Evaluation of Chemical Changes in Laboratory-Induced Colistin-Resistant Klebsiella pneumoniae
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Bacterial Strain and Growth Conditions
3.2. Determination of Minimal Inhibitory Concentration (MIC) of Colistin Against K. pneumoniae ATCC®BAA1705™
3.3. Colistin Resistance Induction
3.4. LPS Extraction
3.5. Determination of Functional Groups in K. pneumoniae Cells and Extracted LPS Using FTIR and Raman Spectroscopies
3.6. Dynamic Light Scattering Assay
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef]
- El Fertas-Aissani, R.; Messai, Y.; Alouache, S.; Bakour, R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol. Biol. 2013, 61, 209–216. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 2014, 20, 821–830. [Google Scholar] [CrossRef] [PubMed]
- E Falagas, M.; A Bliziotis, I.; Kasiakou, S.K.; Samonis, G.; Athanassopoulou, P.; Michalopoulos, A. Outcome of infections due to pandrug-resistant (PDR) Gram-negative bacteria. BMC Infect. Dis. 2005, 5, 24–27. [Google Scholar] [CrossRef]
- Koyama, Y.; Kurosasa, A.; Tsuchiya, A.; Takakuta, K. A new antibiotic “colistin” produced by spore-forming soil bacteria. J. Antibiot. 1950, 3, 457–458. [Google Scholar]
- Komura, S.; Kurahashi, K. Partial purification and properties of L-2, 4-diaminobutyric acid activating enzyme from a Polymyxin E producing organism. J. Biochem. 1979, 86, 1013–1021. [Google Scholar] [CrossRef]
- Paterson, D.; Harris, P. Colistin resistance: A major breach in our last line of defence. Lancet Infect. Dis. 2016, 16, 132–133. [Google Scholar] [CrossRef]
- Ah, Y.-M.; Kim, A.-J.; Lee, J.-Y. Colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2014, 44, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Orsini, F.; Ami, D.; Villa, A.; Sala, G.; Bellotti, M.; Doglia, S. FT-IR microspectroscopy for microbiological studies. J. Microbiol. Methods 2000, 42, 17–27. [Google Scholar] [CrossRef]
- Boschetto, F.; Adachi, T.; Horiguchi, S.; Fainozzi, D.; Parmigiani, F.; Marin, E.; Zhu, W.; McEntire, B.J.; Yamamoto, T.; Kanamura, N.; et al. Monitoring metabolic reactions in Staphylococcus epidermidis exposed to silicon nitride using in situ time-lapse Raman spectroscopy. J. Biomed. Opt. 2018, 23, 056002. [Google Scholar] [CrossRef] [PubMed]
- Kürekçi, C. Applicability of Raman spectroscopy for characterization of three major foodborne pathogens. Harran Univ. Vet. Fak. Derg. 2016, 5, 141–145. [Google Scholar]
- Sandt, C.; Madoulet, C.; Kohler, A.; Allouch, P.; De Champs, C.; Manfait, M.; Sockalingum, G. FT-IR microspectroscopy for early identification of some clinically relevant pathogens. J. Appl. Microbiol. 2006, 101, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Amiali, N.M.; Golding, G.R.; Sedman, J.; Simor, A.E.; Ismail, A.A. Rapid identification of community-associated methicillin-resistant Staphylococcus aureus by Fourier transform infrared spectroscopy. Diagn. Microbiol. Infect. Dis. 2011, 70, 157–166. [Google Scholar] [CrossRef]
- Tareb, R.; Bernardeau, M.; Amiel, C.; Vernoux, J. Usefulness of FTIR spectroscopy to distinguish rough and smooth variants of Lactobacillus farciminis CNCM-I-3699. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hrubanova, K.; Krzyzanek, V.; Nebesarova, J.; Ruzicka, F.; Pilat, Z.; Samek, O. Monitoring Candida parapsilosis and Staphylococcus epidermidis biofilms by a combination of scanning electron microscopy and Raman spectroscopy. Sensors 2018, 18, 4089. [Google Scholar] [CrossRef]
- Łopusiewicz, Ł.; Mazurkiewicz-Zapalowicz, K.; Tkaczuk, C. Chemical changes in spores of the entomopathogenic fungus Metarhizium robertsii after exposure to heavy metals, studied through the use of FTIR spectroscopy. J. Elementology 2019, 25, 487–499. [Google Scholar] [CrossRef]
- Kwiatkowski, P.; Pruss, A.; Wojciuk, B.; Dołęgowska, B.; Wajs-Bonikowska, A.; Sienkiewicz, M.; Mężyńska, M.; Łopusiewicz, Ł. The influence of essential oil compounds on antibacterial activity of mupirocin-susceptible and induced low-level mupirocin-resistant MRSA strains. Molecules 2019, 24, 3105. [Google Scholar] [CrossRef]
- Salman, A.; Tsror, L.; Pomerantz, A.; Moreh, R.; Mordechai, S.; Huleihel, M. FTIR spectroscopy for detection and identification of fungal phytopathogenes. Spectroscopy 2010, 24, 261–267. [Google Scholar] [CrossRef]
- Beringer, P. The clinical use of colistin in patients with cystic fibrosis. Curr. Opin. Pulm. Med. 2001, 7, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Landman, D.; Georgescu, C.; Martin, D.A.; Quale, J. Polymyxins Revisited. Clin. Microbiol. Rev. 2008, 21, 449–465. [Google Scholar] [CrossRef]
- Neonakis, I.; Samonis, G.; Messaritakis, H.; Baritaki, S.; Georgiladakis, A.; Maraki, S.; Spandidos, D. Resistance status and evolution trends of Klebsiella pneumoniae isolates in a university hospital in Greece: Ineffectiveness of carbapenems and increasing resistance to colistin. Chemotherapy 2010, 56, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Kontopidou, F.; Plachouras, D.; Papadomichelakis, E.; Koukos, G.; Galani, I.; Poulakou, G.; Dimopoulos, G.; Antoniadou, A.; Armaganidis, A.; Giamarellou, H. Colonization and infection by colistin-resistant Gram-negative bacteria in a cohort of critically ill patients. Clin. Microbiol. Infect. 2011, 17, E9–E11. [Google Scholar] [CrossRef]
- Suh, J.-Y.; Son, J.S.; Chung, D.R.; Peck, K.R.; Ko, K.S.; Song, J.-H. Nonclonal emergence of colistin-resistant Klebsiella pneumoniae isolates from blood samples in South Korea. Antimicrob. Agents Chemother. 2010, 54, 560–562. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tan, T.Y.; Ng, S.Y. The in-vitro activity of colistin in gram-negative bacteria. Singap. Med. J. 2006, 47, 621–624. [Google Scholar]
- Walkty, A.; DeCorby, M.; Nichol, K.; Karlowsky, J.A.; Hoban, D.J.; Zhanel, G.G. In vitro activity of colistin (polymyxin E) against 3,480 isolates of Gram-negative bacilli obtained from patients in Canadian hospitals in the CANWARD study, 2007–2008. Antimicrob. Agents Chemother. 2009, 53, 4924–4926. [Google Scholar] [CrossRef] [PubMed]
- Halaby, T.; Al Naiemi, N.; Kluytmans, J.; Van Der Palen, J.A.M.; Vandenbroucke-Grauls, C.M.J.E. Emergence of colistin resistance in Enterobacteriaceae after the introduction of selective digestive tract decontamination in an intensive care unit. Antimicrob. Agents Chemother. 2013, 57, 3224–3229. [Google Scholar] [CrossRef]
- Rodrigues, C.; Sousa, C.; Lopes, J.; Novais, Â.; Peixe, L. A front line on Klebsiella pneumoniae capsular polysaccharide knowledge: Fourier transform infrared spectroscopy as an accurate and fast typing tool. mSystems 2020, 5, 00386–00419. [Google Scholar] [CrossRef]
- Kim, S.; Reuhs, B.; Mauer, L. Use of Fourier transform infrared spectra of crude bacterial lipopolysaccharides and chemometrics for differentiation of Salmonella enterica serotypes. J. Appl. Microbiol. 2005, 99, 411–417. [Google Scholar] [CrossRef]
- Hu, Y.; Zhou, H.; Lu, J.; Sun, Q.; Liu, C.; Zeng, Y.; Zhang, R. Evaluation of the IR Biotyper for Klebsiella pneumoniae typing and its potentials in hostpial hygiene management. Microb. Biotechnol. 2020, 1–10. [Google Scholar] [CrossRef]
- Grunert, T.; Jovanovic, D.; Sirisarn, W.; Johler, S.; Weidenmaier, C.; Ehling-Schulz, M.; Xia, G. Analysis of Staphylococcus aureus wall teichoic acid glycoepitopes by Fourier transform infrared spectroscopy provides novel insights into the staphylococcal glycocode. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Devi, A.K.; Balamurugan, K. Lipopolysaccharide of Klebsiella pneumoniae attenuates immunity of Caenorhabditis elegans and evades by altering its supramolecular structure. RSC Adv. 2016, 6, 30070–30080. [Google Scholar] [CrossRef]
- Muñoz, A.J.; Espínola, F.; Moya, M.; Ruiz, E. Biosorption of Pb(II) ions by Klebsiella sp. 3S1 isolated from a wastewater treatment plant: Kinetics and mechanisms studies. BioMed Res. Int. 2015, 2015, 1–12. [Google Scholar] [CrossRef]
- Dieckmann, R.; Hammerl, J.A.; Hahmann, H.; Wicke, A.; Kleta, S.; Dabrowski, P.W.; Nitsche, A.; Stämmler, M.; Al Dahouk, S.; Lasch, P. Rapid characterisation of Klebsiella oxytoca isolates from contaminated liquid hand soap using mass spectrometry, FTIR and Raman spectroscopy. Faraday Discuss. 2015, 187, 353–375. [Google Scholar] [CrossRef] [PubMed]
- Żarnowiec, P.; Czerwonka, G.; Kaca, W. Fourier transform infrared spectroscopy as a tool in analysis of Proteus mirabilis endotoxins. Adv. Struct. Saf. Stud. 2017, 1600, 113–124. [Google Scholar] [CrossRef]
- Meredith, T.C.; Aggarwal, P.; Mamat, U.; Lindner, B.; Woodard, R. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 2006, 1, 33–42. [Google Scholar] [CrossRef]
- Kamnev, A.; Dyatlova, Y.; Kenzhegulov, O.; Vladimirova, A.; Mamchenkova, P.; Tugarova, A. Fourier transform infrared (FTIR) spectroscopic analyses of microbiological samples and biogenic selenium nanoparticles of microbial origin: Sample preparation effects. Molecules 2021, 26, 1146. [Google Scholar] [CrossRef]
- Töpfer, N.; Müller, M.; Dahms, M.; Ramoji, A.; Popp, J.; Slevogt, H.; Neugebauer, U. Raman spectroscopy reveals LPS-induced changes of biomolecular composition in monocytic THP-1 cells in a label-free manner. Integr. Biol. 2019, 11, 87–98. [Google Scholar] [CrossRef]
- Pascut, F.C.; Goh, H.T.; Welch, N.; Buttery, L.D.; Denning, C.; Notingher, I. Noninvasive detection and imaging of molecular markers in live cardiomyocytes derived from human embryonic stem cells. Biophys. J. 2011, 100, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Brauchle, E.; Knopf, A.; Bauer, H.; Shen, N.; Linder, S.; Monaghan, M.G.; Ellwanger, K.; Layland, S.L.; Brucker, S.Y.; Nsair, A.; et al. Non-invasive chamber-specific identification of cardiomyocytes in differentiating pluripotent stem cells. Stem Cell Rep. 2016, 6, 188–199. [Google Scholar] [CrossRef]
- Olaitan, A.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Pragasam, A.K.; Shankar, C.; Veeraraghavan, B.; Biswas, I.; Nabarro, L.E.B.; Inbanathan, F.Y.; George, B.; Verghese, S. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India—A first report. Front. Microbiol. 2017, 7, 2135. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Thompson, P.; Nation, R.L.; Li, J. Structure−activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef] [PubMed]
- Bialvaei, A.Z.; Kafil, H.S. Colistin, mechanisms and prevalence of resistance. Curr. Med Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.A.; Chan, L.; Hancock, R. Evidence for two distinct mechanisms of resistance to polymyxin B in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1984, 26, 539–545. [Google Scholar] [CrossRef]
- Moore, R.A.; Hancock, R. Involvement of outer membrane of Pseudomonas cepacia in aminoglycoside and polymyxin resistance. Antimicrob. Agents Chemother. 1986, 30, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Dzul, S.P.; Thornton, M.M.; Hohne, D.N.; Stewart, E.J.; Shah, A.A.; Bortz, D.M.; Solomon, M.; Younger, J.G. Contribution of the Klebsiella pneumoniae capsule to bacterial aggregate and biofilm microstructures. Appl. Environ. Microbiol. 2011, 77, 1777–1782. [Google Scholar] [CrossRef]
- Al-Farsi, H.M.; Al-Adwani, S.; Ahmed, S.; Vogt, C.; Ambikan, A.T.; Leber, A.; Al-Jardani, A.; Al-Azri, S.; Al-Muharmi, Z.; Toprak, M.S.; et al. Effects of the antimicrobial peptide LL-37 and innate effector mechanisms in colistin-resistant Klebsiella pneumoniae with mgrB insertions. Front. Microbiol. 2019, 10, 2632. [Google Scholar] [CrossRef]
- Nhu, N.T.K.; Riordan, D.W.; Nhu, T.D.H.; Thanh, D.P.; Thwaites, G.; Lan, N.P.H.; Wren, B.W.; Baker, S.; A Stabler, R. The induction and identification of novel colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 2016, 6, 28291. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST). 2020. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 10.0. Available online: http://www.eucast.org (accessed on 15 May 2020).
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides, EXTRACTION with phenol-water and further applications of the procedure. In Methods in Carbohydrate Chemistry; Whistler, R.L., Wolfan, M.L., Eds.; New York Academic Press: New York, NY, USA, 1965; pp. 83–91. [Google Scholar]
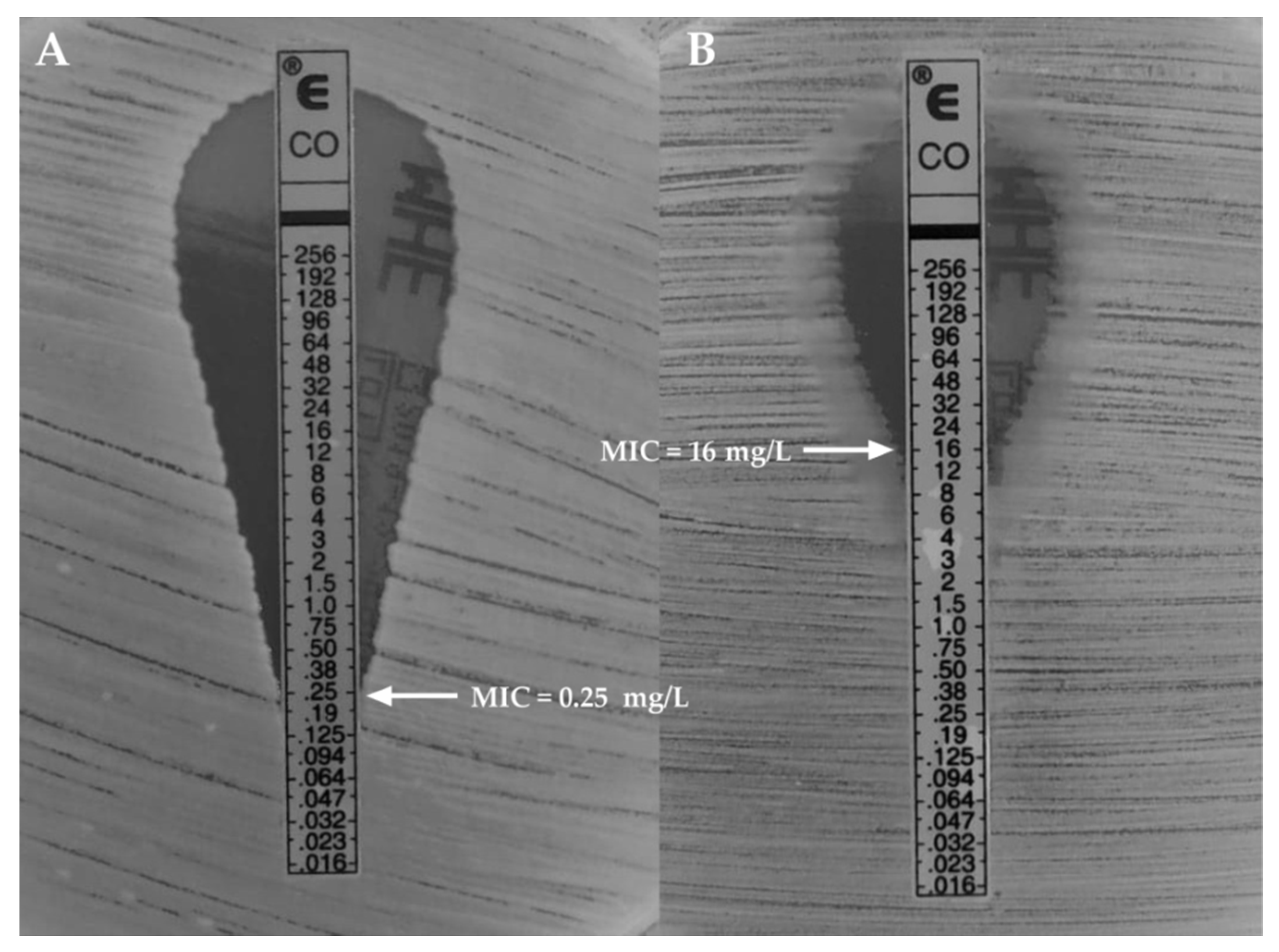
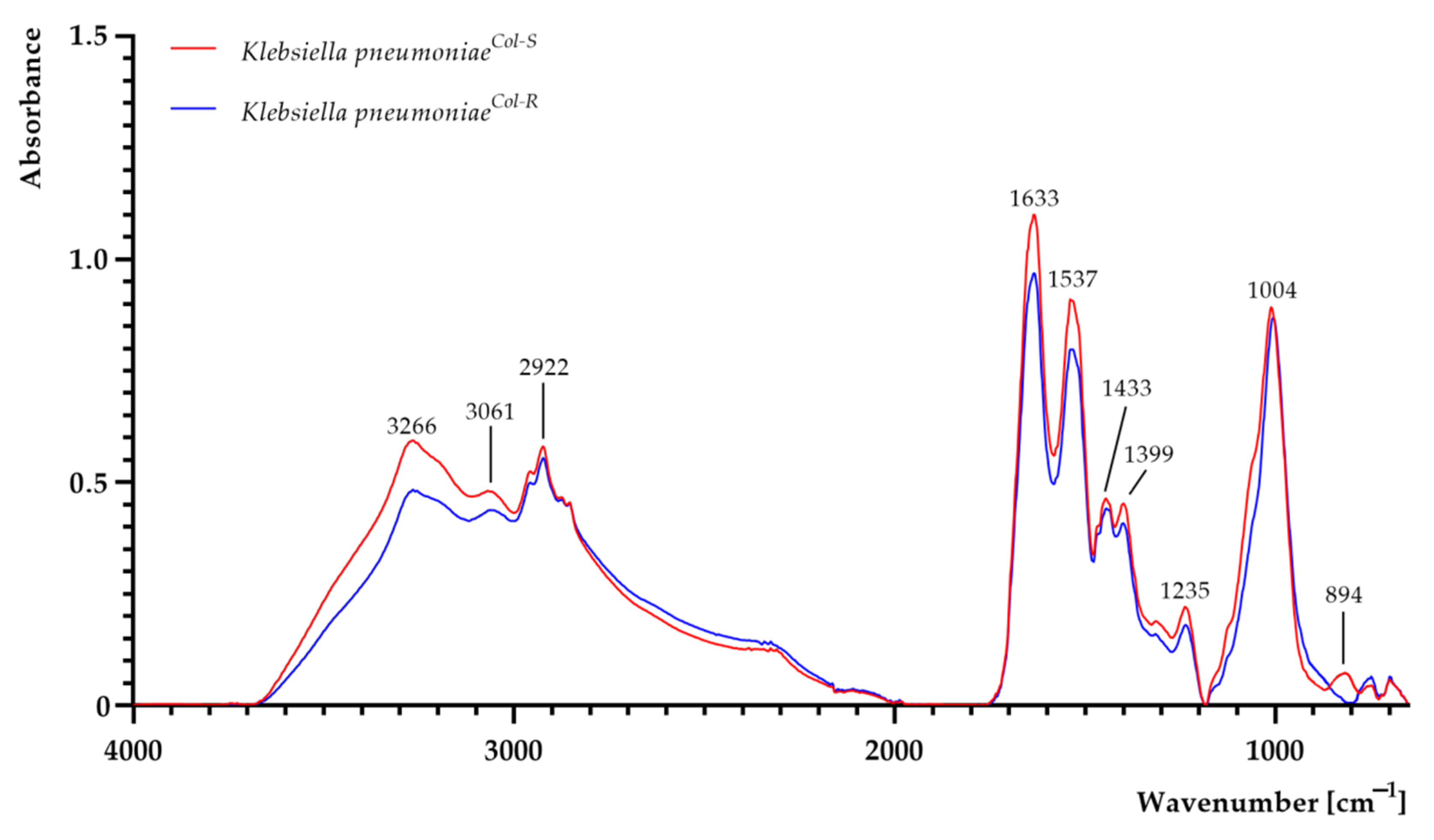
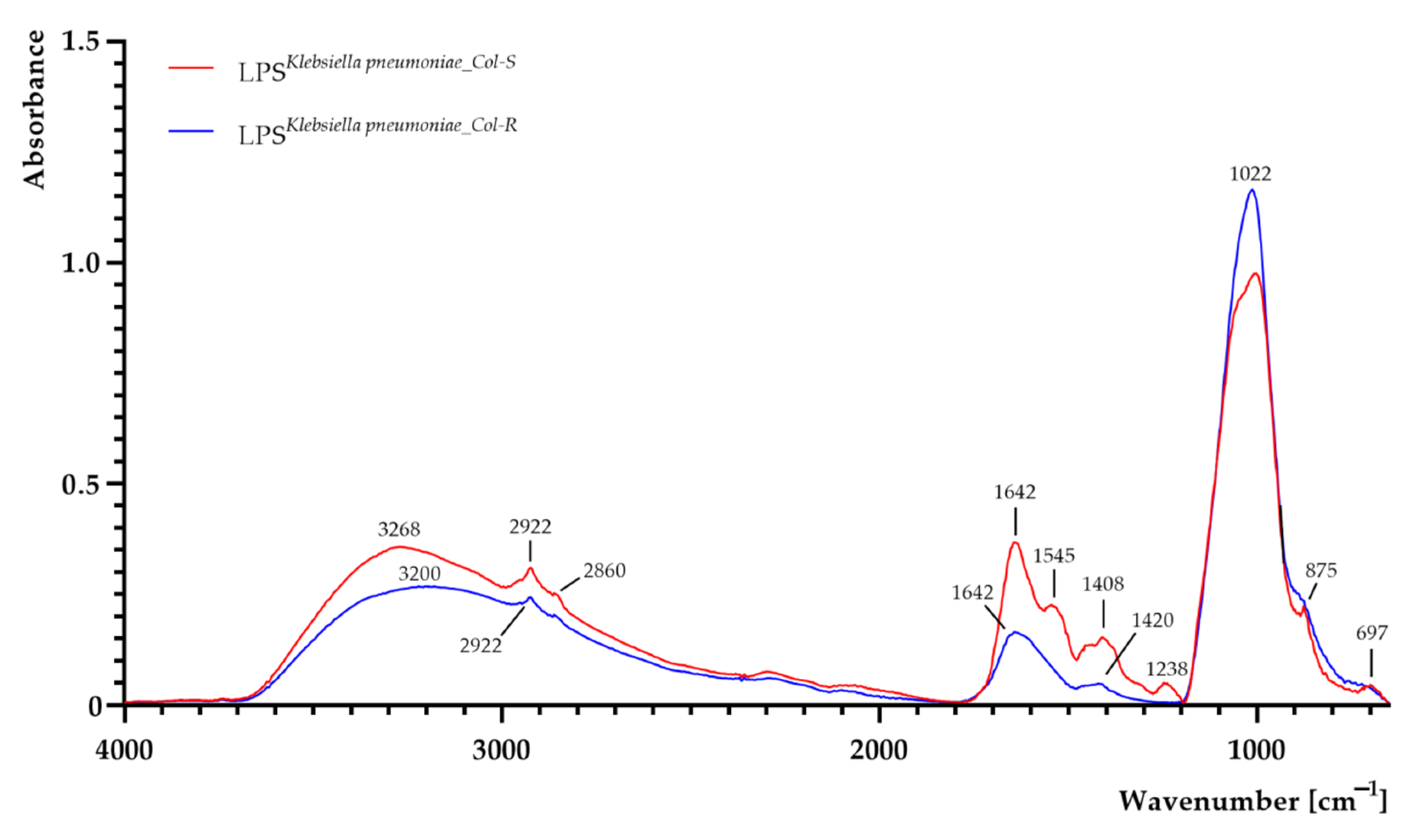

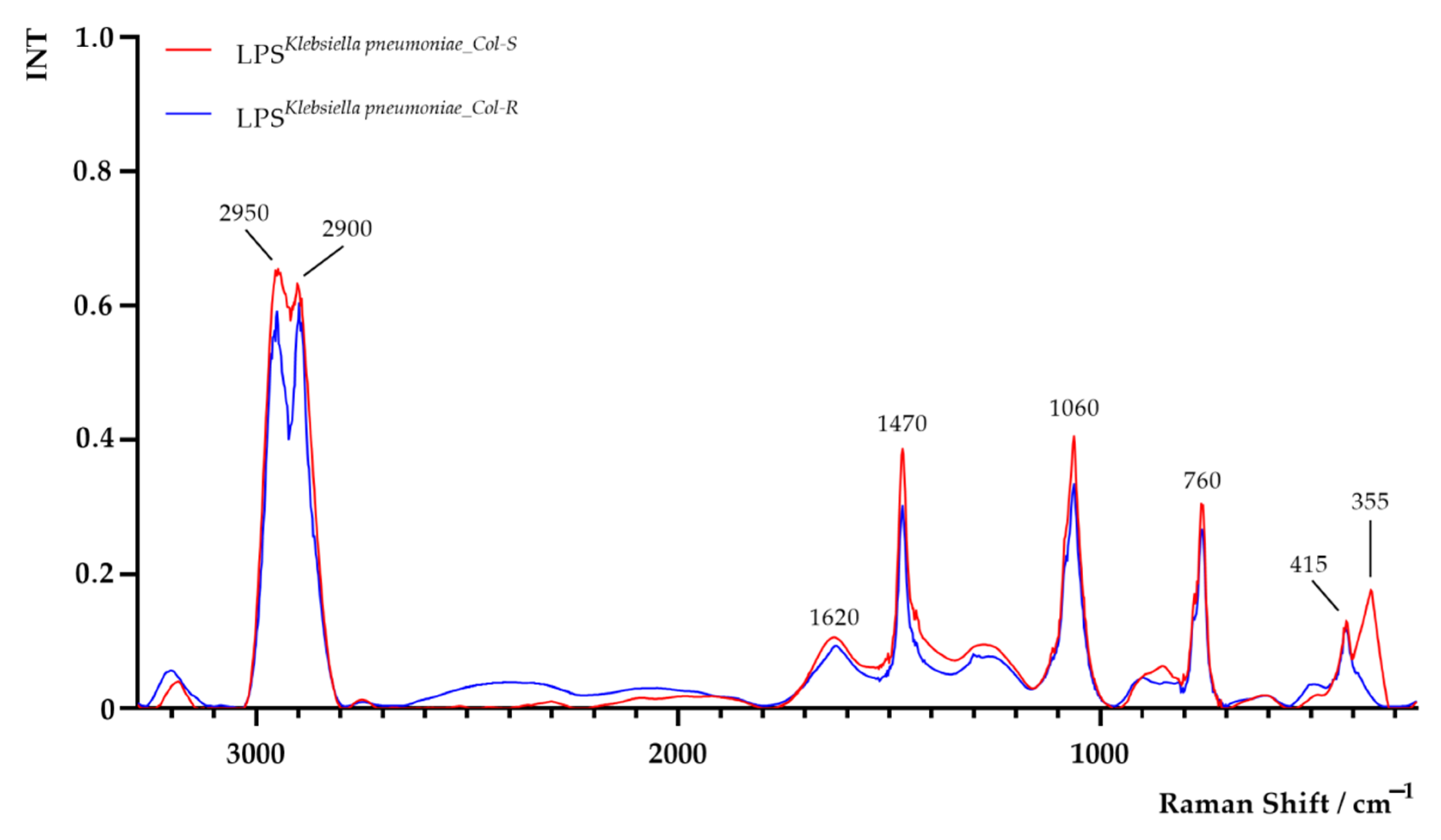
| K. pneumoniae Strains | Hydrodynamic Diameter [nm] | Polydispersity Index (PDI) | ζ-Potential [mV] |
|---|---|---|---|
| K. pneumoniaeCol-S | 2340 ± 40 a | 0.23 ± 0.10 a | −13.3 ± 0.5 a |
| K. pneumoniaeCol-R | 2370 ± 40 a | 0.27 ± 0.13 a | −13.1 ± 0.6 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pruss, A.; Kwiatkowski, P.; Łopusiewicz, Ł.; Masiuk, H.; Sobolewski, P.; Fijałkowski, K.; Sienkiewicz, M.; Smolak, A.; Giedrys-Kalemba, S.; Dołęgowska, B. Evaluation of Chemical Changes in Laboratory-Induced Colistin-Resistant Klebsiella pneumoniae. Int. J. Mol. Sci. 2021, 22, 7104. https://doi.org/10.3390/ijms22137104
Pruss A, Kwiatkowski P, Łopusiewicz Ł, Masiuk H, Sobolewski P, Fijałkowski K, Sienkiewicz M, Smolak A, Giedrys-Kalemba S, Dołęgowska B. Evaluation of Chemical Changes in Laboratory-Induced Colistin-Resistant Klebsiella pneumoniae. International Journal of Molecular Sciences. 2021; 22(13):7104. https://doi.org/10.3390/ijms22137104
Chicago/Turabian StylePruss, Agata, Paweł Kwiatkowski, Łukasz Łopusiewicz, Helena Masiuk, Peter Sobolewski, Karol Fijałkowski, Monika Sienkiewicz, Adam Smolak, Stefania Giedrys-Kalemba, and Barbara Dołęgowska. 2021. "Evaluation of Chemical Changes in Laboratory-Induced Colistin-Resistant Klebsiella pneumoniae" International Journal of Molecular Sciences 22, no. 13: 7104. https://doi.org/10.3390/ijms22137104
APA StylePruss, A., Kwiatkowski, P., Łopusiewicz, Ł., Masiuk, H., Sobolewski, P., Fijałkowski, K., Sienkiewicz, M., Smolak, A., Giedrys-Kalemba, S., & Dołęgowska, B. (2021). Evaluation of Chemical Changes in Laboratory-Induced Colistin-Resistant Klebsiella pneumoniae. International Journal of Molecular Sciences, 22(13), 7104. https://doi.org/10.3390/ijms22137104









