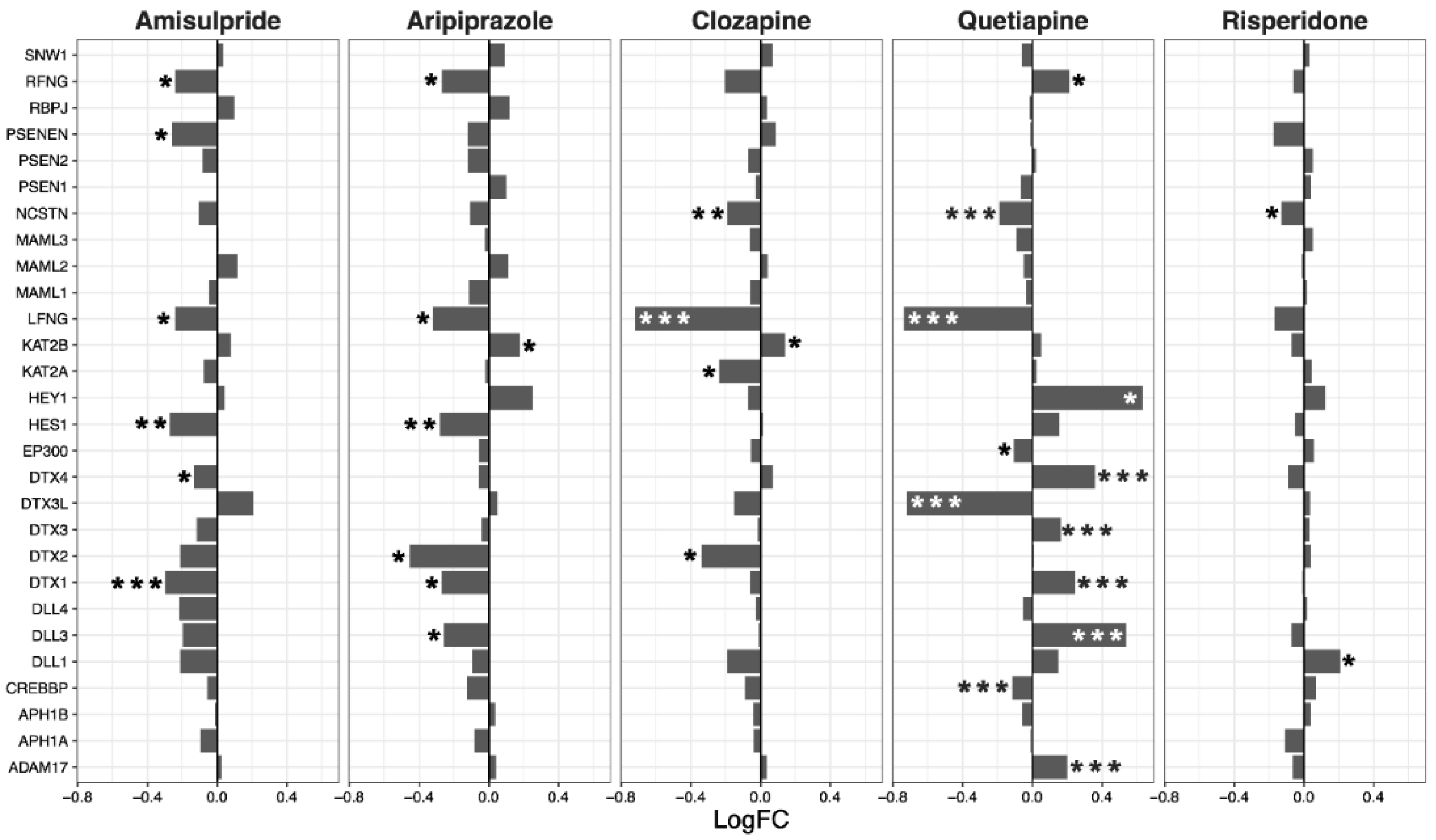
| Drug | ES | NES | p-Value | q-Value |
|---|---|---|---|---|
| Clozapine | −0.61 | −2.09 | 0.00013 | 0.0012 |
| Aripiprazole | −0.54 | −2.04 | 0.00015 | 0.0044 |
| Risperidone | −0.49 | −1.92 | 0.00024 | 0.0049 |
| Quetiapine | −0.60 | −1.79 | 0.00027 | 0.0052 |
| Amisulpride | −0.45 | −1.61 | 0.00085 | 0.016 |
| Lithium | −0.40 | −1.32 | 0.048 | 0.21 |
| Lamotrigine | −0.29 | −1.15 | 0.14 | 0.35 |
| Valproate | 0.31 | 0.89 | 0.67 | 0.66 |
| Wnt | Notch | Hedgehog | TGF-β | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LogFC | p-Value | LogFC | p-Value | LogFC | p-Value | LogFC | p-Value | |||||
| Mean | SEM | Mean | SEM | Mean | SEM | Mean | SEM | |||||
| Amisulpride | −0.13 | 0.04 | 0.0018 | −0.08 | 0.03 | 0.0032 | −0.03 | 0.02 | 0.18 | −0.08 | 0.05 | 0.19 |
| Aripiprazole | −0.21 | 0.04 | 4.80 × 10−6 | −0.07 | 0.03 | 0.043 | −0.04 | 0.02 | 0.07 | −0.05 | 0.1 | 0.66 |
| Clozapine | −0.11 | 0.03 | 0.0013 | −0.08 | 0.03 | 0.0033 | −0.05 | 0.02 | 0.025 | −0.21 | 0.08 | 0.07 |
| Quetiapine | −0.02 | 0.05 | 0.64 | 0.02 | 0.05 | 0.63 | −0.04 | 0.03 | 0.26 | −0.27 | 0.15 | 0.15 |
| Risperidone | −0.06 | 0.02 | 0.02 | −0.01 | 0.02 | 0.72 | 0 | 0.01 | 0.67 | −0.05 | 0.02 | 0.06 |
| Rank | CMap Name | Enrichment | p-Value | Class |
|---|---|---|---|---|
| 1 | Ursolic acid | 0.91 | 0.00008 | Triterpenoid |
| 2 | Levothyroxine sodium | 0.89 | 0.00014 | Thyroid hormones |
| 3 | Ajmaline | 0.94 | 0.00022 | Antiarrhythmic agent |
| 4 | 5707885 | 0.87 | 0.00044 | Unknown |
| 6 | Carbimazole | 0.90 | 0.00182 | Imidazole–thyroid function |
| 7 | 0297417–0002b | 0.88 | 0.00324 | Unknown |
| 9 | Ns-398 | 0.87 | 0.00463 | COX-2 inhibitor (anti-inflammatory) |
| 10 | Cefapirin | 0.78 | 0.00473 | Antibiotic |
| 11 | Estrone | 0.77 | 0.00539 | Estrogen steroid |
| 12 | Strophanthidin | 0.77 | 0.00561 | Cardiac glycoside |
| 17 | Picotamide | 0.68 | 0.00937 | Platelet aggregation inhibitor |
| 20 | Maprotiline | 0.72 | 0.0117 | Tetracyclic antidepressant |
| 22 | Monastrol | 0.53 | 0.01256 | Antimitotic agent |
| 24 | Sr-95639a (Aminopyridazine) | 0.72 | 0.01331 | Muscarinic agonist |
| 28 | Benzethonium chloride | 0.78 | 0.02097 | Antiseptics and Disinfectants |
| 30 | Ciprofibrate | 0.68 | 0.0228 | Fibrate (lipid-lowering agent) |
| 31 | 5255229 | 0.90 | 0.02286 | Unknown |
| 32 | Methazolamide | 0.68 | 0.02316 | Carbonic anhydrase inhibitors |
| 33 | Bumetanide | 0.68 | 0.02379 | Diuretic |
| 37 | Sodium phenylbutyrate | 0.52 | 0.02663 | Urea cycle disorder treatment agents |
| 38 | Tenoxicam | 0.67 | 0.02795 | NSAID (Nonsteroidal anti-inflammatory drug) |
| 40 | Ah-23848 | 0.76 | 0.0291 | Prostanoid EP4 antagonist |
| 41 | Furazolidone | 0.66 | 0.03058 | Oxazolidine–antibiotic agent |
| 42 | Iloprost | 0.75 | 0.03201 | Vasodilator |
| 43 | Minoxidil | 0.60 | 0.03344 | Vasodilator |
| 46 | Hyoscyamine | 0.59 | 0.03475 | Anticholinergic/antispasmodic |
| 45 | Prestwick-642 (Epicatechin) | 0.65 | 0.03475 | Catechin–antioxidant flavonoid |
| 48 | Dihydrostreptomycin | 0.59 | 0.03529 | Antibiotic |
| 49 | Nisoxetine | 0.65 | 0.03539 | Antidepressant -SNRI |
| 50 | Ergocalciferol | 0.65 | 0.03589 | Vitamin D analogue |
| 52 | Ceforanide | 0.65 | 0.03804 | Antibiotic |
| 55 | Pheniramine | 0.58 | 0.04175 | Antihistamine |
| 56 | Ifenprodil | 0.64 | 0.04245 | NMDA receptor antagonist |
| 57 | Pha-00745360 | 0.46 | 0.04312 | Unknown |
| 58 | Clozapine | 0.32 | 0.04422 | Atypical antipsychotic |
| 60 | Diphemanil methylsulfate | 0.57 | 0.04552 | Muscarinic antagonist |
| 63 | Piperacetazine | 0.63 | 0.04729 | Antipsychotic prodrug |
| 66 | Trifluoperazine | 0.63 | 0.04882 | Phenothiazine antipsychotic |
| 68 | Lidoflazine | 0.71 | 0.04974 | Vasodilator |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panizzutti, B.; Bortolasci, C.C.; Spolding, B.; Kidnapillai, S.; Connor, T.; Richardson, M.F.; Truong, T.T.T.; Liu, Z.S.J.; Morris, G.; Gray, L.; et al. Transcriptional Modulation of the Hippo Signaling Pathway by Drugs Used to Treat Bipolar Disorder and Schizophrenia. Int. J. Mol. Sci. 2021, 22, 7164. https://doi.org/10.3390/ijms22137164
Panizzutti B, Bortolasci CC, Spolding B, Kidnapillai S, Connor T, Richardson MF, Truong TTT, Liu ZSJ, Morris G, Gray L, et al. Transcriptional Modulation of the Hippo Signaling Pathway by Drugs Used to Treat Bipolar Disorder and Schizophrenia. International Journal of Molecular Sciences. 2021; 22(13):7164. https://doi.org/10.3390/ijms22137164
Chicago/Turabian StylePanizzutti, Bruna, Chiara C. Bortolasci, Briana Spolding, Srisaiyini Kidnapillai, Timothy Connor, Mark F. Richardson, Trang T. T. Truong, Zoe S. J. Liu, Gerwyn Morris, Laura Gray, and et al. 2021. "Transcriptional Modulation of the Hippo Signaling Pathway by Drugs Used to Treat Bipolar Disorder and Schizophrenia" International Journal of Molecular Sciences 22, no. 13: 7164. https://doi.org/10.3390/ijms22137164
APA StylePanizzutti, B., Bortolasci, C. C., Spolding, B., Kidnapillai, S., Connor, T., Richardson, M. F., Truong, T. T. T., Liu, Z. S. J., Morris, G., Gray, L., Hyun Kim, J., Dean, O. M., Berk, M., & Walder, K. (2021). Transcriptional Modulation of the Hippo Signaling Pathway by Drugs Used to Treat Bipolar Disorder and Schizophrenia. International Journal of Molecular Sciences, 22(13), 7164. https://doi.org/10.3390/ijms22137164






